Abstract
The H/ACA RNAs represent an abundant, evolutionarily conserved and functionally diverse class of non-coding RNAs. Many H/ACA RNAs direct pseudouridylation of rRNAs and snRNAs, while members of the rapidly growing group of ‘orphan' H/ACA RNAs participate in pre-rRNA processing, telomere synthesis and probably, in other nuclear processes. The yeast snR30 ‘orphan' H/ACA snoRNA has long been known to function in the nucleolytic processing of 18S rRNA, but its molecular role remained unknown. Here, we provide biochemical and genetic evidence demonstrating that during pre-rRNA processing, two evolutionarily conserved sequence elements in the 3′-hairpin of snR30 base-pair with short pre-rRNA sequences located in the eukaryote-specific internal region of 18S rRNA. The newly discovered snR30-18S base-pairing interactions are essential for 18S rRNA production and they constitute a complex snoRNA target RNA transient structure that is novel to H/ACA RNAs. We also demonstrate that besides the 18S recognition motifs, the distal part of the 3′-hairpin of snR30 contains an additional snoRNA element that is essential for 18S rRNA processing and that functions most likely as a snoRNP protein-binding site.
Keywords: box H/ACA snoRNA, non-coding RNA, RNA structure, rRNA processing, small nucleolar RNA
Introduction
Eukaryotic cells contain hundreds of small non-coding regulatory RNAs (ncRNAs), which in the form of ribonucleoproteins (RNPs) function in all aspects of gene expression, including regulation of transcription initiation and elongation, post-transcriptional modification and nucleolytic processing of primary RNA transcripts, controlling of mRNA translation, and stability and maintenance of genome integrity (Mattick and Makunin, 2005; Hannon et al, 2006).
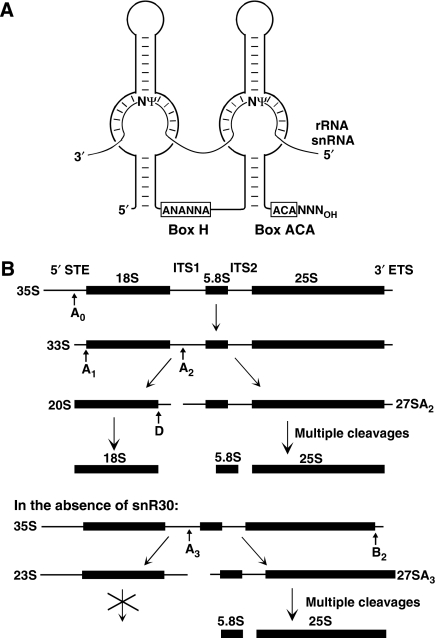
The H/ACA RNAs represent one of the largest and most ancient families of ncRNAs that are present in eukaryotes and archaea (Kiss, 2002; Meier, 2005; Terns and Terns, 2006). All H/ACA RNAs are associated with four evolutionarily conserved RNP proteins, Gar1, Nop10, Nhp2/L7ae and Cbf5p/dyskerin (Reichow et al, 2007). Eukaryotic H/ACA RNAs usually fold into two hairpin structures that are connected and followed by short single-stranded hinge and tail sequences (Balakin et al, 1996; Ganot et al, 1997b) (Figure 1A). The hinge region encompasses the conserved H box (ANANNA, where N stands for any base) and the tail contains the ACA box located three nucleotides upstream of the 3′ end. The H/ACA RNPs accumulate either in the nucleolus (small nucleolar RNPs, snoRNPs) or in the nucleoplasmic Cajal bodies (small Cajal body-specific RNPs, scaRNPs) where they function mainly in pseudouridylation of rRNAs or spliceosomal snRNAs, respectively (Ni et al, 1997; Ganot et al, 1997a; Darzacq et al, 2002; Jády et al, 2003). The H/ACA snoRNAs and scaRNAs function as guide RNAs that select target uridines for pseudouridylation (Figure 1A). Two short antisense elements located in the upper (distal) part of the pseudouridylation loop (also known as pseudouridylation pocket) in the 5′ and/or 3′-hairpin transiently base-pair with sequences flanking the target uridine (Ganot et al, 1997a; Ni et al, 1997). The selected uridine remains unpaired and it is located about 14 nt upstream from the H or ACA box of the guide RNA, providing the essential structural information for the associated Cbf5p/dyskerin pseudouridine synthase (Koonin, 1996; Lafontaine et al, 1998; Zebarjadian et al, 1999).
