Abstract
Krüppel-like transcription factor (KLF)13, previously shown to regulate RANTES expression in vitro, is a member of the Krüppel-like family of transcription factors that controls many growth and developmental processes. To ascertain the function of KLF13 in vivo, Klf13-deficient mice were generated by gene targeting. As expected, activated T lymphocytes from Klf13−/− mice show decreased RANTES expression. However, these mice also exhibit enlarged thymi and spleens. TUNEL, as well as spontaneous and activation-induced death assays, demonstrated that prolonged survival of Klf13−/− thymocytes was due to decreased apoptosis. Microarray analysis suggests that protection from apoptosis-inducing stimuli in Klf13−/− thymocytes is due in part to increased expression of BCL-XL, a potent antiapoptotic factor. This finding was confirmed in splenocytes and total thymocytes by real-time quantitative PCR and Western blot as well as in CD4+CD8− single-positive thymocytes by real-time quantitative PCR. Furthermore, EMSA and luciferase reporter assays demonstrated that KLF13 binds to multiple sites within the Bcl-XL promoter and results in decreased Bcl-XL promoter activity, making KLF13 a negative regulator of BCL-XL.
Exposure of T cells to foreign Ag generates activated effector cells through a process involving activation, proliferation, differentiation, and, finally, lymphocyte death. Changes in gene expression, central to each of these steps, are controlled by transcription factors that specifically bind to promoter elements and act as positive or negative regulators of transcription. Krüppel-like transcription factors (KLFs)3 regulate a large number of genes controlling growth and development that have GC-rich promoters (1). To date, 17 distinct KLFs have been identified, and all contain a highly conserved DNA-binding domain at the C terminus composed of three tandem Cys2His2 zinc-finger motifs, a variable amino terminus that contains activation or repression domains, and nuclear localization sequences adjacent to or within the zinc-finger domain.
We identified KLF13 as the dominant transcription factor controlling expression of the chemokine RANTES in T lymphocytes (2). RANTES, which has been implicated in a myriad of human diseases as diverse as AIDS, cancer, heart disease, asthma, diabetes, and organ transplant rejection, is expressed within hours in activated fibroblasts and epithelial cells, depending only upon Rel proteins for expression (3–5). In contrast, regulation of late expression of RANTES in T lymphocytes is complex (6). Expression cloning identified a number of regulatory proteins controlling RANTES expression in T lymphocytes (2, 7), including p50 and p65 Rel proteins as well as novel factors, designated RANTES factors of late-activated T lymphocytes (RFLAT)-1 (KLF13) (2, 8) and RFLAT-2 (stromelysin-1 platelet-derived growth factor-responsive element binding protein) (9). In T lymphocytes, KLF13 is translationally regulated, providing a rheostat mechanism for rapid expression of RANTES in effector and memory T cells (10). In addition, KLF13 activates the promoters for SV40, γ-globin, and SM22α and represses the promoter for cytochrome P450 CYP1A1 in vitro, indicating that the regulatory activity of KLF13 is in part tissue and/or promoter dependent (11–14).
To more fully understand the in vivo role of KLF13, mice lacking the Klf13 gene were generated by gene targeting. As predicted from the in vitro studies, expression of RANTES is decreased in activated T lymphocytes from Klf13−/− mice. However, these mice also exhibit increased numbers of lymphoid cells, suggesting that KLF13 regulates genes involved in lymphocyte proliferation and/or death. Although no differences in proliferation were observed in bone marrow or thymocytes from Klf13−/− when compared with Klf13−/− mice, both thymocytes and splenocytes of Klf13−/− mice were more resistant to apoptotic stimuli. Expression of BCL-XL, a potent antiapoptotic factor, is elevated in thymocytes and splenocytes from Klf13−/− mice, and in reporter gene assays, KLF13 suppresses transcription driven by the Bcl-xL promoter. Thus, KLF13 is a positive regulator of RANTES and a negative regulator of BCL-XL in thymocytes and splenocytes.
Materials and Methods
Gene targeting
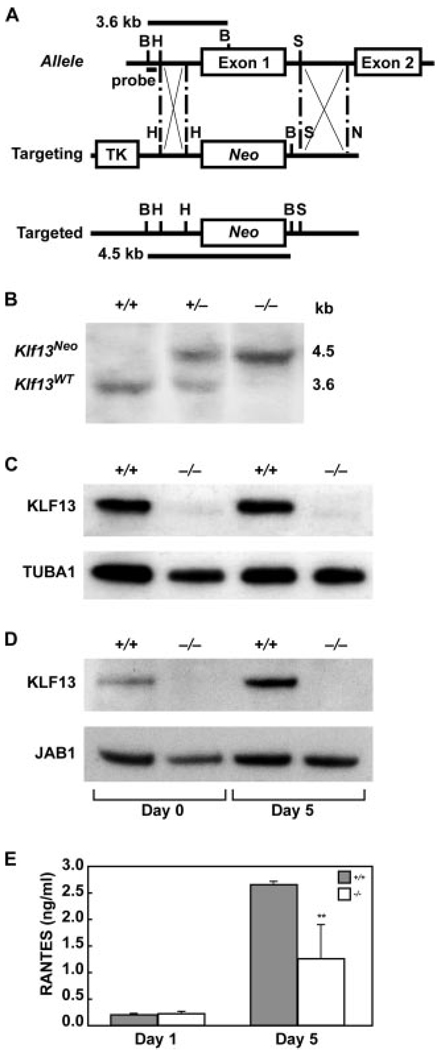
The knockout plasmid was constructed using the pKO Scrambler NTKV-1901 vector as a backbone (Stratagene). A 5.1-kb genomic fragment immediately downstream of Klf13 exon 1 and a 1.9-kb fragment upstream of exon 1 were subcloned either 5′ or 3′ of the Neo gene on the vector to generate a targeting vector (Fig. 1A). This targeting vector was linearized with NotI restriction digestion and electroporated into embryonic stem (ES) cells (129Sv) by the Stanford Transgenic Facility. Neomycin-resistant ES cell clones were screened for homologous recombination by PCR. Primers for amplifying the Klf13 allele were 5′-CTCGGTAATGTCCCGCCCATA-3′ and 5′-AGAGTCGGCCTGTCTTAGGGA-3′; primers for amplifying the replaced Neo gene were 5′-CTCGGTAATGTCCCGCCCATA-3′ and 5′-AAGCCGGTCTTGTCAATCAGGATGATCTGGACG-3′. Recombinant ES cells were confirmed by Southern blot analysis of BamHI-digested genomic DNA hybridized to a 0.9-kb probe (Fig. 1A).
FIGURE 1.
