Abstract
Under free running conditions, FREQUENCY (FRQ) protein, a central component of the Neurospora circadian clock, is progressively phosphorylated, becoming highly phosphorylated before its degradation late in the circadian day. To understand the biological function of FRQ phosphorylation, kinase inhibitors were used to block FRQ phosphorylation in vivo and the effects on FRQ and the clock observed. 6-dimethylaminopurine (a general kinase inhibitor) is able to block FRQ phosphorylation in vivo, reducing the rate of phosphorylation and the degradation of FRQ and lengthening the period of the clock in a dose-dependent manner. To confirm the role of FRQ phosphorylation in this clock effect, phosphorylation sites in FRQ were identified by systematic mutagenesis of the FRQ ORF. The mutation of one phosphorylation site at Ser-513 leads to a dramatic reduction of the rate of FRQ degradation and a very long period (>30 hr) of the clock. Taken together, these data strongly suggest that FRQ phosphorylation triggers its degradation, and the degradation rate of FRQ is a major determining factor for the period length of the Neurospora circadian clock.
Circadian clocks are found in almost all groups of organisms, and they control a wide variety of daily (≈24-hr) endogenous (circadian) rhythms of molecular, physiological, and behavioral activities in these organisms (1). The identification of clock genes in several model systems and the understanding of how these genes are regulated have greatly enhanced our knowledge of how circadian clocks work at the molecular level (1–8). Common themes of clock mechanisms have begun to emerge: clock components comprising a core of the oscillator are part of a network of positive and negative interactions that establish a negative feedback loop that is essential for circadian oscillation (1, 9–16).
The Neurospora circadian oscillator comprises an autoregulatory negative feedback cycle in which frq mRNA and FRQ protein are the central components (1, 17, 18). Mutations at the frq locus cause a variety of clock phenotypes: long and short period length (16–29 hr), arrhythmia, and loss of temperature and nutritional compensation of the clock; the deletion of the frq locus results in loss of normal circadian rhythmicity (18). Both frq mRNA and FRQ protein are rhythmically expressed in a daily fashion, and FRQ protein acts to repress the abundance of its own transcript (17, 19, 20). Importantly, the rhythmic expression of frq is essential for the negative feedback loop as shown by the facts that constitutive expression of frq results in the loss of the overt rhythm and step changes in frq expression reset the phase of the clock (17). Light and temperature, two of the most important environmental signals, reset the Neurospora clock by changing the levels of frq mRNA and FRQ protein (21, 22).
In addition to transcriptional control, frq is subject to several aspects of posttranscriptional regulation (23). First, two forms of FRQ protein are expressed because of initiation at alternative ATGs, a large form of 989 aa (LFRQ) and a smaller form of 890 aa (SFRQ) (19). This mechanism is important for maintaining optimal clock function over a wide range of temperatures (24). Phosphorylation of FRQ is another prominent mode of regulation (20). As soon as either form of FRQ protein is made, it is progressively phosphorylated over time, and its level decreases after it is extensively phosphorylated. Thus, FRQ is differentially phosphorylated at different times of day (20), as is the Drosophila protein PERIOD (3, 25). In Drosophila, the DBT (double-time) protein, a casein kinase I, acts directly or indirectly to phosphorylate the clock protein PER; in a dbt null mutant, PER is constitutively highly expressed and hypophosphorylated, suggesting that the phosphorylation of PER may lead to its degradation (26, 27).
Protein phosphorylation, as one of the most common types of protein modifications, has not only been implicated in determining protein stability (28–30) but has also been shown to regulate the activity, cellular localization, and DNA- or protein-binding ability of different proteins (31–33). In this study, we set out to study the biological function of FRQ phosphorylation and its role in circadian regulation. We have shown that FRQ phosphorylation can be blocked by 6-dimethylaminopurine (6-DMAP), a general kinase inhibitor, and the inhibition of phosphorylation by the drug is correlated with a dramatic decrease in the degradation rate of FRQ protein and lengthening of the period of the clock in a concentration-dependent fashion. To establish the connection between FRQ phosphorylation and effects on the clock, we have identified three phosphorylation sites and determined that phosphorylation at Ser-513 is important for determining the degradation rate of FRQ. These data demonstrate that FRQ phosphorylation is important for regulating FRQ degradation rate, and that stability of FRQ is a major determining factor for the period length of the Neurospora circadian clock.
Materials and Methods
Strains, Culture Conditions, and Race Tube Assay.
Neurospora strains used in this study are bdA (clock wild type), YL15, and different frq mutant strains (20). Strain YL15 expresses only the small FRQ (SFRQ) form containing amino acids (aa) 100–989 (20). The frq null strain 93–4 (bd; frq10 A; his-3) (18) was the host strain for all frq mutation constructs described. Culture conditions were as described previously (17, 21). To monitor the degradation of FRQ protein, a light-to-dark transition (LD) or cycloheximide (CHX) treatment was used. For LD transitions, the Neurospora liquid cultures were first grown for at least one day in constant bright light at 25°C (LL) then transferred into constant darkness at 25°C. When used, CHX (Sigma) was added to cultures at a final concentration of 10 μg/ml after the cultures were grown in LL for at least 24 hr, and the cultures were kept in LL afterward. The cultures were collected at the indicated times.