Figure 1.
Structure and function of H/ACA pseudouridylation guide RNAs and processing of pre-rRNA in S. cerevisiae. (A) Selection of rRNA and snRNA pseudouridylation sites by box H/ACA RNAs. Schematic structure of eukaryotic H/ACA pseudouridylation guide RNAs and the consensus sequences of boxes H and ACA are shown. N stands for any nucleotide and Ψ indicates target uridines selected for pseudouridylation. (B) Major pre-rRNA processing pathways in the presence and absence of snR30. The 18S, 5.8S and 25S rRNAs are synthesized within the 35S pre-rRNA that carries external (5′ETS and 3′ETS) and internal (ITS1 and ITS2)-transcribed spacers. Arrows (A0, A1, A2, A3, B2 and D) indicate nucleolytic-processing sites. Cleavages at the A0, A1 and A2 sites leading to mature 18S rRNA depend on snR30.
Although most H/ACA RNAs function in RNA pseudouridylation, the H/ACA RNAs represent a functionally diverse family of ncRNAs. During the past years, many H/ACA RNAs lacking recognizable complementarity to rRNA and snRNA pseudouridylation sites have been identified (Hüttenhofer et al, 2001; Vitali et al, 2003; Kiss et al, 2004; Gu et al, 2005; Li et al, 2005; Schattner et al, 2006). The function of these ‘orphan' H/ACA RNAs remains elusive. They might direct pseudouridylation of unknown substrate RNAs or more probably, they function in pseudouridylation guide-independent manner. Supporting the latter idea, two ‘orphan' H/ACA RNAs, the vertebrate telomerase scaRNA and the yeast snR30 snoRNA function in telomeric DNA synthesis and pre-rRNA processing (Collins and Mitchell, 2002; Meier, 2005; Terns and Terns, 2006; Matera et al, 2007).
In the nucleoli of yeast cells, the 18S, 5.8S and 25S rRNAs are post-transcriptionally processed by an ordered series of endo- and exonucleolytic cleavages from the RNA polymerase (Pol) I-synthesized 35S pre-rRNA that contains long external (5′ETS and 3′ETS) and internal (ITS1 and ITS2)-transcribed spacers (Fatica and Tollervey, 2002; Boisvert et al, 2007) (Figure 1B). The yeast snR30 H/ACA snoRNA has an essential function in 18S rRNA processing (Bally et al, 1988; Morrissey and Tollervey, 1993). Depletion of snR30 abolishes 18S accumulation by inhibiting cleavages of the 35S pre-rRNA at the A0, A1 and A2 processing sites. In the absence of snR30, the 35S pre-rRNA is cleaved at the A3 site to produce 23S and 27SA3 pre-rRNAs. Although the 27SA3 pre-RNA is further processed to 5.8S and 25S rRNAs, the aberrant 23S RNA is degraded. The molecular mechanism underlying the function of snR30 in 18S rRNA processing remained fully unknown.
To get further insights into the potential molecular roles of ‘orphan' H/ACA RNAs, we have recently embarked on the functional characterization of yeast snR30 (Atzorn et al, 2004). We demonstrated that snR30 is an evolutionarily conserved snoRNA and its 3′-terminal hairpin contains two conserved sequence motifs, termed m1 and m2, that are indispensable for 18S production and cell viability. In this study, we provide biochemical and genetic evidence demonstrating that the m1 and m2 sequences of snR30 function as pre-rRNA recognition elements which base-pair with two short internal 18S rRNA sequences during pre-rRNA processing. We also demonstrate that the 3′-hairpin of snR30 contains a putative snoRNP protein-binding site that, similarly to the m1 and m2 18S recognition elements, is essential for 18S rRNA processing. We propose that snR30 functions as a guide RNA that targets essential processing factor(s) to the maturing pre-rRNA.
