Abstract
Reactive electrophiles generated by lipid peroxidation are thought to contribute to cardiovascular disease and other oxidative stress-related pathologies by covalently modifying proteins and affecting critical protein functions. The difficulty of capturing and analyzing the relatively small fraction of modified proteins complicates identification of the protein targets of lipid electrophiles. We recently synthesized a biotin-modified linoleoylglycerylphosphatidycholine probe called PLPBSO (Tallman et al. Chem. Res. Toxicol. 2007, 7, 227−234), which forms typical linoleate oxidation products and covalent adducts with model peptides and proteins. Supplementation of human plasma with PLPBSO followed by free radical oxidation resulted in covalent adduction of PLPBSO to plasma proteins, which were isolated with streptavidin and identified by liquid chromatography-tandem mass spectrometry (LC-MS−MS). Among the most highly modified proteins was apolipoprotein A1 (ApoA1), which is the core component of high density lipoprotein (HDL). ApoA1 phospholipid adduct sites were mapped by LC-MS−MS of tryptic peptides following mild base hydrolysis to release esterified phospholipid adducts. Several carboxylated adducts formed from phospholipid-esterified 9,12-dioxo-10(E)-dodecenoic acid (KODA), 9-hydroxy, 12-oxo-10(E)-dodecenoic acid (HODA), 7-oxoheptanoic acid, 8-oxooctanoic acid, and 9-oxononanoic acid were identified. Free radical oxidations of isolated HDL also generated adducts with 4-hydroxynonenal (HNE) and other noncarboxylated electrophiles, but these were only sporadically identified in the PLPBSO-adducted ApoA1, suggesting a low stoichiometry of modification in the phospholipid-adducted protein. Both phospholipid electrophiles and HNE adducted His162, which resides in an ApoA1 domain involved in the activation of Lecithin-cholesterol acyltransferase and maturation of the HDL particle. ApoA1 lipid electrophile adducts may affect protein functions and provide useful biomarkers for oxidative stress.
Keywords: Lipid electrophile, protein adduct, affinity probe, apolipoprotein A1
Short abstract
We describe the use of PLPBSO as a probe to identify major protein targets of oxidized phospholipids in human plasma. This approach identified apolipoprotein A1 (ApoA1) as the principal target of PLPBSO oxidation products and led us to map adducts from lipid oxidation products on this protein. We report the identification of adducts from several phospholipid electrophiles, including carboxy-terminal adducts. We further evaluated the sites of ApoA1 adduction by the prototypical lipid electrophile HNE. ApoA1 lipid electrophile adducts may affect protein functions and provide useful biomarkers for oxidative stress.
Introduction
The formation of oxidants is a hallmark of chemical toxicity, inflammation, and other types of environmental stresses.1,2 Oxidative stress and oxidants also are involved in human diseases that account for significant morbidity and mortality, including cancer, atherosclerosis, and neurodegenerative diseases.3−7 Although oxidative stress derives fundamentally from the excessive flux of reduced oxygen species, such as superoxide and hydrogen peroxide, secondary products of lipid oxidation may play critical roles in oxidant-associated molecular pathologies. Lipid electrophiles such as malondialdehyde, hydroxyalkenals, oxoalkenals, epoxyalkenals, and γ-ketoaldehydes readily react with proteins, and the formation of covalent adducts is hypothesized to contribute to oxidant-related diseases and toxicities.8,9 Because they reflect the occurrence of oxidative stress, protein adducts from lipid electrophiles also may serve as biomarkers for oxidant-mediated pathologies.10,11
Most studies of protein modification by lipid electrophiles have examined a few prototypical lipid oxidation products, such as 4-hydroxynonenal (HNE), 4-oxononenal, and γ-ketoaldehydes (levuglandins) using mass spectrometry (MS). This published work has identified relevant protein adduction chemistries with amino acid side chain nucleophiles12−17 and in some cases mapped adducts to specific sequences in modified proteins.17−25 Collectively, the adduct mapping data indicate significant site-specificity in modification, which is further reflected by a recent analysis of the kinetics of competing alkylation reactions of HNE with human serum albumin.26
Polyunsaturated fatty acids in living systems are largely present as phospholipid esters and recent studies have focused on novel biological properties of phospholipids bearing epoxyisoprostane and epoxycyclopentenone substituents27−29 or a reactive α,β-unsaturated carbonyl oxidation product at the sn-2 position.30,31 These products stimulate inflammatory signal transduction pathways in vascular endothelial cells, serve as ligands for the scavenger receptor CD36, and facilitate lipid uptake in macrophages and the formation of foam cells.9,32 Most of the implicated phospholipids have electrophilic substituents and could be expected to form covalent protein adducts. However, the identities of proteins covalently modified by phospholipid electrophiles have not been reported.
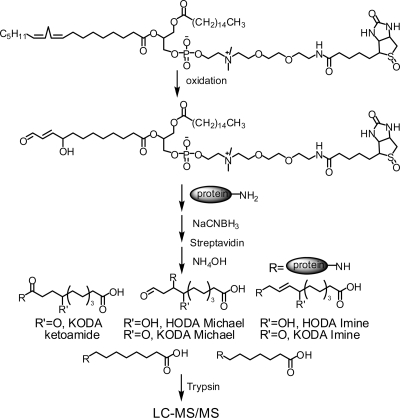
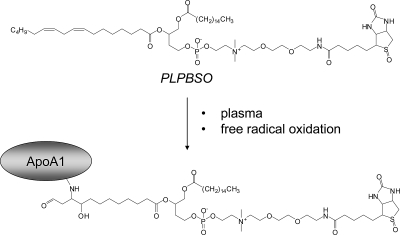
A major challenge in studying protein damage by lipid electrophiles is the sheer diversity of products generated.33,34 For example, the oxidative decomposition of a phospholipid containing linoleate at the sn-2 position would yield several electrophilic phospholipid products. HNE and 4-oxononenal are formed from the ω-end of linoleate ester,35 but other reactive electrophiles contain the carboxy-end of the linoleate ester. Thus, ketooxododecenoate (KODA), hydroxyoxododecenoate (HODA), and other adducts esterified to phospholipid are expected to be among the electrophile adducts formed with proteins from oxidized linoleate-containing phospholipids (Figure 1).
Figure 1.
Structure of PLPBSO and application to identification of protein targets of phospholipid electrophiles. See text for discussion.
Characterization of the protein targets of the family of electrophiles formed from a phospholipid is also complicated by the relatively low levels of adducts compared to unmodified proteins in relevant biological systems. Analysis of the protein targets of a family of phospholipid electrophiles requires a means of high affinity capture of the adducted proteins. Recently, we reported the synthesis of the biotinylated phosphatidylcholine PLPBSO, which is the biotin-sulfoxide analogue of 1-palmitoyl-2-linoleoylglycerylphosphatidyl-choline (PLPC).36 Oxidation of PLPBSO formed KODA, HODA, and other expected linoleate oxidation products and also formed the corresponding electrophile adducts with a model peptide. In that work, we also demonstrated feasibility of streptavidin capture of PLPBSO-adducted human serum albumin from incubation of the protein with oxidized PLPBSO.
Here, we describe the use of PLPBSO as a probe to identify major protein targets of oxidized phospholipids in human plasma. This approach identified apolipoprotein A1 (ApoA1) as the principal target of PLPBSO oxidation products and led us to map adducts from lipid oxidation products on this protein. We report the identification of adducts from several phospholipid electrophiles, including carboxy-terminal adducts. We further evaluated the sites of ApoA1 adduction by the prototypical lipid electrophile HNE. The concordance of ApoA1 site-specific modification between exogenous HNE and endogenous phospholipid electrophiles suggests that the identified targets may be preferred modification sites on ApoA1 in oxidative stress in vivo.