Targeted disruption of the Klf13 gene. A, Schematic representation of the wild-type Klf13 allele, the targeting vector, and the predicted targeted allele are shown. In the targeted Klf13 allele, Neo replaces exon 1 plus 0.7 kb upstream and 0.6 kb downstream of the Klf13 gene. B, BamHI; H, HindIII; S, SacII; N, NotI; TK, thymidine kinase; Neo, neomycin resistance. B, Southern blot analysis of genomic DNA from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice. After BamHI digestion, the wild-type allele (Klf13WT) is detected as a 3.6-kb fragment, whereas the targeted allele (Klf13Neo) is detected as a 4.5-kb fragment using probe as indicated in Fig. 1A. C, Western blot of KLF13 expression in thymocytes from two littermate pairs of wild-type (+/+) and mutant (−/−) mice, with TUBA1 used as an internal control. D, Western blot of KLF13 expression in splenocytes, with JAB1 used as an internal control. Splenocytes from Klf13+/+ and Klf13−/− littermates were left untreated (Day 0) or activated with anti-CD3 and anti-CD28 Abs for 5 days (Day 5). E, KLF13 positively regulates RANTES expression in T lymphocytes. Splenic CD3+ T lymphocytes isolated from Klf13+/+ and Klf13−/− littermates were stimulated with anti-CD3 and anti-CD28 Abs. Supernatants were removed on day 1 and day 5 and RANTES was measured by ELISA. Data presented are the mean ± SD from three pairs of littermates (**, p < 0.01).
Mice
Two Klf13-targeted clones were injected into C57BL/6J blastocysts. Chimeras from these clones were mated with C57BL/6J females (The Jackson Laboratory). Germline transmission of the targeted Klf13 allele was detected and heterozygous mice were backcrossed with C57BL/6J mice for six generations. Mice heterozygous for the mutant gene were then interbred to homozygosity. All mice were analyzed at the age of 11–12 wk, unless otherwise indicated. All studies performed in mice were reviewed and approved by the Stanford University Institutional Review Board.
Western blot
Whole cell extracts from thymi and spleens were prepared using Nonidet P-40 lysis buffer (250 mM NaCl, 1% Nonidet P-40, 0.25 M sodium deoxycholate, 50 mM Tris buffer (pH 8), 2 mM EDTA, 1 mM PMSF, 10 µg/ml leupeptin, and 10 µg/ml aprotinin). Total protein (20 µg) was resolved by 10% SDS-PAGE and transferred to polyvinylidene difluoride membrane. Western blot was performed using Abs specific for BCL-XL (BD Biosciences), KLF13 (2), TUBA1 (Abcam), or JAB1 (Santa Cruz Biotechnology). Protein bands were detected by ECL (Amersham Biosciences).
T lymphocyte isolation and RANTES ELISA
T lymphocytes were isolated by magnetic bead depletion of non-T lymphocytes using a MACS Pan T Cell Isolation kit (Miltenyi Biotec). For ELISA, monoclonal anti-mouse RANTES Ab (2 µg/ml, 100 µl/well, 96-well microtiter plates; R&D Systems) was used as the capture Ab and biotinylated goat anti-mouse RANTES polyclonal Ab (100 ng/ml, 100 µl/ well; R&D Systems) was used as the detecting Ab. Recombinant RANTES was purchased from R&D Systems. HRP-conjugated anti-goat Ab (1/1,000 dilution, 100 µl/well; BD Biosciences) was used to detect bound Ab. After washing, 3,3′, 5,5′-tetramethylbenzene was added and the plate was incubated at room temperature (RT) for 30 min. The reaction was stopped by addition of 50 µl of 2 N H2SO4 and absorbance was read at 450 nm.
Flow cytometric analysis and cell sorting
The following fluorescein (FITC)-, PE-, Cy-Chrome (PE-Cy5)-, or allophycocyanin-coupled Abs were obtained from BD Biosciences: CD3, CD4, CD8, CD45R/B220, NK1.1, and TCRβ. mAb specific for TCRγδ was purchased from Caltag Laboratories. Cells were blocked with unlabeled Abs against CD16 (FcRIII) and CD32 (FcRII) (Caltag Laboratories), stained on ice with optimal amounts of dye-coupled Abs diluted in staining medium (PBS, 2% BSA, 0.05% azide) for 30 min, then washed twice. Stained cells were analyzed by flow cytometry on a FACScan (BD Biosciences). Dead cells were excluded on the basis of scatter characteristics, and 10,000 events were acquired per sample. For cell sorting, total thymocytes were stained with Abs specific for mouse CD4 and CD8, and CD4+CD8− cells were purified by cell sorting on a FACS Vantage (BD Biosciences).
BrdU uptake
In vivo labeling was performed using the FITC BrdU Flow Kit (BD Biosciences) according to the manufacturer’s instructions. Briefly, mice were i.p. injected with 1 mg of BrdU and sacrificed 2 h later. Thymus and bone marrow cells were harvested, stained with FITC anti-BrdU and 7-aminoactinomycin D, and analyzed by flow cytometry on a FACScan (BD Biosciences).
CFSE labeling
Splenocytes, depleted of erythrocytes by hypotonic lysis, were suspended at a density of 2 × 107 cells/ml in PBS. An equal volume of 10 µM carboxy-fluorescein diacetate succinimidyl ester (CFDASE; Molecular Probes) in PBS was added and the cells were gently mixed for 10 min at 37°C. Unbound CFDASE, or the deacetylated form, CFSE, was quenched by the addition of an equal volume of FCS. FACS analysis of cells immediately following CFSE labeling indicated a labeling efficiency exceeding 99%. Labeled cells were cultured in vitro with anti-CD3 (0.2 µg/ml) and anti-CD28 (0.2 µg/ml) Abs (BD Biosciences). At the time of harvest (24, 48, 72, and 96 h), CFSE-labeled splenocytes were washed in cold PBS supplemented with 2% FCS and 0.01% sodium azide, and 106 cells per sample were stained with a combination of PE-conjugated CD4 and PECy5-conjugated CD8 Ab (BD Biosciences). TOPRO-3 (Molecular Probes) was used to distinguish live and dead cells. Four-color flow cytometry was performed on a FACSCalibur dual-laser cytometer: 100,000 events were collected.
Apoptosis assays
TUNEL assay
An In Situ Cell Death Detection Kit (Roche Applied Science) was used to localize 3′ DNA strand breaks following the manufacturer’s protocol. Briefly, 1.5 × 106 thymocytes were washed, permeabilized, fixed, then labeled at 37°C with TUNEL reaction mixture for 1 h. Labeled cells were analyzed by flow cytometry.
Spontaneous and activation-induced thymocyte death assays
Thymocytes were cultured in 48-well plates at 2 × 106 cells/ml in complete medium. Cells were incubated in medium with or without Con A (10 µg/ ml; Sigma-Aldrich). Cells were collected at 0, 3, 20, and 24 h followed by incubation with propidium iodide (4 µg/ml; Sigma-Aldrich) for 5 min at RT. Apoptosis was determined by flow cytometry, gating on the subdiploid population.
Staurosporine treatment
Freshly isolated cells (4 × 105 cells) were incubated at 37°C with 1 µM staurosporine in complete medium for 4 h and analyzed for annexin V and propidium iodide staining as described previously (15).