Race tube assay, densitometric analyses of race tubes, and calculations of period length were performed as described (24).
Plasmid Constructs and Neurospora Transformation.
pKAJ120 (containing the whole frq gene and a his-3 targeting sequence) and pYL15 (a DraIII-SphI deletion of pKAJ120 that allows only the expression of SFRQ) were the parental plasmids for all other frq constructs (18, 20). All the deletions and point mutations of the FRQ ORF were constructed by using the Transformer Site-Directed Mutagenesis kit (CLONTECH). A SphI-EcoRV frq fragment containing most of the FRQ ORF from pKAJ120 was cloned into pUC19, and the resulting plasmid, pUC19Mfrq, was used as the in vitro mutagenic template. The mutagenic primers used were FRQ4 (to delete FRQ aa 435–558), FRQ4A (to delete FRQ aa 435–496), FRQ4B (to delete FRQ aa 500–558), FRQ4C (to delete FRQ aa 500–519), FRQ4D (to delete FRQ aa 531–558), FRQ501A (to mutate Ser-501 to Ala and Ser-503 to Gly), 513R (to mutate Ser-513 to Arg and Thr-514 to Ala), 519A (to mutate Ser-519 to Ala), 513I (to mutate Ser-513 to Ile), 513D (to mutate Ser-513 to Asp). After mutagenesis, the AflII-BssHII fragments from the resulting constructs were inserted into AflII-BssHII-digested pYL15 (which expresses only SFRQ) or pKAJ120 (which expresses both FRQ forms). Final plasmids based on pYL15 are named pSFRQ# (e.g., pSFRQ4); if based on pKAJ120, they are called pFRQ# (e.g., pFRQ501A). All constructs were confirmed by DNA sequencing and were targeted by transformation to the his-3 locus of strain 93–4, as previously described (34). For protein analyses, at least three to four independent transformants were examined. For race tube analysis, at least 20 independent transformants were examined.
Protein Analysis.
Protein extraction, quantification, and Western blot analysis are as previously described (20). Equal amounts of total protein (≈100 μg) were loaded in each protein lane, and after the blots were developed by chemiluminescence (enhanced chemiluminescence, Amersham), they were stained by amido black to verify equal loading of protein (24). Densitometry of the signal was performed by using nih image 1.61.
Results
6-DMAP Can Block FRQ Phosphorylation in Vivo, Slow Down the Degradation Rate of FRQ, and Lengthen the Period of the Clock.
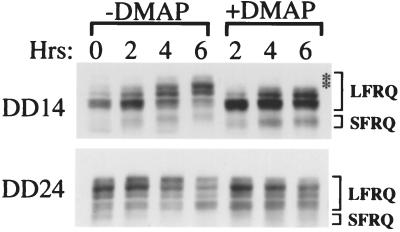
To understand the biological function of FRQ phosphorylation, we tested several kinase inhibitors for their ability to block FRQ phosphorylation in vivo. 6-DMAP, a general kinase inhibitor (35), was found to inhibit FRQ phosphorylation in vivo (Fig. 1). Two different time points about half of a circadian cycle apart were chosen: constant darkness (DD)14 (14 hr in constant darkness, corresponding to subjective morning) and DD24 (corresponding to subjective evening). DD14 is the time at which the phosphorylation level of FRQ protein is low, and protein level is on the rise, whereas at DD24, FRQ is highly phosphorylated and its level is decreasing (20). As can be seen from the control lanes (−DMAP) of Fig. 1, FRQ protein is progressively phosphorylated (note the gradual mobility shifts of FRQ protein and the changes in the distribution of different FRQ phosphorylation forms). These changes in protein mobility are the results of FRQ phosphorylation events, as shown by the fact that when protein extracts are treated with λ-phosphatase, all bands are converted into two, representing the large and small forms of FRQ (20). This progressive phosphorylation is especially apparent for the DD14 samples. However, in the presence of 5 mM 6-DMAP, the mobility shifts of FRQ at both time points are substantially delayed, indicating that phosphorylation is blocked by the drug; compare the phosphorylation patterns at 4 and 6 hr with and without the inhibitor. The top two phosphorylation bands are completely missing in the +DMAP lanes for the DD14 samples.
Figure 1.
6-DMAP blocks FRQ phosphorylation in vivo. At two different circadian time points, DD14 (CT2, Top) and DD24 (CT13, Bottom), half of the cultures were treated with 5 mM 6-DMAP. Cultures were harvested at the indicated times after the treatment. Western blot analysis shows the phosphorylation profiles of FRQ protein. The asterisks indicate missing LFRQ phosphorylation bands in the 6-DMAP treated sample.