Experimental Procedures
Chemicals and Reagents
PLPBSO was synthesized as described recently.36 Plasma samples were collected from healthy human donors as part of an IRB-approved study. 2,2′-azobis[2-(2-imidazolin-2-yl)propane]-dihydrochloride (AIPH) was from Wako Chemicals (Osaka, Japan), streptavidin sepharose beads were from GE Healthcare (Piscataway, NJ), and ammonium hydroxide was from Sigma (St. Louis, MO). ApoA1 antibody (rabbit polyclonal antibody to the RLAEYHAKATEH peptide of ApoA1) was from Cayman Chemical (Ann Arbor, MI). Rabbit polyclonal antibody against HNE histidine Michael adducts was from Calbiochem (San Diego, CA). Alexa Fluor 680-conjugates streptavidin and all secondary antibodies were purchased from Molecular Probes (Carlsbad, CA). Protein G agarose was from Roche Applied Science (Indianapolis, IN). Mass spectrometry grade trypsin (Trypsin Gold) was from Promega (Madison, WI).
Isolation of Plasma
Whole blood from fasting, healthy subjects was collected in a 440 mL ACD blood collection bag (Baxter) containing 2 g of dextrose monohydrate, 1.66 g of sodium citrate dihydrate, 188 mg of anhydrous citric acid, 140 mg of monobasic sodium phosphate monohydrate, and 17.3 mg of adenine. Plasma was subsequently isolated from red blood cells by centrifuging the whole blood bag at 4200 rpm for 10 min at 22 °C.
Supplementation of PLPBSO to Plasma and Isolation of HDL
PLPBSO was dissolved in DMSO (50 mM) and was added to plasma at a final concentration 100 μM (<0.3% v). The same volume of DMSO was added to the control plasma. They were placed in 37 °C oil bath and stirred for 1.5 h. Half of the supplemented and native plasma were aliquoted into eppendorf tubes and stored at −80 °C until further use. HDL was isolated by ultracentrifugation (1.063 g/mL < d < 1.12 g/mL) from both native and supplemented plasma.37
Oxidation of Native and PLPBSO Supplemented Plasma
Native and PLPBSO supplemented plasma (50 μL) were oxidized with 5 mM AIPH overnight at 37 °C. Lipid electrophile adducts were stabilized by reduction with 30 mM NaCNBH3. A total of 50 μL of PBS was added to the sample, and 50 μL of protein-G agarose slurry then was used to remove IgG side chain that nearly comigrates with ApoA1 on SDS-PAGE. The supernatant was collected and filtered through Amicon 10 kDa molecular weight cutoff filter to remove small molecules residual azo initiator and reducing agent. The samples then were diluted to 300 μL with 100 mM ammonium bicarbonate, treated with 10 mM DTT at 50 °C for 30 min, and then with 20 mM IAM in the dark at room temperature.
Streptavidin Capture of Biotinylated Proteins for Protein Identification
Streptavidin sepharose beads (100 μL slurry) were washed with 3 × 500 μL of 6 M urea in PBS. The oxidized, reduced, and alkylated native or PLPBSO supplemented plasma samples were added to the washed streptavidin beads and allowed to rotate for 2 h at room temperature. The supernatant was discarded and the beads were washed with 3 × 500 μL of 6 M urea in PBS with rotation for 15 min at room temperature, then with 3 × 500 μL of 1 M NaCl with rotation for 15 min at room temperature, and finally with 3 × 500 μL of 50 mM ammonium bicarbonate. Elution of biotinylated proteins was done by selective hydrolysis of the PLPBSO acyl ester using 100 μL of 15% NH4OH38 in 3 M urea overnight at room temperature. After hydrolysis, the supernatant was collected and the beads were rinsed with 2 × 100 μL of 50 mM ammonium bicarbonate and combined with the eluate, which was placed under house vacuum for 30 min to remove excess NH3. The pH was adjusted to ∼8 with 30% acetic acid.
Sample Digestion, Preparation, and LC-MS-MS Analysis of Peptides by Gas-Phase Fractionation
Samples were enzymatically digested for 18−24 h at 37 °C using 0.5 μg of trypsin and the solution then was acidified to pH 3 with formic acid to halt the digestion. Then, the samples were evaporated under vacuum, desalted using Zip-Tips (Millipore C18, Millipore, Billerica, MA), and resuspended in 200 μL of 0.1% formic acid for LC-MS−MS analysis.
LC-MS−MS analyses were performed on a Thermo LTQ linear ion trap instrument (Thermo Electron, San Jose, CA) equipped with a Thermo Surveyor HPLC system, nanospray source, and microautosampler. Peptides were resolved on a 100 μm × 11 cm fused silica capillary column (Polymicro Technologies, LLC, Phoenix, AZ) packed with 5 μm, 300 Å Jupiter C18 (Phenomenex, Torrance, CA). Liquid chromatography was carried out at ambient temperature at a flow rate of 0.2 mL min−1 using a gradient mixture of 0.1% (v/v) formic acid in water and 0.1% (v/v) formic acid in acetonitrile. Peptides eluting from the capillary tip were introduced into the LTQ nanoelectrospray source with a capillary voltage of approximately 2 kV. A gas-phase fractionation method39 was used to increase identifications of lower abundance proteins that were present. Each sample was run three times, first with a full scan and precursor selection in the range of m/z 400−600, then m/z 600−900, and finally m/z 900−2000. Five data-dependent MS−MS scans were performed on the selected precursor ions from each full scan. MS−MS spectra were recorded using dynamic exclusion of previously analyzed precursors for 30 s with a repeat of 1 and a repeat duration of 2. MS−MS data were evaluated using the Sequest algorithm40 and the IPI Human database, which included both forward and reverse sequences for all entries. Sequest outputs were organized and analyzed with a custom database utility called CHIPS (Complete Hierarchical Integration of Protein Searches). CHIPS allows the user to filter Sequest outputs based on Sequest output parameters such as XCorr, Rsp, and Sp scores. In this case, both the native oxidized plasma (control) and PLPBSO supplemented oxidized plasma MS−MS data were filtered using Xcorr score thresholds of ≥2 for 1+ peptides, Xcorr ≥2.5 for 2+ peptides, and Xcorr ≥3 for 3+ peptides with Rsp ≤ 5 and Sp ≥ 350. The false positive rate for peptide identification based on reversed sequence identifications with these criteria was 13.4%. The requirement that protein identifications require at least two distinct peptides per protein reduced the false positive rate for protein identification to 0.4%. Proteins that were identified after biotin-streptavidin capture in both the control and PLPBSO supplemented oxidized plasma sample were removed and the remaining identified proteins were considered covalently adducted by PLPBSO.
Confirmation of Biotinylated Lipid Adducts Using Immunoblotting Techniques
Proteins captured on streptavidin beads as described above were eluted using 1 M formic acid in acetonitrile/water (3:2, v/v) overnight at room temperature after washing with the solutions described above. The supernatant was removed and the beads were rinsed with 2 × 100 μL of 50 mM ammonium bicarbonate. The combined elution mixture was evaporated under vacuum. The eluted proteins were resolved by 10% NuPAGE Novex Bis-Tris gel and the resolved proteins then were transferred to a PVDF membrane, which was incubated with Alexa Fluor 680-conjugated streptavidin and imaged using the LiCOR Odyssey imaging system (Licor, Lincoln, NE).
Confirmation of ApoA1 Adduction by Immunoblotting
Both oxidized and unoxidized plasma were rotated with 100 μL of Protein A-Agarose slurry (see above) to remove IgG light chain, which nearly coelutes with ApoA1. The remaining plasma proteins were resolved on a 10% NuPAGE Novex Bis-Tris gel using MES running buffer and the proteins were electrophoretically transferred to a PVDF membrane. The blocked membrane was incubated with rabbit polyclonal antibody raised to the RLAEYHAKATEH peptide of ApoA1 and immunoblot images were acquired with the LiCOR Odyssey imaging system.