Results
In vivo crosslinking of yeast snR30 to 18S rRNA sequences
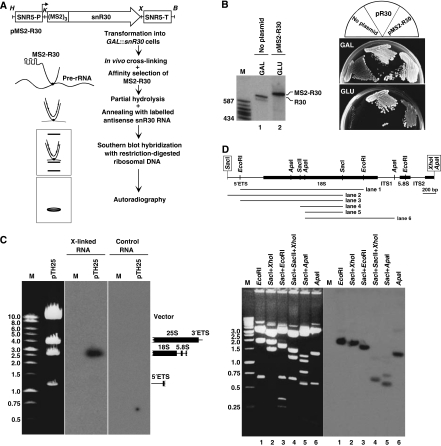
An early study showed that yeast snR30 can be crosslinked in vivo to 35S pre-rRNA, suggesting that snR30 functions in 18S processing through forming direct interaction(s) with pre-rRNA sequences (Morrissey and Tollervey, 1993). To define the region(s) of yeast 35S pre-rRNA that interact with snR30, we performed in vivo psoralen crosslinking and mapping experiments as outlined in Figure 2A. A synthetic DNA encoding three copies of a binding motif for bacteriophage MS2 coat protein was fused to the 5′ end of the coding region of yeast snR30 gene. The tagged MS2-R30 gene was placed under the control of the SNR5 constitutive promoter and transformed into the GAL∷snR30 yeast strain in which the authentic SNR30 promoter had been replaced by the inducible GAL10 promoter (Atzorn et al, 2004). When the transformed cells were grown on glucose-containing medium, only the plasmid-derived MS2-R30 RNA accumulated, as the galactose-dependent GAL10 promoter remained inactive (Figure 2B, lane 2). The MS2-R30 RNA supported cell growth on glucose, indicating that the 5′-terminal MS2-binding motifs did not interfere with snR30 function (Figure 2B).
Figure 2.
In vivo crosslinking of yeast 35S pre-rRNA with snR30. (A) An experimental strategy for localization of the in vivo interaction site of yeast 35S pre-rRNA with snR30. Structure of the pMS2-R30 expression vector used to express an snR30 RNA tagged with three binding elements for bacteriophage MS2 coat protein is shown. The promoter (SNR5-P) and terminator (SNR5-T) regions of yeast SNR5 gene and the relevant restriction sites (H, HindIII; K, KpnI; X, XhoI; B, BamHI) are indicated. For details, see the text. (B) Growth properties of yeast GAL∷snR30 strains not transformed (no plasmid) or transformed with the pR30 and pMS2-R30 expression plasmids on glucose-(GLU) and galactose-containing (GAL) medium. Expression of MS2-R30 and endogenous snR30 was verified by northern blot analysis. Lane M, molecular size markers in nucleotides (HaeIII- and TaqI-digested pBR322). (C) In vivo snR30–pre-rRNA psoralen crosslink localized by Southern blot analysis. The yeast 35S rRNA expression plasmid pTH25 was digested with a mixture of BamHI, XhoI and SalI and separated on a 0.7% agarose gel. The restriction fragments were visualized by ethidium bromide staining, transferred onto a nylon membrane and hybridized with affinity-selected MS2-R30 RNA that had been crosslinked (X-linked) to pre-rRNA and annealed with labelled antisense snR30 RNA. Control hybridization was performed with MS2-R30 RNA that had not been crosslinked to pre-rRNA. Schematic structures of yeast rDNA restriction fragments are shown on the right. Lane M, DNA size markers. (D) Mapping of in vivo interaction of snR30 with 18S rRNA. A physical map of the rDNA insert of pY18S is shown. The boxed restriction sites derived from the pBluescript cloning vector. DNA fragments obtained by restriction digestion of pY18S were separated on a 1% agarose gel, stained with ethidium bromide, transferred onto a nylon membrane and probed with crosslinked and affinity-selected MS2-R30. The hybridizing restriction fragments are indicated under the map.