Our previous results have suggested that the phosphorylation of FRQ protein could be important for regulating its degradation. Rhythmicity is seen not only in the level of the FRQ protein but also in its phosphorylation state. Moreover, FRQ protein always becomes extensively phosphorylated before its level starts to decrease (20). Secondly, after the inhibition of protein synthesis, FRQ protein becomes highly phosphorylated before its degradation (24). Because 6-DMAP can block FRQ phosphorylation, we wondered about the effect of this inhibition on FRQ stability. To examine the degradation of FRQ, two different kinds of treatment were used: either a simple LD transition or the addition of the protein synthesis inhibitor CHX (10 μg/ml) to the culture. An LD transition results in rapid degradation of frq mRNA and FRQ protein [(11, 21), Fig. 2A], and resets the clock to subjective dusk (circadian time 12, CT12). CHX treatment at 10 μg/ml was previously shown to block de novo synthesis of FRQ completely (24, 36). In this study, both methods were used and gave consistent results.
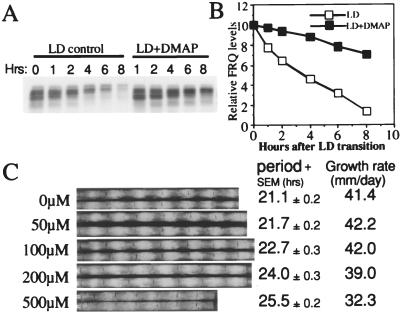
Figure 2.
6-DMAP slows down FRQ degradation and lengthens the period of the Neurospora clock in a dose-dependent manner. (A) Western blot analysis showing the reduction of FRQ degradation after an LD transition in the presence of 5 mM 6-DMAP. Cultures were first grown in LL for 1 day before being transferred into DD. Just before the LD transition, 5 mM 6-DMAP was added to half of the cultures, and cultures were harvested at the indicated times in DD. (B) Densitometric analysis of the Western blot shown in A. (C) Race tube data showing that 6-DMAP lengthens the period of the clock in a dose-dependent manner. (Left) The concentrations of the drug. Two replicate race tubes for each concentration are shown.
As shown in Fig. 2 A and B, FRQ is rapidly degraded after the LD transition. Note the gradual shifts in the phosphorylation bands with time. However, in the presence of 5 mM 6-DMAP, the degradation process is significantly reduced: there is still about 70% of FRQ remaining after 8 hr in DD, compared with only 10–15% for the control. Consistent with the results in Fig. 1, the phosphorylation process was also effectively slowed down by the drug treatment; the shift of phosphorylation bands is much slower than that seen in the control (Fig. 2A). Similar results were obtained with and without 6-DMAP in the CHX-treated culture (data not shown). These results suggest that the inhibition of FRQ phosphorylation leads to a slower degradation rate.
Because 6-DMAP treatment slows down FRQ degradation, we reasoned that the Neurospora circadian clock might also run more slowly (have a longer period length) in constant darkness in the presence of the drug. To test this prediction, different concentrations of 6-DMAP (from 0 to 500 μM) were added to the race tube medium and the circadian conidiation banding rhythms monitored in constant darkness. As predicted, the period length of the conidiation rhythm gradually increases with increasing concentrations of 6-DMAP, from 21.1 hr (−drug) to 25.5 hr (500 μM 6-DMAP) (Fig. 2C). This result suggests that the phosphorylation-induced degradation of FRQ is an important element in determining the period length of the circadian clock in Neurospora. Despite the significant changes in period length, there is only a small reduction of the growth rate in the cultures when the drug concentration is less than 200 μM, suggesting that the period lengthening effect of the drug is not caused by nonspecific effects resulting in toxicity.
Identification of Three Phosphorylation Sites on FRQ.
Although the data above are consistent with a role of FRQ phosphorylation in determining its degradation rate, nonspecific side effects of the drug cannot be excluded. To establish the relationship between phosphorylation and FRQ stability, it is important to determine the locations of the pertinent sites and the individual effects of phosphorylation at those sites. If indeed FRQ phosphorylation is important for triggering its degradation, the elimination of certain FRQ phosphorylation sites should stabilize FRQ. To examine this, we made systematic deletions of ≈100 aa spanning the entire ORF of FRQ. We reasoned that if a region containing phosphorylation sites is deleted, fewer bands should be observed by Western analysis. Because both the large and small forms of FRQ are progressively phosphorylated, and it is difficult to resolve different phosphorylation bands when both forms are present, all the initial deletions were made in the ORF of the SFRQ form [aa 100–989 (20)]. All the constructs were transformed into the frq null strain [frq10 (18)]. To examine the phosphorylation patterns in different strains, cultures were harvested after about 2 days of growth in LL. Under these conditions, FRQ protein is evenly phosphorylated, and all phosphorylation bands can be seen. After analyzing different deletion mutants (data not shown), we found that deletion of aa 435–558 (SFRQ4) causes a significant reduction in the number of bands (Fig. 3 A and B), as well as the expected reduction in molecular weight (compare the SFRQ4 lane to the control lane, SFRQ). This result suggests that some phosphorylation sites reside in this deleted region.