Identification of Biotinylated Lipid Oxidation Induced Adducts on ApoA1
PLPBSO supplemented oxidized plasma was incubated with 100 μL of streptavidin sepharose slurry followed by the stringent washing as described above. The immobilized biotinylated proteins then were eluted from the beads by hydrolysis of the phospholipid fatty acyl ester bonds with 15% NH4OH for 12−15 h at room temperature. The eluted proteins were resolved on a 10% NuPAGE Novex Bis-Tris gel and stained with colloidal blue and the ApoA1 band (∼25−28 kDa) was excised and subjected to in-gel tryptic digestion.41 The eluted tryptic peptide solution was evaporated to dryness and the peptides were resuspended in 50 μL of 0.1% formic acid in water for LC-MS−MS analysis. MS−MS spectra were acquired as described above, except that gas-phase fractionation was not used; precursor ions for MS−MS were selected from full scans over the range m/z 300−2000. The data were analyzed with the P-Mod algorithm42 using ApoA1 tryptic peptide sequences to detect MS−MS spectra corresponding to mass-modified ApoA1 peptides. P-Mod is available for download at http://www.mc.vanderbilt.edu/msrc/bioinformatics/software.php. Additional analyses used a similar algorithm in development in our laboratory, called MonsterMod, which is similar to P-Mod, but includes improvements in the scoring and statistical models for generation and evaluation of sequence-to-spectrum matches (Ham, A. L.; Wernke, G. R.; Tabb, D. L.; Kim, H. Y.; Liebler, D. C., manuscript in preparation).
Identification of HNE Adduction Sites in ApoA1
To characterize the formation of HNE adducts in HNE-treated samples, isolated HDL was treated with HNE at concentrations of 1 μM, 10 μM, 100 μM, and 1 mM. The adducts were stabilized by reduction with 30 mM NaBH4. The HNE treated HDL proteins were resolved by SDS-PAGE on a 10% NuPAGE Novex Bis-Tris gel and stained using colloidal blue followed by tryptic in-gel digestion of the ApoA1 band between ∼20−28 kDa. To determine HNE adducts formed endogenously during HDL oxidation, HDL was incubated with AIPH initiator overnight at concentrations of 0, 100, 1000, and 5000 μM followed by NaBH4 reduction. The HDL proteins then were processed and analyzed as described immediately above. In both cases, the recovered peptides were analyzed by LC-MS−MS and the data then were searched with the P-Mod. Mass shifts of 158 corresponded to NaBH4-reduced Michael adducts.
Results
Strategy for Identification of Proteins Adducted by PLPBSO and Mapping of Adducts
The use of PLPBSO as a probe to identify protein targets of phospholipid electrophiles is depicted in Figure 1. Oxidation of PLPBSO with AIPH yields reactive aldehydes, α,β-unsaturated aldehydes, epoxy-α,β-unsaturated aldehydes, and other products that can form protein adducts that are stabilized with NaBH3CN. Oxidation products in which the electrophile comprises a remnant portion of the linoleate chain at the sn-2 position will covalently link the adducted protein to the biotinylated choline headgroup, which enables capture with streptavidin. Thus, capture of biotinylated proteins following oxidation enables capture and analysis of the protein targets of PLPBSO-derived phospholipid electrophiles.
We used NaBH3CN to stabilize adducts formed with PLPBSO oxidation products in order to detect both HODA and KODA adducts. Although NaBH4 traps Michael adducts more efficiently, it also reduces KODA ketones to hydroxyls. On the other hand, NaBH3CN reduces imines and reduces carbonyls very slowly. The latter reaction was sufficiently slow in our studies to potentially allow redistribution of adducts between Michael and imine forms. Model studies (not shown) with an HNE-adducted peptide indicated that reduction with NaBH4 for 4 h at room temperature yielded only the Michael adduct, whereas reduction with NaBH3CN under the same conditions yielded the reduced Michael adduct and nonreduced aldehyde and imine adducts. Because our product analyses were essentially qualitative in nature, this effect probably did not significantly bias our results. However, this chemistry would pose challenges for quantitative analyses of KODA adducts.
Although digestion and LC-MS−MS analysis of the captured proteins will enable their identification, the adduct peptides would most likely go undetected because the phospholipid would suppress ionization or induce fragmentations in the phospholipid moiety that would compete with peptide fragmentation, thus, precluding identification by database search of MS−MS spectra.
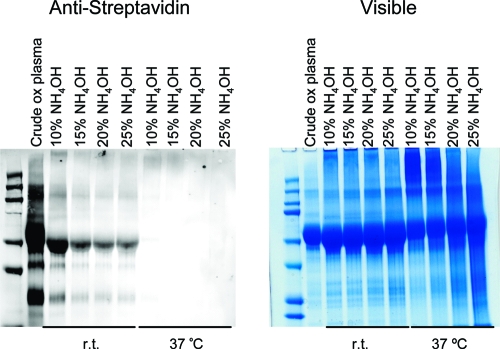
We developed a mild ammonia hydrolysis procedure that cleaves the phospholipid fatty acyl ester bonds with minimal concomitant hydrolysis of the adducted protein (Figure 1). Selective ammonia hydrolysis enabled release of the biotinylated proteins from the streptavidin beads and left protein−lipid electrophile adducts containing a carboxy-terminal group (Figure 2). Stringent wash of the streptavidin beads containing captured proteins with 6 M urea and 1 M NaCl reduced nonspecific binding to the streptavidin. Following a tryptic digest of the released proteins, both adducted and unadducted peptides were detected by LC-MS−MS analysis.
Figure 2.
Base hydrolysis of biotinylated proteins in various concentrations of NH4OH and different temperatures shown in Alexa Fluor 680-conjugated streptavidin Western blot (left) and visible coomassie-stained gel (right). Disappearance of biotin in the Western shows the degree of hydrolysis of the phospholipid headgroup. The visible gel indicates the stability of the proteins under the hydrolysis conditions.
Identification of Proteins Adducted by PLPBSO and Mapping of Adducts
Oxidation of plasma supplemented with PLPBSO was initiated using the water soluble free radical initiator AIPH. The water soluble free radical azo initiator, AIPH, was used to initiate a true free radical chain reaction. The use of the azo initiator allows the rate of oxidation to be readily controlled by adjusting the concentration of AIPH and the temperature of the reaction. Full-scan LC-MS chromatograms of the tryptic digests indicate that the streptavidin beads captured a significant amount of biotinylated protein in the PLPBSO-supplemented, oxidized plasma, as indicated by the strong ion current in the LC-MS of the corresponding tryptic digests (Figure S1A in Supporting Information). In contrast, very little protein was captured on streptavidin from the nonsupplemented, oxidized plasma samples (Figure S1B in Supporting Information). To identify proteins adducted by PLPBSO, these tryptic digests then were subjected to LC-MS−MS analysis using a gas-phase fractionation procedure and the data then were searched against the IPI Human protein database. The experiment was repeated four times and 21 proteins were confidently identified (2+ peptide identifications at 95% confidence). The proteins represented by the highest sequence coverage, reflecting those present in the PLPBSO-labeled protein pool in greatest abundance, are listed in Table 1 (A complete listing of identified proteins is provided in Table S2 in Supporting Information). ApoA1 was the most abundantly represented PLPBSO target.
Table 1. Sequence Coverage of Major Proteins Captured by PLPBSO Following Oxidation in Human Plasma.
| protein | sequence coveragea |
|---|---|
| albumin | 58.6% |
| alpha-1-antitrypsin | 26.9% |
| apolipoprotein A1 | 68.3% |
| apolipoprotein A2 | 49.0% |
| apolipoprotein A4 | 34.4% |
| apolipoprotein E | 30.6% |
| complement C3 | 24.8% |
Sequence coverage is defined as the fraction of the protein sequence accounted for by MS−MS-identified peptides.
