Abstract
Biomolecules labeled with azides can be detected through Cu-free click chemistry with cyclooctyne probes, but their intrinsic hydrophobicity can compromise bioavailability. Here, we report the synthesis and evaluation of a novel azacyclooctyne, 6,7-dimethoxyazacyclooct-4-yne (DIMAC). Generated in nine steps from a glucose analogue, DIMAC reacted with azide-labeled proteins and cells similarly to cyclooctynes. However, its superior polarity and water solubility reduced nonspecific binding, thereby improving the sensitivity of azide detection.
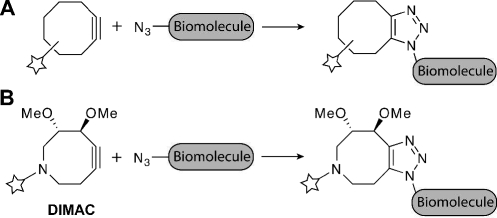
A growing area of reaction methodology focuses on the development of chemical transformations with the selectivity, kinetics, and biocompatibility required to proceed in living systems.(1) Several efforts in this area have converged on the azide as a reagent. Its small size, diverse modes of reactivity, and bioorthogonality make the azide an ideal chemical reporter group for biomolecules.(2) Azides can be incorporated into glycans, lipids, and proteins using metabolic machineries,2a–2d covalent inhibitors,(2e) or enzymatic transfers.(2f)Staudinger ligation with phosphines,(2a) Cu-catalyzed cycloaddition with terminal alkynes (often referred to as click chemistry),(3) or strain-promoted cycloaddition with cyclooctynes(4) allow for modification of the azido biomolecules with diverse probes. The latter reaction, also termed Cu-free click chemistry (Figure 1A), was designed to eliminate the cytotoxic metal catalyst required for conventional click chemistry. Incorporating the alkyne into a strained eight-membered ring system promoted the cycloaddition with azides without compromising selectivity in biological systems.(4) Since the initial report, fluorination(5) and fused phenyl rings(6) have improved the reaction kinetics, and a difluorinated analogue has been employed to image glycans in developing zebrafish.(7)
Figure 1.
Cu-free click chemistry with (A) cyclooctyne reagents or (B) 6,7-dimethoxyazacyclooct-4-yne (DIMAC) reagents.
These achievements reflect optimization of the reaction for use with live cells and simple model organisms. However, we envision applications of Cu-free click chemistry in mammalian disease models where the bioavailability and pharmacokinetic properties of the reagents become important. The cyclooctynes currently employed for Cu-free click chemistry comprise hydrocarbon scaffolds that limit their solubility in aqueous solutions. The hydrophobicity of these cyclooctyne scaffolds could also promote sequestration by membranes or nonspecific binding to serum proteins, thereby reducing their bioavailable concentrations. Consequently, we have focused on designing strained cycloalkynes with enhanced water solubility.
Here, we report the synthesis and biological evaluation of a novel heterocyclic and heteroatom-substituted cyclooctyne. The compound, 6,7-dimethoxyazacyclooct-4-yne (DIMAC, Figure 1B), has a nitrogen atom within the strained ring system that interrupts the hydrophobic surface area and provides a convenient site for probe conjugation. We reasoned that two methoxy groups would enhance the compound’s polarity, and placement of one at the propargylic position would impart reaction kinetics similar to existing simple cyclooctynes.4,5a LogS calculations for DIMAC methyl amide gave a value of ∼−2.7, while that of a parent cyclooctyne methyl amide was ∼−3.1.(8)
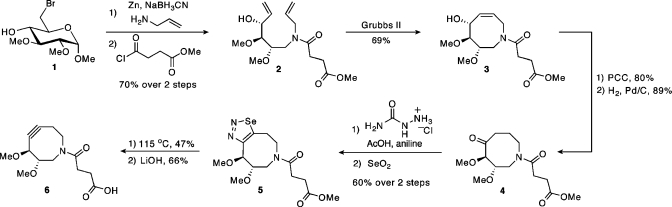
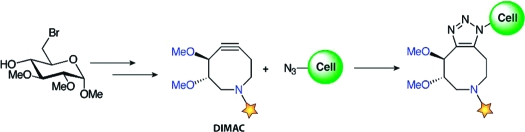
As shown in Scheme 1, DIMAC was synthesized in nine steps beginning from methyl 6-bromoglucopyranoside (1).(9) First, compound 1 was transformed to acyclic diene 2 via a zinc reduction/reductive amination reaction followed by amide formation with methyl succinyl chloride.(10) The eight-membered ring was generated by a Grubbs ring-closing metathesis reaction to yield 3.(11) Allylic alcohol 3 was converted to ketone 4 via oxidation to the enone followed by hydrogenation.
Scheme 1. Synthesis of DIMAC.