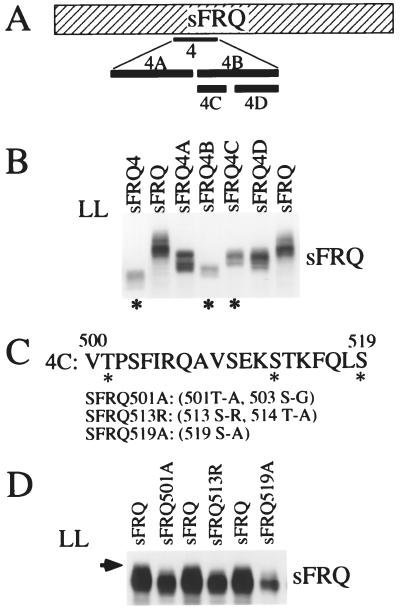
Figure 3.
Identification of three FRQ phosphorylation sites. (A) Schematic diagram of SFRQ deletion constructs. The dashed box represents the entire SFRQ ORF (aa 100–989). The black bars below indicate the locations of the deleted regions in different sFRQ constructs. “4” denotes sFRQ4; 4A-D represent subsections of “4.” (B) FRQ phosphorylation profiles in various sFRQ deletion strains. Cultures were grown in LL for at least 24 hr before harvesting. * marks the strains in which SFRQ is less phosphorylated than the wild-type SFRQ. The various FRQ phosphorylation forms were separated by using a longer than normal electrophoresis time to emphasize the mobility differences caused by phosphorylation. (C) The sequence of the 20-aa region deleted in sFRQ4C. The three potential phosphorylation sites conserved among different fungal frq homologs are indicated by *. The point mutations introduced are described in parentheses. (D) SFRQ phosphorylation profiles of the mutants containing point mutations at the three potential sites. The arrow indicates the phosphorylation band missing in the three mutants.
To narrow down the region further, two smaller deletions were made: SFRQ4A (deletion of aa 435–496) and SFRQ4B (deletion of aa 500–558) (Fig. 3A). Because SFRQ4B was found to have fewer phosphorylation bands (Fig. 3B), two further deletions were made within aa 500–558: SFRQ4C (deletion of aa 500–519) and SFRQ4D (deletion of aa 531–558). As shown in Fig. 3B, SFRQ4C is phosphorylated to a lesser extent than SFRQ4D and the wild-type SFRQ, indicating that some phosphorylation sites are located within this 20-aa region (Fig. 3C).
To identify the phosphorylation sites within this 20-aa region, its sequence was compared with the corresponding regions of all other frq homologs (37, 38), and three conserved putative phosphorylation sites were identified: Thr-501, Ser-513, and Ser-519 (marked by asterisks in Fig. 3C). To determine whether these three sites are indeed phosphorylation sites, point mutations were introduced at each position (Fig. 3C). In the T501A and S513R mutations, an additional Ser or Thr was also mutated. After transformation of these constructs into the frq null strain, their phosphorylation patterns were compared with that of the wild-type SFRQ. As shown in Fig. 3D, the highest molecular weight band is missing in these three mutants, indicating these sites are indeed phosphorylation sites.
FRQ Phosphorylation Site at Ser-513 Is Important for Regulating FRQ Degradation Rate.
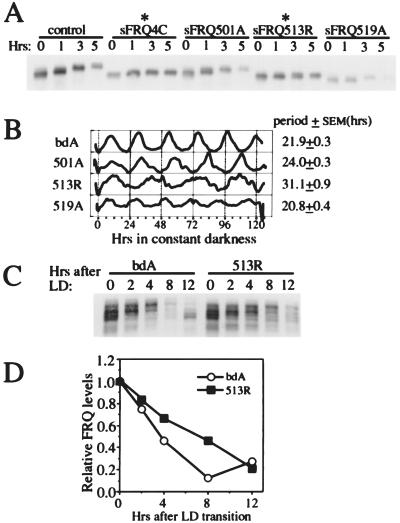
To examine whether these mutations affect degradation of SFRQ, CHX was added and the degradation of SFRQ monitored by Western blot analysis. As shown in Fig. 4A, the deletion of region 4C dramatically reduced the degradation rate of SFRQ. The T501A and S519A mutations did not appear to significantly affect the degradation rate of SFRQ; on the other hand, the S513R mutation resulted in a dramatic reduction in the degradation rate, suggesting that the phosphorylation site at aa 513 is important for regulating FRQ degradation. The different effects of these three mutations clearly indicate that not all FRQ phosphorylation sites are created equal, and certain sites are more important than others for determining the stability of FRQ.