Confirmation of ApoA1 Adduction with Oxidized Biotinylated Lipid
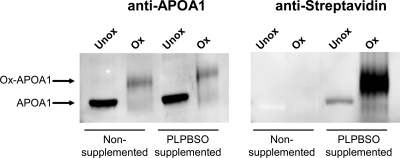
To confirm ApoA1 adduction using immunoblotting techniques, PLPBSO supplemented plasma or native plasma both were oxidized with AIPH. Immunoblotting was done with either a polyclonal antibody raised against ApoA1 peptides or Alexa Fluor 680-conjugated streptavidin. The anti-ApoA1 immunoblot indicated that, after oxidation in both the PLPBSO supplemented plasma and native plasma, the ApoA1 band is shifted to higher apparent molecular weight (Figure 3). The Alexa Fluor 680-conjugated streptavidin immunoblot shows biotin labeling of the higher molecular weight band, which is due to the covalent adduction of phospholipid to the protein.
Figure 3.
Immunoblot confirmation of PLPBSO adducts on ApoA1. Oxidation induces a migration shift of ApoA1 in SDS-PAGE analysis and streptavidin labels the PLPBSO-modified ApoA1 protein. Oxidized plasma was immunoblotted with polyclonal ApoA1 antibody followed by Alexa fluor 680 antirabbit (left) or with antistreptavidin (right).
Identification of Carboxylated Electrophile Adducts in ApoA1
To identify the adducts with carboxy-terminal electrophiles in ApoA1, PLPBSO-supplemented plasma was oxidized with AIPH followed by NaCNBH3 reduction to stabilize the adducts and PLPBSO-adducted proteins were captured with streptavidin beads. After stringent washes to remove nonspecifically bound protein, the beads were treated with 15% ammonium hydroxide to selectively hydrolyze the phospholipid esters. The eluted proteins (enriched in carboxyl-terminal lipid adducts) were resolved by SDS-PAGE and the ApoA1 band was excised for in-gel trypsin digestion followed by LC-MS−MS analysis. Analysis of the MS−MS data with the P-Mod algorithm42 searched ApoA1 tryptic peptide sequences against the MS−MS data to identify peptides that correspond to ApoA1 peptides, including adducts. This approach does not require the user to specify anticipated adduct masses or sequence specificities (e.g., His modification), but instead performs an unbiased search for spectra that match search sequences with or without mass shifts due to adducts. The program then maps adduct positions based on MS−MS fragmentation and calculates probabilities of random matches.42 The ApoA1 search sequences used included miscleaved peptides that may result from lysine adduction by electrophiles. Our criteria for reporting adducts included (1) presence of b-and/or y-ion series consistent with the adducted peptide sequence, (2) localization of adduct mass shifts by P-Mod to H or K residues capable of reacting with the electrophiles to form the indicated adducts, (3) presence of diagnostic water loss fragments formed from hydroxy-containing adducts (e.g., reduced Michael adducts).
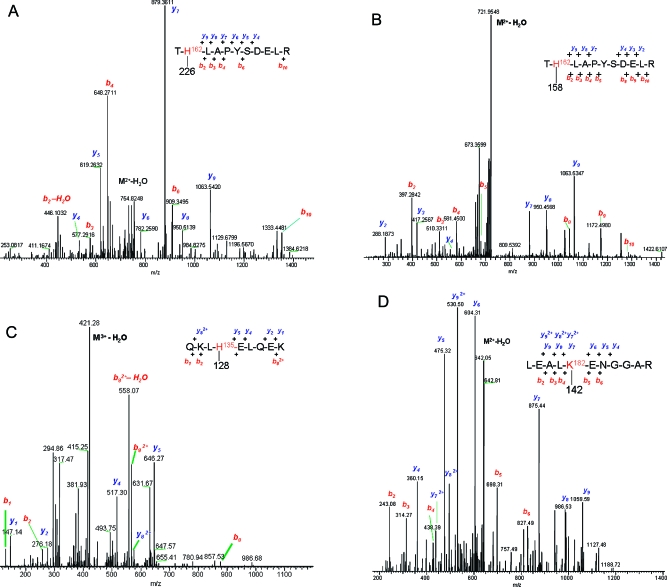
This analysis mapped sites of ApoA1 modification and the detected mass shifts enabled identification of the carboxylated electrophiles (Table 2). Annotated MS−MS spectra of these adducts are provided in Figure 5 and Figure S2 in Supporting Information. The identified residues include both lysine (K45, K106, K188, K195) and histidine residues (H135, H155, H162) and included both imine (Schiff base) and Michael adducts with carboxylated simple aldehydes, as well as carboxylated α,β-unsaturated aldehydes (HODA and KODA). As noted above, the use of NaBH3CN as a reducing agent enabled us to distinguish KODA and HODA adducts. However, we note that KODA adducts corresponding in mass to Michael addition products may also be ketoamide adducts, which were recently described by Sayre and colleagues16 and which would not be distinguished by our MS analyses.
Table 2. Carboxylated Electrophile Adducts Found in ApoA1 from Plasma Supplemented with PLPBSO and Subjected to Oxidation with AIPH.
| sequences modified | positions | mass shift | carboxylated electrophile(s) (adduct type(s)) |
|---|---|---|---|
| QLNLK*LLDNWDSVTSTFSK | K45 | 210 | KODA (imine adduct) |
| QKLH*ELQEK | H135 | 128 | 7-oxoheptanoic acid (imine adduct) |
| AH*VDALRTHLAPYSDELR | H155 | 228 | HODA (Michael adduct nonreduced) |
| TH*LAPYSDELR | H162 | 226 | KODA (Michael adduct, nonreduced or ketoamide adduct) |
| LEALK*ENGGAR | K182 | 158/142 | 8-oxoocatanoic acid (carbinolamine and imine adduct forms) |
| LEALK*ENGGAR | K182 | 128 | 7-oxooctanoic acid (imine adduct) |
| LAEYHAK*ATEHLSTLSEK | K195 | 156 | 9-oxononanoic acid (imine adduct) |
Figure 5.
LC-MS−MS of the T*H162LAPYSDELR tryptic peptide sequence from ApoA1 modified with KODA (A), with HNE (B) at His 162; of the QKL*H135ELQEK tryptic peptide sequence modified with 7-oxoheptanoic acid at His135 (C); and of the LEAL*K182ENGGAR tryptic peptide sequence modified with 8-oxooctanoic acid at Lys182 (D).
We note the formation of the apparently novel imine adducts of 7-oxoheptanoic acid, 8-oxooctanoic acid, and 9-oxononanoic acid in these analyses. The latter is formed by the Hock cleavage reaction that also generates HNE during linoleate oxidation.43 Formation of 8-oxooctanoic acid likely proceeds by β fragmentation of the linoleate 9-hydroperoxide to give the 8-yl-octanoic acid. This radical would react with oxygen to give the 8-hydroperoxyoctanoic acid which, upon dehydration, would give the 8-oxooctanoic acid compound. The mechanism for formation of the 7-oxoheptanoic acid is not obvious.
These analyses also identified some electrophile adducts derived from ω-terminal portion of the phospholipid-bound fatty acyl chain. These detected mass shifts corresponded to Schiff base adducts of hexanal, heptanal, octanal, and nonanal (Table S1 in Supporting Information). Detection of these species was less consistent from experiment-to-experiment than was detection of the carboxylated adducts described above. This may reflect the fact that the streptavidin capture and wash protocol would select against ω-terminal adducts, unless ApoA1 molecules modified by these species also were covalently labeled with the phospholipid.