From cells expressing MS2-R30, spheroplasts were prepared, incubated with AMT psoralen and irradiated with long-wave UV light. Psoralen intercalates into RNA helical regions and on photoactivation, forms covalent crosslinks between pyrimidines (predominantly uridines) on opposite strands (Cimino et al, 1985). From the psoralen-treated and UV-irradiated cells, total RNA was extracted and the MS2-R30 RNA, together with crosslinked pre-rRNAs, was affinity-selected with a recombinant MS2 coat protein that had been fused to maltose-binding protein and immobilized on amylose resin (Zhou et al, 2002). The selected MS2-R30 and the co-selected pre-RNA were partially fragmented by gentle hydrolysis, annealed with internally 32P-labelled antisense snR30 RNA and used as a probe for Southern blot analysis of restriction-digested yeast rDNA. The hybridizing restriction fragments detected by autoradiography represented pre-rRNA sequences crosslinked to MS2-R30.
When a Southern membrane carrying three restriction fragments that covered the entire yeast rDNA was probed with crosslinked and affinity-selected MS2-R30 RNA, only one hybridizing fragment was detected (Figure 2C). The highlighted DNA fragment represented the 35S pre-rRNA from the 5′-terminal region of 18S until the beginning of 25S rRNA. Importantly, a control MS2-R30 RNA selected from psoralen-treated, but not UV-irradiated cells, failed to hybridize to yeast rDNA. To locate the crosslink(s) more precisely, the 5′ portion of yeast pre-rDNA spanning the 5′ETS, 18S, ITS1, 5.8S and ITS2 regions was subcloned, fragmented by multiple restriction digestions and analysed by Southern blot hybridization with crosslinked and affinity-selected MS2-R30 RNA (Figure 2D). In this assay, the shortest efficiently hybridizing restriction fragment was a 600 bp ApaI–SacI fragment that corresponded to the C646–A1246 internal region of 18S rRNA (lane 5). As this ApaI–SacI fragment of the 18S rDNA was included into all hybridizing restriction fragments (lanes 1–4 and 6), we concluded that in living yeast cells, snR30 interacts with pre-rRNA sequences located between the C646 and A1246 residues of 18S rRNA.
Potential base-pairing interactions of snR30 and 18S rRNA
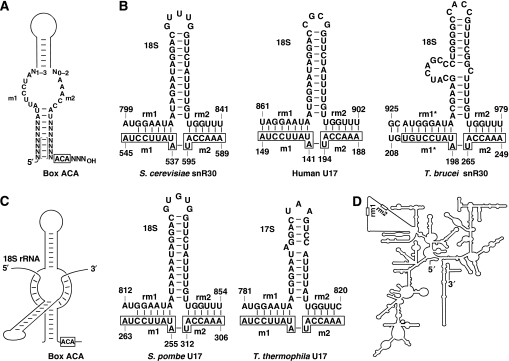
We have previously demonstrated that the 3′-hairpin of snR30 contains two evolutionarily conserved sequence motifs, m1 and m2, that are essential for 18S production (Atzorn et al, 2004). A consensus secondary structure accommodating the 3′-hairpins of all known snR30 (U17) snoRNAs is shown in Figure 3A. The unpaired m1 and m2 sequences are located on the opposite strands of an internal loop that is highly reminiscent of the pseudouridylation loop of H/ACA modification guide RNAs. Therefore, we hypothesized that similarly to the antisense elements of pseudouridylation guide RNAs, the m1 and m2 sequences of snR30 function as pre-rRNA-docking sites which base-pair with ribosomal target sequences located between the C646 and A1246 residues of 18S rRNA.