Microarray analysis
Total RNA was isolated using RNeasy Minikit (Qiagen) and genomic DNA was removed using the RNase-Free DNase Set (Qiagen). Methods for all procedures are detailed in protocols provided by the Stanford Functional Genomics Facility (http://www.microarray.org/sfgf/).
Real-time quantitative PCR (rt-qPCR)
cDNA was generated using the QuantiTect Reverse Transcription kit (Qiagen) from 1 µg of total RNA. rt-qPCR was performed in triplicate on 0.05 µg of cDNA using QuantiTect Primer Assays (Qiagen) and SYBR Green PCR Master Mix (Applied Biosystems). GusB expression was used as an endogenous control. rt-qPCR was performed on an ABI PRISM 7900-HT Sequence Detection System (Applied Biosystems). The expression of mRNA is represented as fold increase (2−ΔΔCt), where ΔΔCt = [ΔCt(−/−)] − [ΔCt(+/+)], and ΔCt = [Ct(sample)] − [Ct(Gusb)].
EMSA
32P-end-labeled probes corresponding to sequential 300-bp regions of the murine Bcl-xL promoter from −1800 to −1 bp relative to the translational start site were prepared using T4 PNK (New England Biolabs). These 300-bp regions were PCR amplified from mouse genomic DNA using the following primers designated by their position relative to the Bcl-xL translation start site: upstream primers (−1820 bp, 5′-ACGCGTCGCCTGAGACTCCTCAAATCGC-3′; −1499 bp, 5′-ACGCGTAGGTGAGAGCGCCGCCTC-3′; −1199 bp, 5′-ACGCGTCTCCAGAAGGCCGCCTTGG-3′; −897 bp, 5′-ACGCGTCTGTCTTCCCCCTGTCCGC-3′; −602 bp, 5′-AC GCGTGCAGGTTCCTAAGCTTCGCAATTCC-3′; −302 bp 5′-ACGCGTCGGATGAAACAATTCAAAGCTGGCTGG-3′) and downstream primers (−1500 bp, 5′-AGATCTGCGAGCCCCAGAGAGCCGT-3′; −1200 bp, 5′-AGATCTCCCCCTGCAGGGCTCGA-3′; −898 bp, 5′-AGATCT CCTCGGATCTGTGATCTGATTTCGG-3′; −600 bp, 5′-AGATCTTGCCCCCTCACAACCTGGTTC-3′; −300 bp, 5′-AGATCTCCTCCTCTTTTTACTACAACCACCC-3′; −1 bp, 5′-CTGAGATCTTTTTATAATAGAGATGGGCTCAACCAGTCC-3′). Recombinant GST-tagged KLF13 expression plasmid (Klf13/pGEX4T-1) was constructed by sub-cloning full-length Klf13 from RFLAT-1/pcDNA3.1(+) (2) into pGEX4T-1 (Amersham Biosciences). Recombinant GST derived from empty vector and GST-KLF13 was purified using glutathione Sepharose 4B according to the manufacturer’s protocol. For EMSA, probe was incubated at RT for 30 min with 0.3 µg of purified GST derived from empty pGEX4T-1 vector or recombinant GST-tagged KLF13 in 20 mM HEPES (pH 7.9), 1 mM MgCl2, 0.08 mM EGTA, 0.4 mM DTT, 40 mM KCl, 5% Ficoll, 1.25 mg/ml BSA, and 0.015 mg/ml poly(dI:dC) (Sigma-Aldrich). Supershift was performed by adding 1 µl of anti-KLF13 polyclonal rabbit antisera (2) or anti-p53 Ab (Santa Cruz Biotechnology). DNA-protein complexes were resolved on 4% nondenaturing acrylamide gels in 0.25× TBE buffer at 4°C. Gels were dried and exposed to film overnight.
Luciferase reporter assay
A series of luciferase reporter constructs comprised of nested deletions and sequential 300-bp regions of the murine Bcl-xL promoter were PCR-amplified and subcloned into the MluI and BglII sites of pGL3 basic (Promega). These were designated as −1,800/−1, −1,500/−1, −1,200/−1, −900/−1, −600/−1, −300/−1, −1,800/−1,500, −1,500/−1,200, −1,200/−900, −900/−600, and −600/−1 by the approximate region of upstream promoter relative to the translational start site present in each construct. The exact position and sequence of the primers used for PCR amplification of the deletion constructs were as described above for the EMSA probes. Murine KLF13 expression plasmid was constructed as described previously (2). NIH 3T3 cells (2.5 × 105) were transfected in triplicate using FuGENE 6 reagent (Roche Applied Science) and 2 µg of DNA comprised of 250 ng of specific muBcl-xL/pGL3 reporter construct and 1.75 µg of either murine KLF13 expression plasmid or pcDNA3.1(+) vector as a control. Cell extracts were harvested at 24 h and assayed for luciferase by luminometry using the luciferase reporter assay system (Promega). Luciferase activity was normalized by total cellular protein content assayed using a BCA protein quantitation kit (Pierce). Data ± SD are an average of three independent experiments.
Statistical analysis
ANOVA was used to determine differences of resistance to spontaneous and activation-induced cell death of thymocytes from Klf13+/+ and Klf13−/− mice. Other variables were determined using unpaired Student’s t test. Significance is considered at p < 0.05.
Results
Generation of Klf13-deficient mice
Homologous recombination in murine ES cells was used to produce a targeted mutation of the Klf13 gene (Fig. 1A). The resultant null mutation deleted exon 1, encoding two-thirds of the entire Klf13 gene, confirmed by PCR (data not shown) and Southern blotting (Fig. 1B). Klf13+/− ES cell clones were used to produce chimeric mice, which transmitted the targeted allele through the germline. Heterozygous mice were grossly normal and were backcrossed to the C57BL/6J strain for five generations to produce Klf13−/− mice on a pure genetic background.
In Klf13+/+ mice, KLF13 is constitutively expressed in thymocytes (Fig. 1C, lanes 1 and 3). In resting splenocytes of Klf13+/+ mice, KLF13 is expressed at low levels but increases after activation (Fig. 1D, lanes 1 and 3). In Klf13−/− mice, no Klf13 mRNA was detectable by PCR (data not shown) and no KLF13 protein was identified by Western blot in either thymocytes (Fig. 1C, lanes 2 and 4) or splenocytes (Fig. 1D, lanes 2 and 4). T lymphocytes isolated from spleens of Klf13−/− mice expressed less RANTES 5 days after activation than did cells from Klf13+/+ mice (Fig. 1E).