Although adducts were found throughout the protein, the adduction of H162 is of interest because this residue is within a sequence known to interact with Lecithin-cholesterol acyltransferase (LCAT), an enzyme that contributes to HDL particle maturation.44
Analysis of HNE Adducts in ApoA1 Following Oxidation of HDL Particles with AIPH or Treatment with HNE
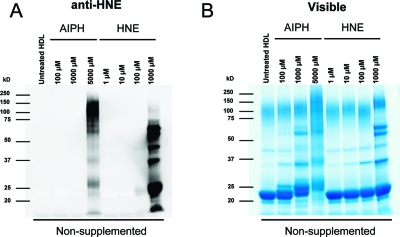
Our analyses of the streptavidin-captured ApoA1 (see above) identified relatively few adducts corresponding to modification by noncarboxylated electrophiles originating from the ω-terminus of the fatty acid (e.g., HNE). To determine if HNE adducts are formed on ApoA1 upon oxidation of native HDL associated lipids, HDL particles were isolated from plasma and then oxidized overnight with different concentrations of AIPH or treated with HNE. Adducts from both treatments were stabilized by reduction with NaBH4. Anti-HNE immunoblotting revealed immunoreactivity of the ApoA1 band (∼20−26 kDa) from HDL particles oxidized with 5 mM AIPH or treated with either 100 μM or 1 mM HNE (Figure 4A). The visible gel in Figure 4B shows that a molecular weight shift and band broadening for ApoA1 (similar to that observed in Figure 2, above) was induced by as little as 100 μM AIPH, but not by HNE treatment. This suggests that the ApoA1 migration is affected perhaps by multiple modifications of the ApoA1 protein that affect protein charge, shape and/or SDS binding, but that HNE adduction alone does not produce these effects.
Figure 4.
Isolated HDL treated with HNE and oxidized with AIPH in different concentrations. They are visualized with anti-HNE (left) or colloidal blue (right). Note the migration shift seen in the colloidal blue stained gel in the ApoA1 band between 20−26 kDa is induced by oxidation, but not by HNE.
HNE adduction sites on ApoA1 were mapped by LC-MS−MS analysis of in-gel tryptic digests of the ApoA1 band from AIPH-oxidized and HNE-treated samples described above. MS−MS spectra were acquired by data-dependent scanning mode, which automatically samples intact peptide ion signals for MS−MS.45 The MS−MS spectra were searched against a human protein sequence database with the Sequest algorithm with allowance for variable modifications on histidine and lysine residues for borohydride-reduced HNE Schiff base adducts (+140 amu) or Michael adducts (+158 amu). As a complementary approach, the sequences of ApoA1 tryptic peptides were searched against the MS−MS data with the P-Mod algorithm to enable detection of spectra corresponding to modified peptides. P-Mod hits corresponding to the expected +140 and +158 mass shifts on histidine or lysine residues were recorded. All putative spectra of adducts were subjected to manual inspection to verify the quality of sequence matches. Ten sites of adduction were identified, including K12, K23, K94, K96, K107, H135, H155, H162, H193, H199, K208, and K238 (Table 3). All of these modifications were identified as borohydride-reduced HNE Michael adducts. Lysine adducts were found on missed tryptic cleavage sites. Accurate mass measurements for the adduct precursor ions with a Thermo LTQ-Orbitrap instrument confirmed calculated values to within 3−6 ppm (Table 3).
Table 3. Sites of HNE Adduction on ApoA1 Following Treatment of HDL with HNE or Oxidation of Plasma with AIPHa.
| AIPH (μM) |
HNE (μM) |
|||||||
|---|---|---|---|---|---|---|---|---|
| sequence | 100 | 1000 | 5000 | 1 | 10 | 100 | 1000 | |
| VK*DLATVYVDVLK | K12 | 1/3b | ||||||
| DLATVYVDVLK*DSGR | K23 | 1/3 | ||||||
| DLEEVK*AK | K94 | 2/3 | 3/3 | |||||
| AK*VQPYLDDFQK | K96 | 1/3 | 3/3 | |||||
| K*WQEEMELYR | K107 | 3/3 | ||||||
| LH*ELQEK | H135 | 3/3 | 3/3 | |||||
| AH*VDALR | H155 | 1/3 | 2/3 | 3/3 | 3/3 | 3/3 | 3/3 | |
| TH*LAPYSDELR | H162 | 2/3 | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 | 3/3 |
| LAEYH*AK | H193 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| ATEH*LSTLSEK | H199 | 1/3 | 3/3 | 3/3 | 3/3 | |||
| AK*PALEDLR | K208 | 1/3 | 3/3 | 3/3 | ||||
| VSFLSALEEYTK*K | K238 | 1/3 | ||||||
Mass error (ppm) for measurements of the precursor ions of the observed peptide adducts were analyzed on a Thermo LTQ-Orbitrap instrument and were as follows: K12, 3.2 ppm; K23, 3.8 ppm; K94, 3.9 ppm; K96, 3.8 ppm; K107, 3.7; H135, 4.0 ppm; H155, 3.2 ppm; H162, 2.9 ppm; H193, 3.7; H199, 3.3; K208, 3.8 ppm; K238, 5.9.
Denotes that the adducts were detected in either 1 of 3, 2 of 3, or 3 of 3 independent experiments, respectively.
In replicate experiments in which HDL was treated with either HNE or oxidized with AIPH, histidines in the sequences TH162LAPYSDELR and LAEYH193AK were repeatedly found to be modified even with exposure to as little as 1 μM HNE (Table 3). His162, which lies within the LCAT-interacting domain of ApoA1,44 was a target for modification by both HNE and the carboxyated electrophile KODA, representative MS−MS spectra for which are both shown in Figure 5.
Discussion
Although lipid electrophiles generated during oxidative stress are widely thought to play critical roles in injury and adaptation to stress, relatively little is known about the protein targets of these species. Target identification is complicated by the great diversity of potential targets, the low abundance of modifications, and the difficulty of selectively capturing and analyzing the modified proteins. We have approached this problem by employing affinity-tagged lipid substrates that form oxidation products representative of the same lipid class, but that also enable high affinity capture of the lipid electrophile adducts. The PLPBSO probe generates oxidation products similar to the corresponding phosphatidylcholine analogue and also forms adducts to proteins.36 Use of PLPBSO enabled detection of ApoA1 as a major protein target of phospholipid electrophiles in plasma. Subsequent analysis of ApoA1 from oxidized plasma identified novel lipid electrophile adducts derived from phospholipid oxidation products. Moreover, adduct mapping studies revealed adduction in a functionally critical region of the protein.
ApoA1 is the core protein component of HDL particle, which mediates reverse cholesterol transport from tissues to the liver and also exerts antiatherogenic effects.44,46,47 ApoA1 and plasma HDL cholesterol also are inversely correlated with coronary artery disease.48 Recent work indicates that HDL function is impaired in cardiovascular disease and systemic inflammatory disorders.47 Functional impairment of HDL has been attributed to oxidative modifications of ApoA1 by myeloperoxidase (49−54) or reactive products of lipid oxidation.55,56 Thus, our results indicating that ApoA1 is a major target of reactive phospholipid electrophiles led us to focus attention on the types and site-specificity of ApoA1 adduction.
The biotin tag on the choline headgroup in PLPBSO allows affinity capture of protein adducts that contain the phospholipid headgroup. These adducts are derived from electrophiles esterified to the phospholipid and form carboxylated adducts upon ammonia hydrolysis. Nevertheless, not all adducts identified from streptativin-captured protein are necessarily derived from PLPBSO. Some of the identified adducts may originate from endogenous ApoA1 lipid esters and are formed on ApoA1 that is also adducted with PLPBSO. The formation of carboxylated lipid electrophile adducts has been studied to only a limited extent. Williams and colleagues characterized KODA and HODA adducts formed by decomposition of linoleate hydroperoxide in the presence of purified cytochrome c.17 We also observed adducts with 7-oxoheptanoic acid and 8-oxooctanoic acid on ApoA1 lysine residues. Although the formation of these imine adducts is not unexpected, this is the first report of the direct detection of these adducts, to our knowledge. The covalent binding of aldehyde-containing phospholipids has been shown previously to generate antigenic components of oxidized low density lipoprotein (LDL).57,58 These changes, in turn, promote recognition of oxidized LDL by C-reactive protein59 and functional alterations in monocytes exposed to oxidized LDL.60
A notable characteristic of ApoA1 subjected to free radical oxidation is the alteration of its migration characteristics in SDS-PAGE (Figure 3). This may reflect the formation of carboxylated adducts on histidine and lysine residues, which would generate a variety of ApoA1 forms with modified charge characteristics and possibly altered SDS binding. In contrast, treatment with HNE does not produce this effect, even when treatment produced comparable levels of HNE adduction.