We explored two routes for conversion of the ketone to the corresponding alkyne: vinyl triflate formation followed by syn-elimination of triflic acid(5) and formation of a selenadiazole followed by fragmentation to the alkyne.(12) Although successful in our previous cyclooctyne syntheses,(5) the vinyl triflate method proved too harsh for the target compound. Thus, we condensed compound 4 with semicarbazide and oxidized the resulting intermediate to yield selenadiazole 5. Subsequent thermal decomposition of the selenadiazole followed by saponification of the methyl ester produced DIMAC (6).
The reactivity of DIMAC was tested in a model 1,3-dipolar cycloaddition reaction with benzyl azide (Scheme S2, Supporting Information). The reaction proceeded cleanly with a second-order rate constant of 3.0 × 10−3 M−1 s−1 (Figure S1, Supporting Information), slightly higher than that for the parent cyclooctynes (1−2 × 10−3 M−1s−1).4,5a This slight enhancement in rate might reflect added ring strain due to the shorter C−N bond length or the sp2 character of the amide nitrogen.(13)
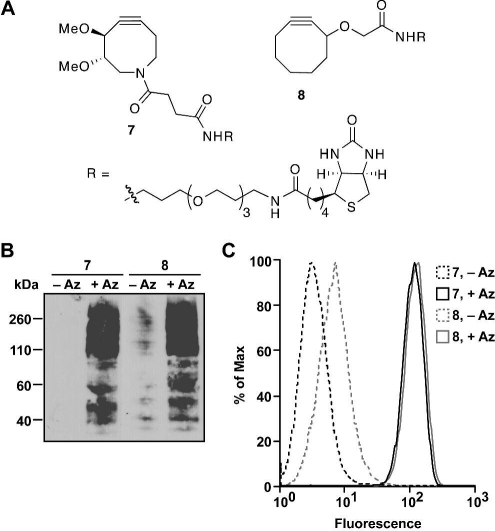
Next, we tested the ability of DIMAC to label glycan-associated azides within cell lysates and on the surface of live cells. DIMAC was first conjugated to biotin (Figure 2A; Scheme S3, Supporting Information), providing the means to detect its cycloadducts. For comparative purposes, we performed parallel experiments with previously reported cyclooctyne−biotin conjugate 8.(5a) The difference in water solubility of these two reagents was apparent during the preparation of 2.5 mM stock solutions. DIMAC−biotin 7 was readily soluble in aqueous buffer, whereas cyclooctyne−biotin 8 required an organic cosolvent (30% DMF).
Figure 2.
(A) Biotin conjugates of DIMAC (7) and a parent cyclooctyne (8).(5a) (B) Western blot of Jurkat cell lysates. Cells were treated with (+Az) or without (−Az) 25 μM Ac4ManNAz for 3 d, and lysates (35 μg total protein) were reacted with 250 μM 7 or 8 overnight at 25 °C. Equal protein loading was confirmed by Ponceau S staining (Figure S2, Supporting Information). The blot was probed with HRP-conjugated α-biotin. (C) Flow cytometry analysis of Jurkat cells. Cells metabolically labeled as in (B) were reacted with 250 μM 7 or 8 for 1 h at 25 °C and then stained with FITC−avidin. Similar data were obtained in three replicate experiments.
Jurkat cells were grown in the presence (+Az) or absence (−Az) of 25 μM N-azidoacetylmannosamine (Ac4ManNAz) for 3 days, resulting in the metabolic labeling of cell-surface glycans with N-azidoacetyl sialic acid (SiaNAz) residues.(2a) Cell lysates were incubated overnight with 250 μM 7 or 8 and then analyzed by Western blot probing with horseradish peroxidase (HRP)-conjugated α-biotin. Both DIMAC−biotin (7) and the control cyclooctyne 8 labeled glycoproteins from the lysate in an azide-dependent manner (Figure 2B). However, compound 8 produced a greater extent of nonspecific protein labeling.
To test DIMAC’s reactivity with azides on live cells, Jurkat cells were again incubated with 25 μM Ac4ManNAz for 3 days and then reacted with 0 or 250 μM 7 or 8. The cells were stained with fluorescein isothiocyanate (FITC)-conjugated avidin and analyzed by flow cytometry. DIMAC−biotin (7) and cyclooctyne−biotin 8 selectively labeled cell surfaces in an azide-dependent manner with comparable efficiencies (Figure 3C).(14) Compound 8, however, showed significantly higher nonspecific background labeling. The DIMAC reagent labeled cells with a signal/background ratio of 26:1, whereas the ratio for cyclooctyne 8 was 14:1. Indeed, the fluorescence of cells lacking azides but treated with DIMAC−biotin followed by FITC–avidin was indistinguishable from those treated with FITC–avidin alone (Figure S4, Supporting Information). Importantly, DIMAC did not show any cytotoxicity during cell labeling (Figures S5−7, Supporting Information). DIMAC also exhibited time- and dose-dependent labeling (Figure S8, Supporting Information).