Figure 4.
The effects of deletion and mutation of the FRQ phosphorylation sites on FRQ degradation and the circadian clock. (A) Western blot analysis showing that deletion of region 4C (aa 500–519) and mutation of S513 slow down the degradation of SFRQ. After the cultures were grown in LL for a day, 10 μg/ml of CHX was added at time 0. The appearance of more compact FRQ signals (compared with FRQ signals in Fig. 3 B and D) was the result of a shorter electrophoresis time. (B) The densitometric analysis of the race tube data showing that S513R mutations dramatically increase the period of the clock in DD. In contrast to the strains used in A, these strains are derived from a wild-type frq gene and produce both LFRQ and SFRQ forms. (C and D) Western blot and densitometric analyses showing that S513R mutations slow down FRQ degradation after an LD transition.
The slow degradation of FRQ has been predicted to have a major role in determining the long period of the circadian oscillation (19). If so, we would predict that the slow degradation of FRQ in the S513R mutant would result in a strain having a substantially longer period than that of the wild type. To test this prediction, these mutations were introduced into constructs that contain both FRQ forms (because both forms are required for an optimally running clock) (24). The constructs were transformed into the frq null strain, and the resultant clock phenotypes monitored by race tube assays (Fig. 4B). As predicted, the period length of the S513R mutant (≈31 hr) is about 50% longer than that of the wild type. Consistent with these results, the degradation rate of FRQ after an LD transition is slower in the 513R mutant than in the wild type (Fig. 4 C and D). Note that after ≈8 hr in DD, the wild-type FRQ has already reached the trough level, whereas FRQ in the 513R mutant is still in the middle of the degradation process. After 12 hr, the newly synthesized and less phosphorylated wild-type FRQ has begun to appear, whereas the mutant protein is still in the process of decay (Fig. 4C). The T501A and S519A mutations also have period effects, but because of the sensitivity of the Western blot analysis and the small period differences, it is unclear whether these effects are related to their influence on FRQ degradation.
Although the above data strongly suggest that the phosphorylation site at Ser-513 is important for triggering FRQ degradation, it is not clear whether Ser-513 is indeed responsible for the effects because the S513R mutant has two point mutations, 513S-R and 514T-A [Thr-514 is not conserved among different frq homologs (38)] (Fig. 3C). To clarify this matter and also to show the involvement of phosphorylation in FRQ degradation, two different mutations were made at Ser-513. One mutation changes Ser-513 to an isoleucine (I), and another mutation changes it to an acidic aa (aspartic acid, D), because it has been shown that acidic aa can mimic the effect of phosphorylation in some cases (30). If indeed Ser-513 is a phosphorylation site responsible for controlling FRQ stability, the S513I and S513R mutations should exhibit similar phenotypes, and the FRQ degradation rate should be slower in S513I mutants than in the S513D mutant if the aspartic acid can mimic a phosphorylated aa.
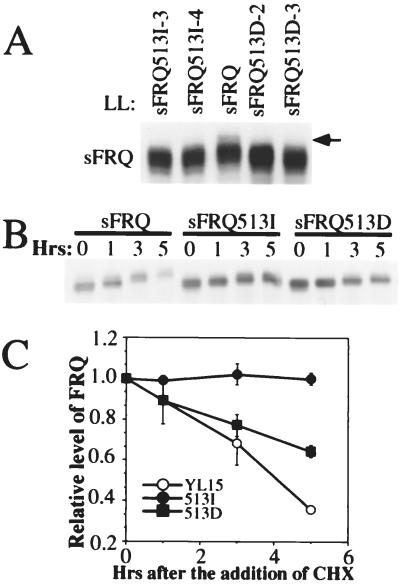
As shown in Fig. 5A, the disappearance of the top phosphorylation band in both SFRQ513I and SFRQ513D strains indicates that Ser-513 is indeed a phosphorylation site. As predicted, the degradation rate of SFRQ513I after the addition of CHX is dramatically reduced (Fig. 5 B and C), a result that is very similar to what has been shown for the SFRQ513R mutant (Fig. 4A). Additionally, as we predicted, the introduction of an acidic residue is able to mimic the effect of phosphorylation to some degree, because the degradation rate of SFRQ513D is faster than that of SFRQ513I. However, SFRQ513D still degrades more slowly than the wild-type SFRQ (Fig. 5 B and C). This experiment was carried out three times with similar results, which are summarized in Fig. 5C.
Figure 5.
Two different mutations at S513 result in differential responses of FRQ degradation. (A) Western blot analysis showing that the S513I and S513D mutations lead to the loss of the top SFRQ phosphorylation band. The arrow indicates the missing band in the mutants. (B) Western blot analysis showing that the S513I mutation dramatically slows down the degradation of SFRQ. Cultures were grown in LL for a day before CHX was added at time 0. (C) Grouped data showing densitometric analysis of Western blots from three independent experiments as in B. Bars = SD.