Our analyses of ApoA1 adducts from PLPBSO-supplemented, oxidized plasma that was captured with streptavidin identified several carboxylated adducts, but relatively few adducts corresponding to electrophiles derived from the ω-terminus of the fatty acid (e.g., HNE). On the other hand, ApoA1 was modified by HNE formed during oxidation of isolated HDL with AIPH and by exogenously added HNE (Table 3). In both cases, the distribution of HNE adducts was similar. This confirms that ApoA1 can undergo modification both by diffusible HNE-like electrophiles and phospholipid-bound electrophiles. The somewhat sporadic detection of HNE and other noncarboxylated adducts in PLPBSO-supplemented plasma following streptavidin capture probably reflects loss of the nonphospoholipid adducted ApoA1 proteins during the wash steps to remove nonbiotinylated proteins. This also implies that, under the conditions of our experiments, most ApoA1 molecules bear a total of one or two adducts and that these are distributed between all the observed sites. Had the protein been more heavily modified, one would have expected to consistently observe HNE adducts and other noncarboxylated adducts as well as carboxylated adducts in the streptavidin-captured protein.
We identified adducts in two target sequences that have been identified previously in studies of oxidized ApoA1. Lys195 was modified by 9-oxononanoic acid and the nearby His193 was adducted by HNE. These sites are immediately adjacent to Tyr192, which was identified by the Hazen and Heinecke groups as sites of nitration and chlorination in ApoA1.49,53,54,61 The latter reaction is catalyzed by myeloperoxidase, which binds to this sequence in ApoA1 in HDL.44 We also mapped KODA and HNE adducts to His162, which resides within a sequence recently shown to be critical for interaction with LCAT.44 Tyr166 undergoes chlorination and nitration in atheromas in vivo and this modification blocks the activation of LCAT by ApoA1, which is an obligatory step in HDL particle maturation.44 His162 is also a solvent-exposed residue in the LCAT-activating loop that contains Tyr166 (44) and this raises the possibility that His162 adduction may have functional consequences similar to those reported for Tyr166 modifications.
Our studies demonstrate that His 162 and His193 are among the most easily detected adducts at low HNE exposure concentrations in HDL (Table 3). It might be tempting to conclude that this indicates high kinetic reactivity of these residues with electrophiles. However, detection in LC-MS−MS analyses depends not only on relative levels of the adducts, but also on ionization efficiency of the adducted peptides. In a recent study of the kinetics of human albumin adduction at different sites by HNE, we noted that the rate constant for formation of the most readily detected adduct (His67) was over 10-fold lower than that for the most reactive site (His242), which was less readily detected in data-dependent LC-MS−MS analyses.26 Thus, we are planning additional studies of the kinetics of ApoA1 modification in plasma and HDL to characterize the hierarchy of reactivity with lipid electrophiles.
Recent studies have highlighted the utility and potential clinical significance of oxidatively modified ApoA1.44,49,61 Our results suggest that covalent adducts with lipid electrophiles can mechanistically link lipid peroxidation, oxidative stress and disease mechanisms in a similar manner. In addition, lipid electrophile adducts of ApoA1 merit evaluation as potential biomarkers of oxidative stress in vivo.
Acknowledgments
This work was supported by National Institutes of Health Grants ES013125 and ES000267.
Supporting Information Available
Tables S1 and S2 and Figures S1 and S2. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Ames B. N. Dietary carcinogens and anticarcinogens. Oxygen radicals and degenerative diseases. Science 1983, 221, 1256–1264. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Free radicals, antioxidants, and human disease: curiosity, cause, or consequence? [see comments]. Lancet 1994, 344(8924), 721–724. [DOI] [PubMed] [Google Scholar]
- Berliner J. A.; Watson A. D. A role for oxidized phospholipids in atherosclerosis. N. Engl. J. Med. 2005, 353(1), 9–11. [DOI] [PubMed] [Google Scholar]
- Butterfield D. A. Amyloid beta-peptide (1−42)-induced oxidative stress and neurotoxicity: implications for neurodegeneration in Alzheimer’s disease brain. A review. Free Radical Res. 2002, 36(12), 1307–1313. [DOI] [PubMed] [Google Scholar]
- Perry G.; Nunomura A.; Hirai K.; Zhu X.; Perez M.; Avila J.; Castellani R. J.; Atwood C. S.; Aliev G.; Sayre L. M.; Takeda A.; Smith M. A. Is oxidative damage the fundamental pathogenic mechanism of Alzheimer’s and other neurodegenerative diseases. Free Radical Biol. Med. 2002, 33(11), 1475–1479. [DOI] [PubMed] [Google Scholar]
- Beckman K. B.; Ames B. N. The free radical theory of aging matures. Physiol. Rev. 1998, 78(2), 547–581. [DOI] [PubMed] [Google Scholar]
- Ames B. N. DNA damage from micronutrient deficiencies is likely to be a major cause of cancer. Mutat. Res. 2001, 475(1−2), 7–20. [DOI] [PubMed] [Google Scholar]
- Marnett L. J.; Riggins J. N.; West J. D. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J. Clin. Invest. 2003, 111(5), 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berliner J. A.; Zimman A. Future of toxicology-lipidomics, an important emerging area for toxicologists: focus on lipid oxidation products. Chem. Res. Toxicol. 2007, 20(6), 849–853. [DOI] [PubMed] [Google Scholar]
- Uchida K. Future of toxicology-lipid peroxidation in the future: from biomarker to etiology. Chem. Res. Toxicol. 2007, 20(1), 3–5. [DOI] [PubMed] [Google Scholar]
- Kadiiska M. B.; Gladen B. C.; Baird D. D.; Germolec D.; Graham L. B.; Parker C. E.; Nyska A.; Wachsman J. T.; Ames B. N.; Basu S.; Brot N.; Fitzgerald G. A.; Floyd R. A.; George M.; Heinecke J. W.; Hatch G. E.; Hensley K.; Lawson J. A.; Marnett L. J.; Morrow J. D.; Murray D. M.; Plastaras J.; Roberts L. J.; Rokach J.; Shigenaga M. K.; Sohal R. S.; Sun J.; Tice R. R.; Van Thiel D. H.; Wellner D.; Walter P. B.; Tomer K. B.; Mason R. P.; Barrett J. C. Biomarkers of Oxidative Stress Study II: Are oxidation products of lipids, proteins, and DNA markers of CCl(4) poisoning. Free Radical Biol. Med. 2005, 38(6), 698–710. [DOI] [PubMed] [Google Scholar]
- Doorn J. A.; Petersen D. R. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem. Res. Toxicol. 2002, 15(11), 1445–1450. [DOI] [PubMed] [Google Scholar]