Enlarged lymphoid organs in Klf13−/− mice
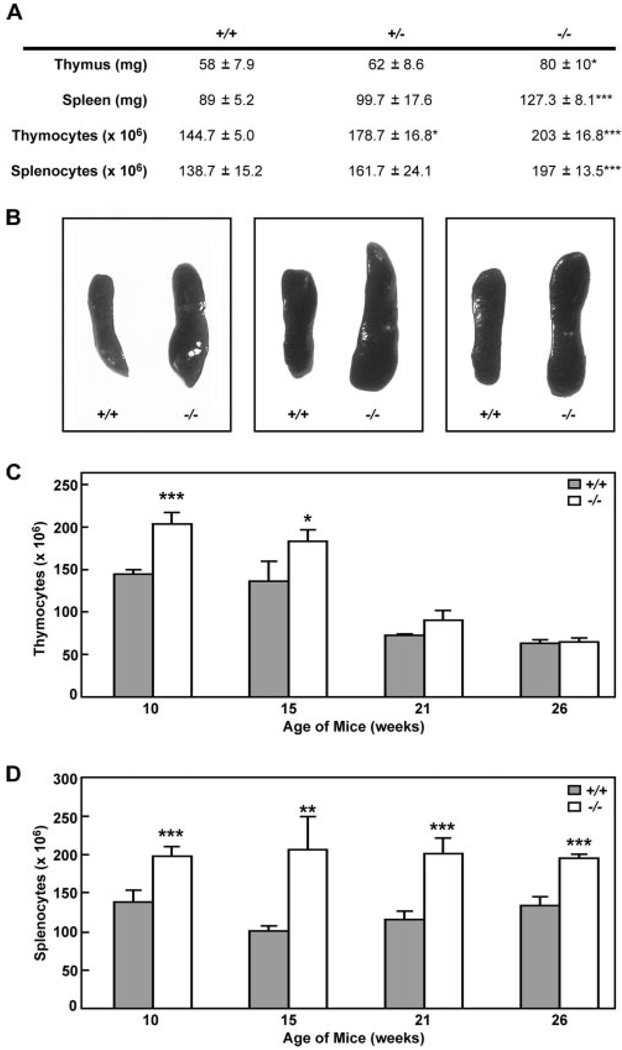
Klf13−/− mice were grossly normal but, upon pathological examination, were found to have enlarged thymi and spleens, as well as increased total thymocytes and splenocytes (Fig. 2, A and B), suggesting a defect not directly related to RANTES expression. Increased numbers of thymocytes in Klf13−/− mice compared with Klf13+/+ littermates was most pronounced in young mice and decreased over time (Fig. 2C), whereas the total number of splenocytes was increased in Klf13−/− mice at all ages examined compared with Klf13+/+ littermates (Fig. 2D). In comparison to wild-type mice, the percentage of CD4+CD8+ cells in the thymus were consistently increased in Klf13−/− mice (Table I), indicating that these cells contribute to the increased number of total thymocytes. In addition, TCRβ+ thymocytes were decreased in Klf13−/− mice, whereas no significant differences of the percentages of other lymphocyte subsets from blood, peritoneum, or spleen were observed (Table I). Pathological examination showed normal organization of thymus and spleen in Klf13−/− mice, suggesting that the enlargement of both spleen and thymus is not due to lymphocyte infiltration from nonlymphoid organs (data not shown). Although KLF13 activates the promoters for γ-globin and SM22α and represses the promoter for cytochrome P450 CYP1A1 in vitro (11–14), no gross defects in hemopoietic or smooth muscle cells were detected in Klf13−/− mice.
FIGURE 2.
Phenotypic comparison of thymus and spleen from Klf13+/+, Klf13+/−, and Klf13+/− littermates. A, Increased mass of lymphoid organs and increased numbers of thymocytes and splenocytes in Klf13−/− mice. Three sets of littermates were subjected to pathological examination and all other parameters examined were normal. B, Enlarged spleens of Klf13−/− mice from three pairs of littermates. C, Increased numbers of thymocytes in Klf13−/− mice at different ages as indicated. All values are the mean ± SD from three pairs of littermates (*, p < 0.05 and ***, p < 0.001). D, Increased numbers of splenocytes in Klf13−/− mice at different ages as indicated. All values are the mean ± SD from three pairs of littermates (**, p < 0.01 and ***, p < 0.001).
Table I.
FACS analysis of lymphocyte subsets in Klf13+/+ and Klf13−/− micea
| % of Cells | |||
|---|---|---|---|
| Subsets | Klf13+/+ | Klf13−/− | |
| Thymus | SP (CD4+) | 8.01 ± 1.3 | 5.9 ± 1.4 |
| SP (CD8+) | 2.3 ± 0.8 | 1.5 ± 0.2 | |
| DP (CD4+ CD8+) | 86.6 ± 1.3 | 89.9 ± 1.4* | |
| DN (CD4−CD8−) | 2.5 ± 1.4 | 1.9 ± 0.9 | |
| TCRβ | 38.28 ± 3.7 | 25.84 ± 4.29* | |
| Blood | T cells (CD3+) | 30.9 ± 2.7 | 29.0 ± 0.55 |
| B cells (B220+) | 23.7 ± 10.1 | 22.2 ± 7.9 | |
| NK cells (NK1.1+) | 7.5 ± 2.6 | 5.1 ± 1.0 | |
| Peritoneum | T cells (CD3+) | 18.5 ± 6.3 | 12.7 ± 2.3 |
| B cells (B220+) | 31.0 ± 12.0 | 35.7 ± 7.6 | |
| NK cells (NK1.1+) | 0.8 ± 0.2 | 1.2 ± 0.4 | |
| Spleen | T cells (CD3+) | 38.3 ± 4.9 | 31.4 ± 11.0 |
| (CD3+CD4+) | 23.7 ± 4.5 | 17.3 ± 3.8 | |
| (CD3+CD8+) | 14.7 ± 0.4 | 14.2 ± 7.5 | |
| B cells (B220+) | 39.2 ± 10.2 | 36.4 ± 11.0 | |
| NK cells (NK1.1+) | 9.5 ± 3.9 | 6.9 ± 2.7 | |
| TCRβ | 34.7 ± 10.0 | 30.4 ± 8.5 | |
| TCRγδ | 1.1 ± 0.4 | 1.4 ± 0.4 | |
Three pairs of mice were examined at 11–12 wk of age. Values are the mean ± SD. DP, Double positive; DN, double negative.
p < 0.05.