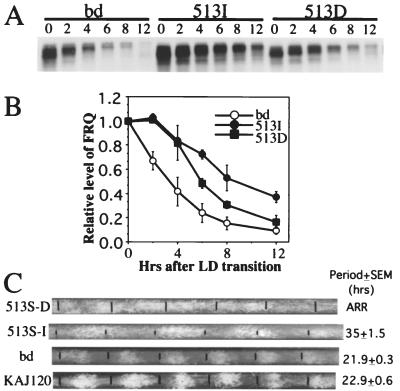
To examine the clock phenotypes of these mutations, they were introduced into constructs that encode both FRQ forms, and after transformation into a frq null strain, their rhythms were monitored in DD. Similar to the results shown in Fig. 5 B and C, the degradation rate of FRQ after an LD transition is much slower in the S513I mutant than in both the wild-type and the 513D strain (Fig. 6 A and B). As expected, the period of the S513I mutant (≈35 hr) is much longer than that of the wild-type and KAJ120 (bearing a wild-type frq gene transformed into the his-3 locus of a frq10 strain) (Fig. 6C); in fact, this mutant has the longest period of all of the frq mutants known to date (39). However, to our surprise, the conidiation process in the 513D mutant appears to be arrhythmic. The loss of rhythmicity in the 513D strain must be caused by the loss of proper regulation of the FRQ negative feedback process and suggests the possibility that this part of FRQ may be involved in aspects of function beyond regulating stability.
Figure 6.
The S513I mutation results in significantly slower FRQ degradation and in a very long circadian period. (A) Western blot analysis of FRQ degradation after an LD transition in the wild-type and the two S513 mutant strains. The numbers above the blot indicate the number of hours after the LD transition. (B) Densitometric analysis of Western blots from three independent experiments as in A. Bars = SD. (C) Race tube data of the wild-type, KAJ120, and S513 mutant strains. The first black bar on each race tube indicates the time of the LD transfer. The subsequent black bars indicate the growth fronts of the cultures every 24 hr. The periods of the strains are shown on the right. ARR, arrhythmic.
Taken together, the data presented here demonstrate that the phosphorylation of FRQ is important for regulating its degradation, and the stability of FRQ is an important factor in determining the period length of the Neurospora clock.
Discussion
Previous studies have shown that the frq gene is one of the central components of a Neurospora negative feedback loop that determines the period length and temperature compensation characteristics of the circadian clock (17, 18, 40). In this study, we investigated the biological role of FRQ phosphorylation, and several lines of evidence were presented consistent with phosphorylation regulating FRQ stability. First, we showed that a kinase inhibitor, 6-DMAP, can slow FRQ phosphorylation in vivo; it also slows FRQ degradation and lengthens the period of the Neurospora circadian clock. Second, by systematic mutagenesis of the FRQ ORF, three FRQ phosphorylation sites were identified. Subsequent elimination of the phosphorylation site at Ser-513 leads to a dramatic reduction in the rate of FRQ degradation and a significantly increased period. Finally, we have also shown that an acidic aa at residue 513 speeds the degradation of FRQ probably by mimicking the phosphorylation. Taken together, these data strongly suggest that phosphorylation triggers the degradation of FRQ, and that the degradation rate is a determining factor for the period length of the Neurospora circadian clock. Despite the existence of additional phosphorylation sites, Ser-513 appears to be one of the main sites for determining the degradation rate of FRQ.
Interestingly, phosphorylation appears to play a similar role for a Drosophila clock protein, PER, which is also progressively phosphorylated over time. DBT, a casein kinase I homolog, leads either directly or indirectly to the phosphorylation and degradation of PER, because PER is hypophosphorylated in a dbt null strain where its level is constitutively high (26, 27). Neurospora contains at least two casein kinase I homologs, which are presently being examined for roles in FRQ phosphorylation.