- Brame C. J.; Salomon R. G.; Morrow J. D.; Roberts L. J. Identification of extremely reactive gamma-ketoaldehydes (isolevuglandins) as products of the isoprostane pathway and characterization of their lysyl protein adducts. J. Biol. Chem. 1999, 274(19), 13139–13146. [DOI] [PubMed] [Google Scholar]
- Brame C. J.; Boutaud O.; Davies S. S.; Yang T.; Oates J. A.; Roden D.; Roberts L. J. Modification of proteins by isoketal-containing oxidized phospholipids. J. Biol. Chem. 2004, 279(14), 13447–13451. [DOI] [PubMed] [Google Scholar]
- Lin D.; Lee H. G.; Liu Q.; Perry G.; Smith M. A.; Sayre L. M. 4-Oxo-2-nonenal is both more neurotoxic and more protein reactive than 4-hydroxy-2-nonenal. Chem. Res. Toxicol. 2005, 18(8), 1219–1231. [DOI] [PubMed] [Google Scholar]
- Zhu X.; Sayre L. M. Long-lived 4-oxo-2-enal-derived apparent lysine michael adducts are actually the isomeric 4-ketoamides. Chem. Res. Toxicol. 2007, 20(2), 165–170. [DOI] [PubMed] [Google Scholar]
- Williams M. V.; Wishnok J. S.; Tannenbaum S. R. Covalent adducts arising from the decomposition products of lipid hydroperoxides in the presence of cytochrome c. Chem. Res. Toxicol. 2007, 20(5), 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Minkler P. E.; Sayre L. M. Mass spectroscopic characterization of protein modification by 4-hydroxy-2-(E)-nonenal and 4-oxo-2-(E)-nonenal. Chem. Res. Toxicol. 2003, 16(7), 901–911. [DOI] [PubMed] [Google Scholar]
- Bolgar M. S.; Yang C. Y.; Gaskell S. J. First direct evidence for lipid/protein conjugation in oxidized human low density lipoprotein. J. Biol. Chem. 1996, 271(45), 27999–28001. [DOI] [PubMed] [Google Scholar]
- Carbone D. L.; Doorn J. A.; Kiebler Z.; Sampey B. P.; Petersen D. R. Inhibition of Hsp72-mediated protein refolding by 4-hydroxy-2-nonenal. Chem. Res. Toxicol. 2004, 17(11), 1459–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone D. L.; Doorn J. A.; Kiebler Z.; Petersen D. R. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem. Res. Toxicol. 2005, 18(8), 1324–1331. [DOI] [PubMed] [Google Scholar]
- Carbone D. L.; Doorn J. A.; Kiebler Z.; Ickes B. R.; Petersen D. R. Modification of heat shock protein 90 by 4-hydroxynonenal in a rat model of chronic alcoholic liver disease. J. Pharmacol. Exp. Ther. 2005, 315(1), 8–15. [DOI] [PubMed] [Google Scholar]
- Alderton A. L.; Faustman C.; Liebler D. C.; Hill D. W. Induction of redox instability of bovine myoglobin by adduction with 4-hydroxy-2-nonenal. Biochemistry 2003, 42, 4398–4405. [DOI] [PubMed] [Google Scholar]
- Sampey B. P.; Carbone D. L.; Doorn J. A.; Drechsel D. A.; Petersen D. R. 4-Hydroxy-2-nonenal adduction of extracellular signal-regulated kinase (Erk) and the inhibition of hepatocyte Erk-Est-like protein-1-activating protein-1 signal transduction. Mol. Pharmacol. 2007, 71(3), 871–883. [DOI] [PubMed] [Google Scholar]
- Suman S. P.; Faustman C.; Stamer S. L.; Liebler D. C. Proteomics of lipid oxidation-induced oxidation of porcine and bovine oxymyoglobins. Proteomics 2007, 7(4), 628–640. [DOI] [PubMed] [Google Scholar]
- Szapacs M. E.; Riggins J. N.; Zimmerman L. J.; Liebler D. C. Covalent adduction of human serum albumin by 4-hydroxy-2-nonenal: kinetic analysis of competing alkylation reactions. Biochemistry 2006, 45(35), 10521–10528. [DOI] [PubMed] [Google Scholar]
- Watson A. D.; Subbanagounder G.; Welsbie D. S.; Faull K. F.; Navab M.; Jung M. E.; Fogelman A. M.; Berliner J. A. Structural identification of a novel pro-inflammatory epoxyisoprostane phospholipid in mildly oxidized low density lipoprotein. J. Biol. Chem. 1999, 274(35), 24787–24798. [DOI] [PubMed] [Google Scholar]
- Subbanagounder G.; Leitinger N.; Schwenke D. C.; Wong J. W.; Lee H.; Rizza C.; Watson A. D.; Faull K. F.; Fogelman A. M.; Berliner J. A. Determinants of bioactivity of oxidized phospholipids. Specific oxidized fatty acyl groups at the sn-2 position. Arterioscler., Thromb., Vasc. Biol. 2000, 20(10), 2248–2254. [DOI] [PubMed] [Google Scholar]
- Subbanagounder G.; Wong J. W.; Lee H.; Faull K. F.; Miller E.; Witztum J. L.; Berliner J. A. Epoxyisoprostane and epoxycyclopentenone phospholipids regulate monocyte chemotactic protein-1 and interleukin-8 synthesis. Formation of these oxidized phospholipids in response to interleukin-1beta. J. Biol. Chem. 2002, 277(9), 7271–7281. [DOI] [PubMed] [Google Scholar]
- Podrez E. A.; Poliakov E.; Shen Z.; Zhang R.; Deng Y.; Sun M.; Finton P. J.; Shan L.; Febbraio M.; Hajjar D. P.; Silverstein R. L.; Hoff H. F.; Salomon R. G.; Hazen S. L. A novel family of atherogenic oxidized phospholipids promotes macrophage foam cell formation via the scavenger receptor CD36 and is enriched in atherosclerotic lesions. J. Biol. Chem. 2002, 277(41), 38517–38523. [DOI] [PubMed] [Google Scholar]
- Podrez E. A.; Poliakov E.; Shen Z.; Zhang R.; Deng Y.; Sun M.; Finton P. J.; Shan L.; Gugiu B.; Fox P. L.; Hoff H. F.; Salomon R. G.; Hazen S. L. Identification of a novel family of oxidized phospholipids that serve as ligands for the macrophage scavenger receptor CD36. J. Biol. Chem. 2002, 277(41), 38503–38516. [DOI] [PubMed] [Google Scholar]
- Podrez E. A.; Byzova T. V.; Febbraio M.; Salomon R. G.; Ma Y.; Valiyaveettil M.; Poliakov E.; Sun M.; Finton P. J.; Curtis B. R.; Chen J.; Zhang R.; Silverstein R. L.; Hazen S. L. Platelet CD36 links hyperlipidemia, oxidant stress and a prothrombotic phenotype. Nat. Med. 2007, 13(9), 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter N. A.; Caldwell S. E.; Mills K. A. Mechanisms of free radical oxidation of unsaturated lipids. Lipids 1995, 30(4), 277–290. [DOI] [PubMed] [Google Scholar]
- Roberts L. J.; Salomon R. G.; Morrow J. D.; Brame C. J. New developments in the isoprostane pathway: identification of novel highly reactive gamma-ketoaldehydes (isolevuglandins) and characterization of their protein adducts. FASEB J. 1999, 13(10), 1157–1168. [DOI] [PubMed] [Google Scholar]
- Lee S. H.; Oe T.; Blair I. A. Vitamin C-induced decomposition of lipid hydroperoxides to endogenous genotoxins. Science 2001, 292(5524), 2083–2086. [DOI] [PubMed] [Google Scholar]
- Tallman K. A.; Kim H. Y.; Ji J. X.; Szapacs M. E.; Yin H.; McIntosh T. J.; Liebler D. C.; Porter N. A. Phospholipid-protein adducts of lipid peroxidation: synthesis and study of new biotinylated phosphatidylcholines. Chem. Res. Toxicol. 2007, 20(2), 227–234. [DOI] [PubMed] [Google Scholar]
- Schumaker V. N.; Puppione D. L. Sequential flotation ultracentrifugation. Methods Enzymol. 1986, 128, 155–170. [DOI] [PubMed] [Google Scholar]
- England P. M.; Lester H. A.; Dougherty D. A. Mapping disulfide connectivity using backbone ester hydrolysis. Biochemistry 1999, 38(43), 14409–14415. [DOI] [PubMed] [Google Scholar]