Lymphoid cells from Klf13+/+ and Klf13−/− mice exhibit similar patterns of cell proliferation, whereas thymocytes from Klf13−/− mice are more resistant to apoptosis
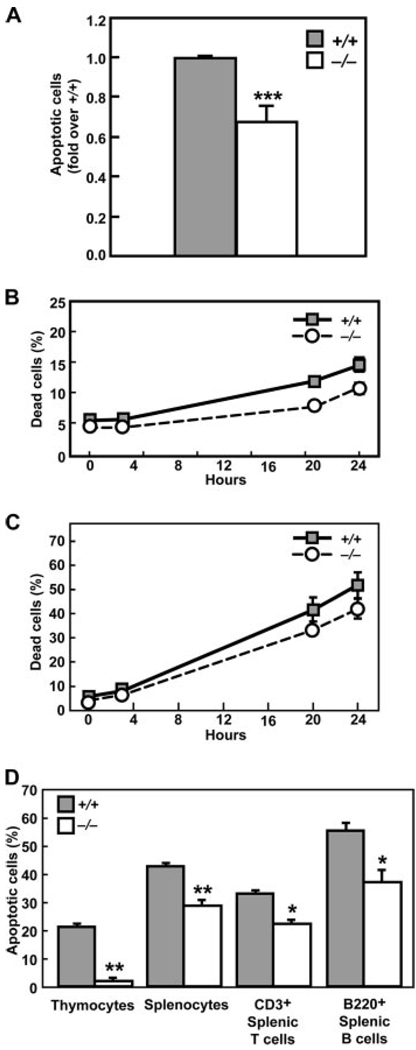
Potential mechanisms contributing to the enlarged lymphoid organs in Klf13−/− mice include increased cell proliferation or decreased cell death. To differentiate between these possibilities, proliferation and survival were evaluated in cells from bone marrow and thymus from Klf13+/+ and Klf13−/− mice. No differences in cell proliferation, as measured by BrdU uptake, were detected in these cells (Fig. 3A). Cell division of CFSE-labeled CD4+ and CD8+ splenocytes was similar in Klf13+/+ and Klf13−/− mice (Fig. 3B). Several approaches were taken to investigate whether reduced apoptosis contributed to thymic enlargement. TUNEL assay was used to measure the spontaneous cell death rate of thymocytes without in vitro culture. The percentage of apoptotic cells in thymocytes from Klf13−/− mice was reduced compared with Klf13+/+ littermates (Fig. 4A). In addition, thymocytes from Klf13−/− mice showed prolonged cell survival compared with cells from Klf13+/+ littermates under conditions approximating spontaneous or activated cell death (Fig. 4, B and C). When cultured with staurosporine, an apoptosis-inducing agent, both thymocytes and splenocytes from Klf13−/− mice were more resistant to apoptosis than those from wild-type littermates (Fig. 4D). This result was also true for CD3+ T cells and B220+ B cells isolated from spleens. Thus, thymocytes and splenocytes from Klf13−/− mice are more resistant to cell death than their Klf13+/+ counterparts.
FIGURE 3.
Normal proliferation and cell division in cells from Klf13−/− mice. A, Incorporation of BrdU into bone marrow cells (BM, top panels) and thymocytes (T, bottom panels) from Klf13+/+ and Klf13−/− mice. B, Cell division in activated Klf13+/+ and Klf13−/− CD4+ or CD8+ splenocytes. CFSE-labeled cells were cultured in vitro with anti-CD3 and anti-CD28 Abs and analyzed by flow cytometry at the indicated times. For both experiments, three littermate pairs were tested and data from one littermate pair are presented.
FIGURE 4.
Thymocytes in Klf13−/− mice exhibit defective cell death. A, TUNEL assay. Fold decrease in apoptotic cells of thymocytes in Klf13−/− mice in comparison to Klf13+/+ mice. Data are the mean ± SD from three pairs of littermates (***, p < 0.001). B, Klf13−/− thymocytes are more resistant to spontaneous cell death when cultured in vitro. Data are the mean ± SD from three pairs of littermates. Differences between Klf13+/+ and Klf13−/− mice were analyzed by ANOVA and shown to be significant (p < 0.0001). C, Klf13−/− thymocytes are more resistant to activation-induced cell death than Klf13+/+ thymocytes when cultured in vitro with Con A (10 µg/ml). Data are the mean ± SD from three pairs of littermates. Differences between Klf13+/+ and Klf13−/− mice were analyzed by ANOVA and shown to be significant (p < 0.0017). D, Cells from Klf13−/− mice are more resistant to staurosporine-induced apoptosis compared with Klf13+/+ mice. Three littermate pairs were tested in triplicate and data from one littermate pair are presented as the mean ± SD (**, p < 0.01 and ***, p < 0.001).
KLF13 inhibits expression of BCL-XL
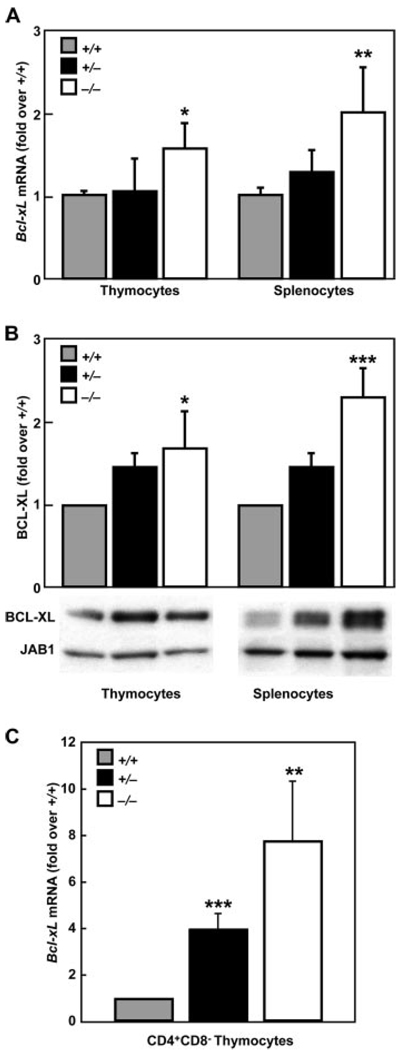
Because Klf13−/− mice have enlarged lymphoid organs and a corresponding decrease in lymphoid cell apoptosis, cDNA microarrays were used to identify molecule(s) that might be involved in this unexpected phenotype and rt-qPCR was used to confirm these data. Both microarray and rt-qPCR showed that the expression of the antiapoptotic molecule, Bcl-xL, was elevated in Klf13−/− thymocytes compared with Klf13+/+ littermates (Table II and Fig. 5A). We also examined other molecules involved in apoptosis, including Bcl-2 and caspase family members as well as Apaf1, but did not find any other differences that were detected by both methods (Table II). Western blot analyses confirmed the observation that there was a significant increase in expression in BCL-XL in thymocytes and splenocytes from Klf13−/− mice compared with Klf13+/+ littermates (Fig. 5B). BCL-XL expression in thymocytes and splenocytes from Klf13+/− mice was also examined by rt-qPCR and Western blot, and the levels were found to be intermediate between Klf13+/+ mice and Klf13−/− mice (Fig. 5, A and B). Although the majority of thymocytes are CD4+CD8+, it is the single-positive (SP) thymocytes that eventually emigrate to peripheral lymphoid tissues, including the spleen where they reside as mature T cells. Therefore, we sorted CD4+CD8− thymocytes and examined Bcl-xL expression. Due to the limited number of sorted CD4+CD8− cells obtained from each thymus, we were only able to assay Bcl-xL expression by rt-qPCR and not by Western blot. The even smaller population of CD4−CD8+ thymocytes did not allow examination of this cell type for BCL-XL expression by either method. For CD4+CD8− thymocytes, there was a significant increase in Bcl-xL expression in Klf13−/− mice compared with their Klf13+/+ littermates (Fig. 5C), suggesting that mature T cells in the periphery may be more resistant to apoptosis. This observation is consistent with the fact that CD3+ splenic T cells from Klf13−/− mice are more resistant to apoptosis than those from Klf13+/+ littermates (Fig. 4D). In addition, the fold increase in Bcl-xL mRNA observed in CD4+CD8− cells was much greater than that seen in total thymocytes, which may be due to a masking effect by the total cell population.