In summary, DIMAC represents a new class of heterocyclic substrates for Cu-free click chemistry. Its superior polarity and water solubility reduce nonspecific protein and cell binding and improve the sensitivity of azide detection. The synthetic route can be adapted to various carbohydrate starting materials and is amenable to functional group manipulations directed at tuning reactivity, solubility, and perhaps biodistribution. DIMAC and its congeners are therefore promising reagents for Cu-free click chemistry in mammalian systems.
Acknowledgments
We thank J. M. Baskin, S. T. Laughlin, and J. A. Codelli at UC Berkeley for helpful discussions. This work was supported by a grant to C.R.B. from the National Institutes of Health (GM058867).
Supporting Information Available
Experimental procedures, protocols, and spectral data for all new compounds as well as Figures S1−S8 and Schemes S1−S3. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Prescher J. A.; Bertozzi C. R. Nat. Chem. Biol. 2005, 1, 13–21. [DOI] [PubMed] [Google Scholar]
- a Saxon E.; Bertozzi C. R. Science 2000, 287, 2007–2010. [DOI] [PubMed] [Google Scholar]; b Dieterich D. C.; Link A. J.; Graumann J.; Tirrell D. A.; Schuman E. M. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 9482–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Hang H. C.; Geutjes E. J.; Grotenbreg G.; Pollington A. M.; Bijlmakers M. J.; Ploegh H. L. J. Am. Chem. Soc. 2007, 129, 2744–2755. [DOI] [PubMed] [Google Scholar]; d Sawa M.; Hsu T-L.; Itoh T.; Sugiyama M.; Hanson S. R.; Vogt P. K.; Wong C-H. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 12371–12376. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Speers A. E.; Adam G. C.; Cravatt B. F. J. Am. Chem. Soc. 2003, 125, 4686–4687. [DOI] [PubMed] [Google Scholar]; f Fernandez-Suarez M.; Baruah H.; Martinez-Hernandez L.; Xie K. T.; Baskin J. M.; Bertozzi C. R.; Ting A. Y. Nat. Biotechnol. 2007, 25, 1483–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. Angew. Chem., Int. Ed. 2002, 41, 2596–2599. [DOI] [PubMed] [Google Scholar]; b Breinbauer R.; Kohn M. ChemBioChem 2003, 4, 1147–1149. [DOI] [PubMed] [Google Scholar]
- Agard N. J.; Prescher J. A.; Bertozzi C. R. J. Am. Chem. Soc. 2004, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- a Agard N. J.; Baskin J. M.; Prescher J. A.; Lo A.; Bertozzi C. R. ACS Chem. Biol. 2006, 1, 644–648. [DOI] [PubMed] [Google Scholar]; b Baskin J. M.; Prescher J. A.; Laughlin S. T.; Agard N. J.; Chang P. V.; Miller I. A.; Lo A.; Codelli J. A.; Bertozzi C. R. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning X.; Guo J.; Wolfert M. A.; Boons G-J. Angew. Chem., Int. Ed. 2008, 47, 2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin S. T.; Baskin J. M.; Amacher S. L.; Bertozzi C. R. Science 2008, 320, 664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calculated for compounds 7 and 8 where R = CH3 using logS software developed by Tetko et al.,www.vcclab.org/lab/alogps/. More negative values indicate poorer water solubility.; a Tetko I. V.; Tanchuk V. Y.; Kasheva T. N.; Villa A. E. J. Chem. Inf. Comput. Sci. 2001, 41, 246–252. [DOI] [PubMed] [Google Scholar]; b Tetko I. V.; Tanchuk V. Y.; Kasheva T. N.; Villa A. E. P. J. Chem. Inf. Comput. Sci. 2001, 41, 1488–1493. [DOI] [PubMed] [Google Scholar]
- a Jones K.; Wood W. W. Carbohydr. Res. 1986, 155, 217–222. [Google Scholar]; b Scheme S1, Supporting Information
- Sletten E. M.; Liotta L. J. J. Org. Chem. 2006, 71, 1335–1343. [DOI] [PubMed] [Google Scholar]
- Miller S. J.; Kim S. H.; Chen Z. R.; Grubbs R. H. J. Am. Chem. Soc. 1995, 117, 2108–2109. [Google Scholar]
- a Meier H.; Voigt E. Tetrahedron 1972, 28, 187–198. [Google Scholar]; b Meier H.; Stavridou E.; Storek C. Angew. Chem., Int. Ed. 1986, 9, 809–810. [Google Scholar]; c Petersen H.; Kolshorn H.; Meier H. Angew. Chem., Int. Ed. 1978, 17, 461–462. [Google Scholar]
- Meier H.; Hanold N.; Molz T.; Bissinger H. J.; Kolshorn H.; Zountsas J. Tetrahedron 1986, 42, 1711–1719. [Google Scholar]
- For a comparison of DIMAC to a difluorinated cyclooctyne as well as a triarylphosphine for reaction via the Staudinger ligation, see Figure S3, Supporting Information.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.