- Spahr C. S.; Davis M. T.; McGinley M. D.; Robinson J. H.; Bures E. J.; Beierle J.; Mort J.; Courchesne P. L.; Chen K.; Wahl R. C.; Yu W.; Luethy R.; Patterson S. D. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. I. Profiling an unfractionated tryptic digest. Proteomics 2001, 1(1), 93–107. [DOI] [PubMed] [Google Scholar]
- Eng J. K.; McCormack A. L.; Yates J. R. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino-Acid-Sequences in A Protein Database. J. Am. Soc. Mass Spectrom. 1994, 5(11), 976–989. [DOI] [PubMed] [Google Scholar]
- Ham A. J.; Caprioli R. M.; Gross M. L., Proteolytic Digestion Protocols. In The Encyclopedia of Mass Spectrometry, Volume 2 Biological Applications Part A: Peptides and Proteins. Elsevier Ltd.: Kidlington, Oxford, U.K., 2005; pp 10−17. [Google Scholar]
- Hansen B. T.; Davey S. W.; Ham A. J.; Liebler D. C. P-Mod: an algorithm and software to map modifications to peptide sequences using tandem MS data. J. Proteome Res. 2005, 4(2), 358–368. [DOI] [PubMed] [Google Scholar]
- Schneider C.; Tallman K. A.; Porter N. A.; Brash A. R. Two distinct pathways of formation of 4-hydroxynonenal. Mechanisms of nonenzymatic transformation of the 9- and 13-hydroperoxides of linoleic acid to 4-hydroxyalkenals. J. Biol. Chem. 2001, 276(24), 20831–20838. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Wagner M. A.; Zheng L.; Parks J. S.; Shy J. M. III; Smith J. D.; Gogonea V.; Hazen S. L. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 2007, 14(9), 861–868. [DOI] [PubMed] [Google Scholar]
- Stahl D. C.; Swiderek K. M.; Davis M. T.; Lee T. D. Data-controlled automation of liquid chromatography/tandem mass spectrometry analysis of peptide mixtures. J. Am. Soc. Mass Spectrom. 1995, 7, 532–540. [DOI] [PubMed] [Google Scholar]
- Barter P. J.; Rye K. A. Relationship between the concentration and antiatherogenic activity of high-density lipoproteins. Curr. Opin. Lipidol. 2006, 17(4), 399–403. [DOI] [PubMed] [Google Scholar]
- Kontush A.; Chapman M. J. Functionally defective high-density lipoprotein: a new therapeutic target at the crossroads of dyslipidemia, inflammation, and atherosclerosis. Pharmacol. Rev. 2006, 58(3), 342–374. [DOI] [PubMed] [Google Scholar]
- Assmann G.; Gotto A. M. Jr. HDL cholesterol and protective factors in atherosclerosis. Circulation 2004, 109(23 Suppl. 1), III8−14. [DOI] [PubMed] [Google Scholar]
- Zheng L.; Nukuna B.; Brennan M. L.; Sun M.; Goormastic M.; Settle M.; Schmitt D.; Fu X.; Thomson L.; Fox P. L.; Ischiropoulos H.; Smith J. D.; Kinter M.; Hazen S. L. Apolipoprotein A-I is a selective target for myeloperoxidase-catalyzed oxidation and functional impairment in subjects with cardiovascular disease. J. Clin. Invest. 2004, 114(4), 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls S. J.; Hazen S. L. Myeloperoxidase and cardiovascular disease. Arterioscler., Thromb., Vasc. Biol. 2005, 25(6), 1102–1111. [DOI] [PubMed] [Google Scholar]
- Nicholls S. J.; Zheng L.; Hazen S. L. Formation of dysfunctional high-density lipoprotein by myeloperoxidase. Trends. Cardiovasc. Med. 2005, 15(6), 212–219. [DOI] [PubMed] [Google Scholar]
- Peng D. Q.; Wu Z.; Brubaker G.; Zheng L.; Settle M.; Gross E.; Kinter M.; Hazen S. L.; Smith J. D. Tyrosine modification is not required for myeloperoxidase-induced loss of apolipoprotein A-I functional activities. J. Biol. Chem. 2005, 280(40), 33775–33784. [DOI] [PubMed] [Google Scholar]
- Shao B.; Bergt C.; Fu X.; Green P.; Voss J. C.; Oda M. N.; Oram J. F.; Heinecke J. W. Tyrosine 192 in apolipoprotein A-I is the major site of nitration and chlorination by myeloperoxidase, but only chlorination markedly impairs ABCA1-dependent cholesterol transport. J. Biol. Chem. 2005, 280(7), 5983–5993. [DOI] [PubMed] [Google Scholar]
- Shao B.; Oda M. N.; Bergt C.; Fu X.; Green P. S.; Brot N.; Oram J. F.; Heinecke J. W. Myeloperoxidase impairs ABCA1-dependent cholesterol efflux through methionine oxidation and site-specific tyrosine chlorination of apolipoprotein A-I. J. Biol. Chem. 2006, 281(14), 9001–9004. [DOI] [PubMed] [Google Scholar]
- Shao B.; Fu X.; McDonald T. O.; Green P. S.; Uchida K.; O’Brien K. D.; Oram J. F.; Heinecke J. W. Acrolein impairs ATP binding cassette transporter A1-dependent cholesterol export from cells through site-specific modification of apolipoprotein A-I. J. Biol. Chem. 2005, 280(43), 36386–36396. [DOI] [PubMed] [Google Scholar]
- Shao B.; O’Brien K, D.; McDonald T. O.; Fu X.; Oram J. F.; Uchida K.; Heinecke J. W. Acrolein modifies apolipoprotein A-I in the human artery wall. Ann. N.Y. Acad. Sci. 2005, 1043, 396–403. [DOI] [PubMed] [Google Scholar]
- Friedman P.; Horkko S.; Steinberg D.; Witztum J. L.; Dennis E. A. Correlation of antiphospholipid antibody recognition with the structure of synthetic oxidized phospholipids. Importance of Schiff base formation and aldol concentration. J. Biol. Chem. 2002, 277(9), 7010–7020. [DOI] [PubMed] [Google Scholar]
- Chang M. K.; Bergmark C.; Laurila A.; Horkko S.; Han K. H.; Friedman P.; Dennis E. A.; Witztum J. L. Monoclonal antibodies against oxidized low-density lipoprotein bind to apoptotic cells and inhibit their phagocytosis by elicited macrophages: evidence that oxidation-specific epitopes mediate macrophage recognition. Proc. Natl. Acad. Sci. U.S.A. 1999, 96(11), 6353–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M. K.; Binder C. J.; Torzewski M.; Witztum J. L. C-reactive protein binds to both oxidized LDL and apoptotic cells through recognition of a common ligand: Phosphorylcholine of oxidized phospholipids. Proc. Natl. Acad. Sci. U.S.A. 2002, 99(20), 13043–13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson A. D.; Leitinger N.; Navab M.; Faull K. F.; Horkko S.; Witztum J. L.; Palinski W.; Schwenke D.; Salomon R. G.; Sha W.; Subbanagounder G.; Fogelman A. M.; Berliner J. A. Structural identification by mass spectrometry of oxidized phospholipids in minimally oxidized low density lipoprotein that induce monocyte/endothelial interactions and evidence for their presence in vivo. J. Biol. Chem. 1997, 272(21), 13597–13607. [DOI] [PubMed] [Google Scholar]
- Zheng L.; Settle M.; Brubaker G.; Schmitt D.; Hazen S. L.; Smith J. D.; Kinter M. Localization of nitration and chlorination sites on apolipoprotein A-I catalyzed by myeloperoxidase in human atheroma and associated oxidative impairment in ABCA1-dependent cholesterol efflux from macrophages. J. Biol. Chem. 2005, 280(1), 38–47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.