A major implication of this study is that the degradation of a central clock component is essential for the proper functioning of the clock. If we look at a clock following the expression of its central components, we can view one circadian cycle as two halves (19). One half is the activation and accumulation of clock transcripts and proteins, and the other half is the inhibition and the degradation of those transcripts and proteins. Each process is essential for proper clock function, and the speed of each process is what determines the period length of the clock. In Neurospora, the activation process possibly involves the products of wc-1 and wc-2, which initiate every new cycle by activating the expression of frq mRNA and FRQ protein. The other half of the cycle is a combination of the negative feedback on frq transcript levels by its own protein and the degradation process of frq mRNA and FRQ protein (1, 11, 20). In this study, we have shown that the degradation of FRQ protein is indeed one of the key factors affecting the Neurospora circadian system, and the rate of FRQ degradation is a major determinant for the period length of the clock. The noncircadian fluctuations or oscillations that exist in the absence of FRQ (e.g., 18, 42, 43) are rendered circadian in nature by coupling to the FRQ/WC feedback loop, and these data on FRQ phosphorylation reemphasize the essential role of the FRQ/WC loop in establishing the circadian characteristics of this rhythmic system. In similar studies, Price and coworkers have shown that two alleles of Drosophila dbt (dbtS and dbtL) change the period of the clock presumably by affecting the degradation of PER (26, 27). Similarly, in the dinoflagellate Gonyaulax polyedra, various protein kinase inhibitors have been shown to affect circadian rhythmicity (35, 41). Interestingly, the same kinase inhibitor we have used, 6-DMAP, has been shown to have a period-lengthening effect, suggesting phosphorylation also plays a role in the Gonyaulax clock. However, because of the lack of specificity of the drug and its unknown effects on the clock components of Gonyaulax, we cannot be sure that a mechanism similar to the one we have shown in Neurospora also exists in Gonyaulax.
As can be seen from its phosphorylation pattern, FRQ is phosphorylated at multiple sites. The three sites identified in this study represent only a portion of the sites, because when the region containing all three sites is deleted, multiple FRQ phosphorylation bands can still be seen (Fig. 3B). Although it is clear that Ser-513 is one of the major phosphorylation sites responsible for triggering FRQ degradation, it cannot be that Ser-513 is the only important site for degradation because the clock still runs, albeit aberrantly, in the 513I mutant strain. Furthermore, it is also quite possible that the phosphorylation of other sites is involved in other aspects of FRQ function, such as nuclear localization (44) or protein activity. The slightly shorter period of the 519A strain suggests that the phosphorylation site at Ser-519 is not responsible for triggering FRQ degradation, although this site may be involved in other types of regulation (Fig. 4 A and B).
Another implication of our results is that, despite the progressive appearance of the FRQ phosphorylation pattern suggesting that the phosphorylation events might be sequential (i.e., the phosphorylation of one site leads to the phosphorylation of another), the phosphorylation patterns of our mutants suggest this is not the case for all of these phosphorylation sites. The elimination of some sites does not appear to affect the phosphorylation of others (Figs. 3D and 5A); therefore, phosphorylation events at different positions in the protein are likely to be independent of one another.
Circadian clocks control a wide variety of cellular activities with a remarkable degree of precision under constant conditions. To achieve such precision, various types of posttranscriptional and posttranslational regulation are required. In Neurospora, the phosphorylation of FRQ and its subsequent degradation are important for generating the circadian oscillations in the abundance of frq mRNA and FRQ proteins, which are essential for normal circadian rhythmicity. Together with other types of transcriptional and posttranscriptional regulation, they provide a variety of facets where regulatory elements might influence the operation of the clock.
Acknowledgments
We thank Hildur Colot, Deanna Denault, Jon Best, and other members of the laboratory for comments and help on the manuscript. This work was supported by grants from the National Institutes of Health (GM 34985 and MH01186 to J.C.D, MH44651 to J.C.D. and J.J.L., and 1F32GM19230 to Y.L.), the National Science Foundation (MCB-9307299 to J.J.L.), and the Norris Cotton Cancer Center core grant at Dartmouth Medical School.
Abbreviations
- 6-DMAP
6-dimethylaminopurine
- CHX
cycloheximide
- LD
light to dark
- LL
constant light
- CT
circadian time
- DD
constant dark
- aa
amino acid
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dunlap J C. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 2.Young M W. Annu Rev Biochem. 1998;67:135–152. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- 3.Rosbash M, Allada R, Dembinska M, Guo W Q, Le M, Marrus S, Qian Z, Rutila J, Yaglom J, Zeng H. Cold Spring Harbor Symp Quant Biol. 1996;61:265–278. [PubMed] [Google Scholar]