Table II.
Analysis of gene expression in thymocytes from Klf13+/+ and Klf13−/− micea
| Microarray |
rt-qPCR |
||
|---|---|---|---|
| Gene Symbol | GenBank Accession Number | Fold change over Klf13+/+ | Fold change over Klf13+/+ |
| Apaf1 | NM_009684 | 0.52 ± 0.05*** | 1.16 ± 0.25 |
| Bad | NM_007522 | 1.36 ± 0.31 | 1.13 ± 0.13 |
| Bag-1 | NM_009736 | 1.03 ± 0.53 | 1.15 ± 0.34 |
| Bag-3 | NM_013863 | 0.50 ± 0.12*** | 1.15 ± 0.14 |
| Bag-4 | NM_026121 | 0.45 ± 0.05** | 1.28 ± 0.30 |
| Bak1 | NM_007523 | 1.98 ± 0.23 | 0.93 ± 0.25 |
| Bax | NM_007527 | 1.41 ± 0.60 | 1.2 ± 0.02*** |
| Bbc3 | NM_133234 | 0.58 ± 0.17** | 1.49 ± 0.54 |
| Bcl2 | NM_177410 | 1.54 ± 0.75 | 1.22 ± 0.25 |
| Bcl-XL | NM_009743 | 2.38 ± 0.35*** | 1.61 ± 0.31** |
| Bcl2l2 | NM_007537 | 1.29 ± 0.70 | 1.32 ± 0.27** |
| Bcl2l10 | NM_013479 | N/A | N/A |
| Bcl2l11 | NM_207680 | 0.57 ± 0.08 | 1.05 ± 0.29 |
| Bid | NM_007544 | N/A | 1.55 ± 0.37** |
| Bid3 | NM_007545 | 0.39 ± 0.13*** | N/A |
| Bik | NM_007546 | N/A | 1.06 ± 0.19 |
| Bnip2 | NM_016787 | N/A | 0.92 ± 0.26 |
| Bnip3 | NM_009760 | N/A | 1.12 ± 0.42 |
| Bnip3L | NM_009761 | 2.2 ± 0.27 | 1.09 ± 0.24 |
| Bok | NM_016778 | 1.90 ± 0.70 | 0.99 ± 0.34 |
| Mcl1 | NM_008562 | 2.44 ± 0.29 | 0.82 ± 0.14** |
| Casp1 | NM_009807 | 0.46 ± 0.12*** | 1.17 ± 0.84 |
| Casp2 | NM_007610 | 2.16 ± 0.20*** | 1.01 ± 0.28 |
| Casp3 | NM_009810 | 2.33 ± 0.21*** | 1.36 ± 0.37 |
| Casp4 | NM_007609 | N/A | 0.82 ± 0.27 |
| Casp6 | NM_009811 | 1.98 ± 0.49 | 0.76 ± 0.19** |
| Casp7 | NM_007611 | 1.20 ± 0.17 | 1.62 ± 0.59 |
| Casp8 | NM_009812 | 1.28 ± 0.72 | 0.81 ± 0.18 |
| Casp9 | NM_015733 | 1.31 ± 0.46 | 0.96 ± 0.26 |
| Casp12 | NM_009808 | N/A | 1.21 ± 0.56 |
| Casp14 | NM_009809 | 0.59 + 0.04 | N/A |
Three pairs of mice were examined at 11–12 wk of age. Values are the mean ± SD.
p < 0.01
p < 0.001.
FIGURE 5.
Increased expression of BCL-XL in thymocytes and splenocytes from Klf13−/− mice. A, rt-qPCR analysis. Fold increase in Bcl-xL expression in thymocytes and splenocytes of Klf13−/− and Klf13+/− mice compared with Klf13+/+ mice was measured by rt-qPCR and normalized by Gusb. Values are the mean ± SD for three sets of littermates (*, p < 0.05 and **, p < 0.01). B, Western blot analysis. Fold increase in BCL-XL expression in thymocytes and splenocytes of Klf13−/− and Klf13+/− mice compared with Klf13+/+ mice was detected in whole cell lysates and normalized to JAB1. Quantitation was performed by densitometry and values are graphed as the mean ± SD from three sets of littermates (*, p < 0.05 and ***, p < 0.001). Blots shown are representative of one littermate set. C, rt-qPCR analysis of CD4+CD8− thymocytes. Fold increase in Bcl-xL expression in CD4+CD8− cells of Klf13−/− and Klf13+/− mice compared with Klf13+/+ mice was measured by rt-qPCR and normalized by Gusb. Values are the mean ± SD for three sets of littermates (*, p < 0.05 and **, p < 0.01).
The murine Bcl-xL promoter contains a number of potential KLF13 binding sites (Fig. 6A and Ref. 16). EMSA demonstrates that KLF13 can bind to multiple sites of the Bcl-xL promoter (Fig. 6B). To assess the effect of KLF13 on mouse Bcl-xL expression and to determine whether these multiple KLF13 binding sites are functional, a series nested deletions and sequential 300-bp regions from ~−1,800 to −1 bp relative to the translational start site were cloned into a luciferase reporter construct (Fig. 6C). These constructs were cotransfected with KLF13 into NIH 3T3 cells. KLF13 caused a significant decrease in reporter gene activity of the Bcl-xL promoter from each of the deletion constructs tested, suggesting that at least one putative binding site in each 300-bp region is functional. In addition, the greatest basal activity of the Bcl-xL promoter was found in the region from −1,800 to −1,500 bp, and deletion of this region caused a significant decrease in overall activity.
FIGURE 6.
KLF13 is a negative regulator of Bcl-xL expression. A, Schematic representation of the Bcl-xL promoter showing KLF13 consensus binding sites (❙). Numbers are relative to the +1 Bcl-xL translational start site. B, EMSA of recombinant GST-KLF13 binding to the Bcl-xL promoter was performed using probes corresponding to sequential 300-bp regions of the Bcl-xL promoter from −1,800 to −1 bp relative to the translational start site. Purified GST from empty pGEX4T-1 vector was used as a negative control. Supershift using anti-KLF13 Ab or a negative control Ab (p53 Ab) is indicated. C, KLF13 represses Bcl-xL promoter activity. NIH3T3 cells were cotransfected with a series of luciferase reporter constructs containing nested deletions or sequential 300-bp regions of the Bcl-xL promoter from −1,800 to −1 bp relative to the translational start site as indicated, and either pcDNA3.1(+) or mouse KLF13 expression plasmid. Luciferase activity was measured in cell lysates from transfected cells and normalized by total cellular protein concentration. Values are the mean ± SD for three independent transfections (*, p < 0.05, **, p < 0.01, and ***, p < 0.001).