- 4.Hall J C. Trends Neurosci. 1995;18:230–240. doi: 10.1016/0166-2236(95)93908-g. [DOI] [PubMed] [Google Scholar]
- 5.Golden S S, Johnson C H, Kondo T. Curr Opin Microbiol. 1998;1:669–673. doi: 10.1016/s1369-5274(98)80113-6. [DOI] [PubMed] [Google Scholar]
- 6.Bell-Pedersen D, Garceau N, Loros J J. J Genet. 1996;75:387–401. [Google Scholar]
- 7.Wilsbacher L D, Takahashi J S. Curr Opin Genet Dev. 1998;8:595–602. doi: 10.1016/s0959-437x(98)80017-8. [DOI] [PubMed] [Google Scholar]
- 8.Loros J J. Curr Opin Microbiol. 1998;6:698–706. doi: 10.1016/s1369-5274(98)80118-5. [DOI] [PubMed] [Google Scholar]
- 9.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Anderson C R, Tanabe A, Golden S S, Johnson C H, Kondo T. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 10.Darlington T K, Wager-Smith K, Ceriani M F, Stankis D, Gekakis N, Steeves T, Weitz C J, Takahashi J, Kay S A. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 11.Crosthwaite S C, Dunlap J C, Loros J J. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 12.Rutila J E, Suri V, Le M, So W V, Rosbash M, Hall J C. Cell. 1998;93:805–813. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 13.Allada R, White N E, So W V, Hall J C, Rosbash M. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 14.King D, Zhao Y, Sangoram A, Wilsbacher L, Tanaka M, Antoch M, Steeves T, Vitaterna M, Kornhauser J, Lowrey P, Turek F, Takahashi J. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gekakis N, Stankis D, Nguyen H B, Davis F C, Wilsbacher L D, King D P, Takahashi J S, Weitz C J. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z Y, Tobin E M. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 17.Aronson B, Johnson K, Loros J J, Dunlap J C. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 18.Aronson B D, Johnson K A, Dunlap J C. Proc Natl Acad Sci USA. 1994;91:7683–7687. doi: 10.1073/pnas.91.16.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merrow M, Garceau N, Dunlap J C. Proc Natl Acad Sci USA. 1997;94:3877–3882. doi: 10.1073/pnas.94.8.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garceau N, Liu Y, Loros J J, Dunlap J C. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- 21.Crosthwaite S C, Loros J J, Dunlap J C. Cell. 1995;81:1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Merrow M M, Loros J J, Dunlap J C. Science. 1998;281:825–829. doi: 10.1126/science.281.5378.825. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Heintzen C, Loros J, Dunlap J C. Cell Mol Life Sci. 1999;55:1195–1205. doi: 10.1007/s000180050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Garceau N, Loros J J, Dunlap J C. Cell. 1997;89:477–486. doi: 10.1016/s0092-8674(00)80228-7. [DOI] [PubMed] [Google Scholar]
- 25.Edery I, Zweibel L, Dembinska M, Rosbash M. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price J L, Blau J, Rothenfluh A, Adodeely M, Kloss B, Young M W. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 27.Kloss B, Price J L, Saez L, Blau J, Rothenfluh A, Young M W. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 28.Lin W C, Desiderio S. Science. 1993;260:953–959. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- 29.Yaglom J, Linskens M H, Sadis S, Rubin D M, Futcher B, Finley D. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z J, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 31.Komeili A, O'Shea E K. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 32.Ciceri P, Gianazza E, Lazzari B, Lippoli G, Genga A, Hoscheck G, Schmidt R J, Viotti A. Plant Cell. 1997;9:97–108. doi: 10.1105/tpc.9.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaudet R, Savage J R, McLaughlin J N, Willardson B M, Sigler P B. Mol Cell. 1999;3:649–660. doi: 10.1016/s1097-2765(00)80358-5. [DOI] [PubMed] [Google Scholar]
- 34.Bell-Pedersen D, Dunlap J C, Loros J J. Mol Cell Biol. 1996;16:513–521. doi: 10.1128/mcb.16.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Comolli J, Taylor W R, Hastings J W. J Biol Rhythms. 1994;9:13–26. doi: 10.1177/074873049400900102. [DOI] [PubMed] [Google Scholar]
- 36.Dunlap J C, Feldman J F. Proc Natl Acad Sci USA. 1988;85:1096–1100. doi: 10.1073/pnas.85.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merrow M, Dunlap J C. EMBO J. 1994;13:2257–2266. doi: 10.1002/j.1460-2075.1994.tb06507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis M, Feldman J F. Mol Biol Evol. 1997;13:1233–1241. doi: 10.1093/oxfordjournals.molbev.a025689. [DOI] [PubMed] [Google Scholar]
- 39.Dunlap J C. Annu Rev Physiol. 1993;55:683–728. doi: 10.1146/annurev.ph.55.030193.003343. [DOI] [PubMed] [Google Scholar]
- 40.Dunlap J C, Loros J J, Liu Y, Crosthwaite S K. Genes Cells. 1999;4:1–10. doi: 10.1046/j.1365-2443.1999.00239.x. [DOI] [PubMed] [Google Scholar]
- 41.Comolli J C, Hastings J W. J Biol Rhythms. 1999;14:11–19. doi: 10.1177/074873099129000399. [DOI] [PubMed] [Google Scholar]
- 42.Loros J J, Feldman J F. J Biol Rhythms. 1986;1:187–198. doi: 10.1177/074873048600100302. [DOI] [PubMed] [Google Scholar]
- 43.Merrow M, Bruner M, Roenneberg T. Nature (London) 1999;399:584–586. doi: 10.1038/21190. [DOI] [PubMed] [Google Scholar]
- 44.Luo C, Loros J J, Dunlap J C. EMBO J. 1998;17:1228–1235. doi: 10.1093/emboj/17.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]