Discussion
We demonstrate here that disruption of the Klf13 gene results in mice with enlarged lymphoid organs. This phenotype is associated with decreased apoptosis and increased expression of BCL-XL. Activated T lymphocytes from Klf13−/− mice produce less RANTES than cells from Klf13+/+ littermates. Thus, KLF13 is a positive regulator of RANTES and a negative regulator of BCL-XL in vivo.
Previous experiments suggest a dual role for KLF13, as well as family members KLF9 (BTEB-1), KLF1 (EKLF), KLF4 (GKLF), and KLF2 (LKLF), as both activators and repressors in vitro (1, 7, 11–13, 17). Leiden and colleagues (18, 19) generated Klf2−/−, Rag2−/− mice with peripheral T lymphocyte defects, including decreased numbers of peripheral T lymphocytes and abnormal cell surface phenotypes (CD44high, CD69high, L selectinlow), suggesting that peripheral T lymphocytes were spontaneously activated. They showed that Klf2−/− lymphocytes expressed increased Fas ligand and suggested that they were deleted by this mechanism. Thus, the Klf2−/− phenotype, in many ways the opposite of the phenotype observed in Klf13−/− mice, does not involve BCL-2 family members.
Results from in vitro studies suggest possible mechanisms by which KLF13 can repress and activate transcription. Some KLFs (KLF3 (BKLF), KLF8 (BKLF3), and KLF12 (AP-2rep)) repress transcription by binding to the cellular protein CtBP (C-terminal binding protein) (20–22); other KLFs suppress transcription by interacting with corepressors mSin3a and histone deacetylase (23– 26); whereas KLF2 binds an E3 ubiquitin ligase (27). In all of these cases, transcriptional repression is mediated by interaction with corepressors. Kaczynski et al. (13, 14) identified three repression domains in the KLF13 amino terminus that interact with the mSin3A histone deacetylase corepressor complex, repressing CYP1A1. Thus, KLF13 may decrease expression of Bcl-xL through corepressors that deacetylate histones, changing histone structure and blocking transcription. In contrast, transcriptional activation by KLF13 is associated with acetylation, the likely process regulating RANTES expression. Song et al. (28) demonstrated that the histone acetyltransferases, p300/cyclic AMP response element binding protein (CBP) and p300/CBP-associated factor (PCAF), interact in regulation of KLF13. Contingent upon the cellular context and kinetics of binding, these factors result in assembly or disassembly of the complex depending upon the acetylation state. We recently reported that KLF13 serves as a lynchpin for recruitment of a large molecular complex to the RANTES promoter that includes the MAPK Nemo-like kinase, p300/CBP, PCAF, and the ATPase BRG-1 (Brahma-related gene 1) (29). This “enhancesome” regulates gene expression via acetylation and phosphorylation. The ensuing effects of this dynamic complex on chromatin structure, at least in part, explain the ability of KLF13 to function as either a repressor or activator of gene expression.
Others have published widely on the role of BCL-XL as an inhibitor of apoptosis both in vitro and in vivo (30, 31). Lymphocytes from transgenic mice overexpressing BCL-XL exhibit increased cell survival and resistance to apoptotic stimuli (30, 31), analogous to the phenotype we describe here. However, in contrast to studies in Bcl-xL transgenic mice, we see minimal effects on cell phenotype or on positive or negative selection. There is no evidence for tumorigenesis or autoimmunity in Klf13−/− mice. Thus, overexpression of BCL-XL in transgenic mice is functionally different from the increased levels of these proteins induced by Klf13 deletion.
BCL-XL expression is regulated by various proteins including the STAT and Rel/NF-κB transcription factor families that play distinct roles in many different cell types (32). Mice that lack STAT3 in T cells have decreased IL-6-induced proliferation due to impairment of IL-6-mediated survival (33). Fetal liver cells derived from p50/p60 double-knockout mice are blocked in development at an early stage and are unable to generate B and T cells, suggesting an essential role of NF-κB in lymphocyte development (34). However, in our system, the levels of mRNA expression of STAT family members (STAT-1, -3, and -5) and NF-κB did not change in Klf13−/− thymocytes and splenocytes, although BCL-XL expression was increased in these cells compared with those of their Klf13+/+ littermates (data not shown). This result suggests that additional factors contribute to the regulation of BCL-XL expression in lymphocytes. KLF13 appears to be one of these factors and serves as a negative regulator of BCL-XL in thymocytes and splenocytes.
The potential functional consequences of KLF13 increasing RANTES expression and decreasing BCL-XL expression are as follows: 1) chemokine attraction of a robust inflammatory infiltrate composed of lymphocytes, monocytes, and eosinophils, and 2) increased susceptibility of the chemokine-producing cells to apoptotic signals. Thus, KLF13 contributes to inflammation by recruiting an immune infiltrate but then turns off this response via apoptosis.
Footnotes
This work was supported by a grant from the National Institutes of Health (DK35008). A.M.K. is the Shelagh Galligan Professor of Pediatrics.
Abbreviations used in this paper: KLF, Krüppel-like transcription factor; RFLAT, RANTES factors of late-activated T lymphocyte; ES, embryonic stem; RT, room temperature; rt-qPCR, real-time quantitative PCR; Ct, cycle threshold; SP, single positive.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song A, Chen YF, Thamatrakoln K, Storm TA, Krensky AM. RFLAT-1: a new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity. 1999;10:93–103. doi: 10.1016/s1074-7613(00)80010-2. [DOI] [PubMed] [Google Scholar]
- 3.Nelson PJ, Pattison JM, Krensky AM. Gene expression of RANTES. Methods Enzymol. 1997;287:148–162. doi: 10.1016/s0076-6879(97)87012-7. [DOI] [PubMed] [Google Scholar]
- 4.Schall TJ, Jongstra J, Dyer BJ, Jorgensen J, Clayberger C, Davis MM, Krensky AM. A human T cell-specific molecule is a member of a new gene family. J. Immunol. 1988;141:1018–1025. [PubMed] [Google Scholar]
- 5.Miyamoto NG, Medberry PS, Hesselgesser J, Boehlk S, Nelson PJ, Krensky AM, Perez HD. Interleukin-1β induction of the chemokine RANTES promoter in the human astrocytoma line CH235 requires both constitutive and inducible transcription factors. J. Neuroimmunol. 2000;105:78–90. doi: 10.1016/s0165-5728(00)00195-8. [DOI] [PubMed] [Google Scholar]
- 6.Nelson PJ, Ortiz BD, Pattison JM, Krensky AM. Identification of a novel regulatory region critical for expression of the RANTES chemokine in activated T lymphocytes. J. Immunol. 1996;157:1139–1148. [PubMed] [Google Scholar]
- 7.Song A, Nikolcheva T, Krensky AM. Transcriptional regulation of RANTES expression in T lymphocytes. Immunol. Rev. 2000;177:236–245. doi: 10.1034/j.1600-065x.2000.17610.x. [DOI] [PubMed] [Google Scholar]
- 8.Scohy S, Gabant P, Van Reeth T, Hertveldt V, Dreze PL, Van Vooren P, Riviere M, Szpirer J, Szpirer C. Identification of KLF13 and KLF14 (SP6), novel members of the SP/XKLF transcription factor family. Genomics. 2000;70:93–101. doi: 10.1006/geno.2000.6362. [DOI] [PubMed] [Google Scholar]
- 9.Rekdal C, Sjottem E, Johansen T. The nuclear factor SPBP contains different functional domains and stimulates the activity of various transcriptional activators. J. Biol. Chem. 2000;275:40288–40300. doi: 10.1074/jbc.M006978200. [DOI] [PubMed] [Google Scholar]
- 10.Nikolcheva T, Pyronnet S, Chou SY, Sonenberg N, Song A, Clayberger C, Krensky AM. A translational rheostat for RFLAT-1 regulates RANTES expression in T lymphocytes. J. Clin. Invest. 2002;110:119–126. doi: 10.1172/JCI15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asano H, Li XS, Stamatoyannopoulos G. FKLF-2: a novel Kruppel-like transcriptional factor that activates globin and other erythroid lineage genes. Blood. 2000;95:3578–3584. [PubMed] [Google Scholar]
- 12.Martin KM, Cooper WN, Metcalfe JC, Kemp PR. Mouse BTEB3, a new member of the basic transcription element binding protein (BTEB) family, activates expression from GC-rich minimal promoter regions. Biochem. J. 2000;345:529–533. [PMC free article] [PubMed] [Google Scholar]
- 13.Kaczynski JA, Conley AA, Fernandez Zapico M, Delgado SM, Zhang JS, Urrutia R. Functional analysis of basic transcription element (BTE)-binding protein (BTEB) 3 and BTEB4, a novel Sp1-like protein, reveals a subfamily of transcriptional repressors for the BTE site of the cytochrome P4501A1 gene promoter. Biochem. J. 2002;366:873–882. doi: 10.1042/BJ20020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaczynski J, Zhang JS, Ellenrieder V, Conley A, Duenes T, Kester H, van Der Burg B, Urrutia R. The Sp1-like protein BTEB3 inhibits transcription via the basic transcription element box by interacting with mSin3A and HDAC-1 co-repressors and competing with Sp1. J. Biol. Chem. 2001;276:36749–36756. doi: 10.1074/jbc.M105831200. [DOI] [PubMed] [Google Scholar]
- 15.Dong C, Li Q, Lyu SC, Krensky AM, Clayberger C. A novel apoptosis pathway activated by the carboxyl terminus of p21. Blood. 2005;105:1187–1194. doi: 10.1182/blood-2004-06-2188. [DOI] [PubMed] [Google Scholar]
- 16.Grillot DA, Gonzalez-Garcia M, Ekhterae D, Duan L, Inohara N, Ohta S, Seldin MF, Nunez G. Genomic organization, promoter region analysis, and chromosome localization of the mouse bcl-x gene. J. Immunol. 1997;158:4750–4757. [PubMed] [Google Scholar]
- 17.Song A, Patel A, Thamatrakoln K, Liu C, Feng D, Clayberger C, Krensky AM. Functional domains and DNA-binding sequences of RFLAT-1/KLF13, a Kruppel-like transcription factor of activated T lymphocytes. J. Biol. Chem. 2002;277:30055–30065. doi: 10.1074/jbc.M204278200. [DOI] [PubMed] [Google Scholar]
- 18.Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 20.Schuierer M, Hilger-Eversheim K, Dobner T, Bosserhoff AK, Moser M, Turner J, Crossley M, Buettner R. Induction of AP-2α expression by adenoviral infection involves inactivation of the AP-2rep transcriptional corepressor CtBP1. J. Biol. Chem. 2001;276:27944–27949. doi: 10.1074/jbc.M100070200. [DOI] [PubMed] [Google Scholar]
- 21.van Vliet J, Turner J, Crossley M. Human Kruppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res. 2000;28:1955–1962. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Bieker JJ. Unanticipated repression function linked to erythroid Kruppel-like factor. Mol. Cell. Biol. 2001;21:3118–3125. doi: 10.1128/MCB.21.9.3118-3125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellenrieder V, Zhang JS, Kaczynski J, Urrutia R. Signaling disrupts mSin3A binding to the Mad1-like Sin3-interacting domain of TIEG2, an Sp1-like repressor. EMBO J. 2002;21:2451–2460. doi: 10.1093/emboj/21.10.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol. Cell. Biol. 2003;23:3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved α-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol. Cell. Biol. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conkright MD, Wani MA, Lingrel JB. Lung Kruppel-like factor contains an autoinhibitory domain that regulates its transcriptional activation by binding WWP1, an E3 ubiquitin ligase. J. Biol. Chem. 2001;276:29299–29306. doi: 10.1074/jbc.M103670200. [DOI] [PubMed] [Google Scholar]
- 28.Song CZ, Keller K, Murata K, Asano H, Stamatoyannopoulos G. Functional interaction between coactivators CBP/p300, PCAF, and transcription factor FKLF2. J. Biol. Chem. 2002;277:7029–7036. doi: 10.1074/jbc.M108826200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahn YT, Huang B, McPherson L, Clayberger C, Krensky AM. Dynamic interplay of transcriptional machinery and chromatin regulates “late” expression of the chemokine RANTES in T lymphocytes. Mol. Cell. Biol. 2007;27:253–266. doi: 10.1128/MCB.01071-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chao DT, Korsmeyer SJ. BCL-2 family: regulators of cell death. Annu. Rev. Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 31.Chao DT, Korsmeyer SJ. BCL-XL-regulated apoptosis in T cell development. Int. Immunol. 1997;9:1375–1384. doi: 10.1093/intimm/9.9.1375. [DOI] [PubMed] [Google Scholar]
- 32.Grad JM, Zeng XR, Boise LH. Regulation of Bcl-xL: a little bit of this and a little bit of STAT. Curr. Opin. Oncol. 2000;12:543–549. doi: 10.1097/00001622-200011000-00006. [DOI] [PubMed] [Google Scholar]
- 33.Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol. 1998;161:4652–4660. [PubMed] [Google Scholar]
- 34.Horwitz BH, Scott ML, Cherry SR, Bronson RT, Baltimore D. Failure of lymphopoiesis after adoptive transfer of NF-κB-deficient fetal liver cells. Immunity. 1997;6:765–772. doi: 10.1016/s1074-7613(00)80451-3. [DOI] [PubMed] [Google Scholar]