Figure 3.
A proposed interaction of snR30/U17 with 18S rRNA. (A) A consensus structure for the 3′-terminal hairpins of snR30/U17 snoRNAs. The conserved m1, m2 and box ACA sequences are shown. The bracketed base-pairing interactions are not conserved. (B) Putative base-pairing interactions of snR30/U17 snoRNAs with cognate 18S/17S rRNAs. The m1 and m2 motifs selecting ribosomal rm1 and rm2 sequences are boxed. For sequences of human, S. cerevisiae, S. pombe and T. thermophila snR30/U17 snoRNAs, see Atzorn et al (2004) and references therein. T. brucei snR30 has been reported by Barth et al (2005). Sequences of human (U13369), S. cerevisiae (J01353), S. pombe (X58056), T. thermophila (M10938) and T. brucei (AJ009142) 18S/17S rRNAs are from the GenBank. (C) A schematic representation of the predicted interaction of the 3′-hairpin of snR30/U17 snoRNAs with 18S/17S rRNA sequences. (D) A schematic two-dimensional structure of yeast 18S rRNA. A central domain of 18S that is missing from E. coli 16S rRNA is unfolded (Gutell, 1993). Positions of the rm1 and rm2 sequences are indicated.
Sequence examination of this region of yeast 18S rRNA identified two short motifs, called rm1 (G802–A806) and rm2 (U836–U841) that are able to form six base pair perfect helices with the m1 and m2 motifs of snR30, respectively (Figure 3B). The rm1 and rm2 motifs are located in the eukaryote-specific expansion segment ES6 of 18S rRNA. The rm1 and rm2 sequences are conserved in human (U861–A868 and U897–U902), S. pombe (G814–A819 and U849–U854) and Tetrahymena thermophila (G783–A788 and U815–U819) 18S rRNAs and are able to base-pair with the invariant m1 and m2 elements of cognate snR30/U17 snoRNAs (Figure 3B). The recently reported Trypanosoma brucei snR30 RNA (Barth et al, 2005), although has an invariant m2 motif that can interact with the conserved rm2 motif of T. brucei 18S rRNA (U974–U979), carries an altered m1 element (m1*). However, the base-pairing capacity of the mutant m1* motif of T. brucei snR30 with 18S rRNA is maintained by compensatory base changes in the rm1* motif of 18S rRNA. Moreover, the m1* and rm1* regions of T. brucei snR30 and 18S RNAs can form an extended interaction (10 bp) compared with the canonical m1 and rm1 elements in the other species (6–8 bp). A schematic structure of the proposed, evolutionarily conserved interaction of snR30 and 18S rRNA is shown in Figure 3C. Recent structural analyses of the ES6 elements of yeast, wheat and mouse 18S rRNAs showed that the conserved rm1 and rm2 sequences are located in a stem-loop structure (Alkemar and Nygard, 2006). Therefore, base pairing of the m1 and m2 sequences in the proximal part of the internal loop of the 3′-hairpin of snR30/U17 with the distantly located rm1 and rm2 sequences that are folded together with a stem loop of 18S/17S rRNAs results in a complex snoRNA target rRNA structure that is novel to box H/ACA RNAs.
The rm1 and rm2 sequences are required for yeast 18S rRNA processing
The rm1 and rm2 motifs are located within an internal domain of 18S rRNA that is specific for eukaryotic 18S rRNAs and that, at least on the primary sequence, is far from pre-rRNA-processing sites (Figure 3D). Therefore, we first investigated whether the rm1 and rm2 sequences are essential for pre-rRNA processing and 18S rRNA accumulation. We utilized the pTH25 rRNA expression construct that carries the yeast Saccharomyces cerevisiae 35S pre-rRNA gene under the control of the galactose-inducible GAL7 promoter (Beltrame and Tollervey, 1992; Henry et al, 1994) (Figure 4A). To facilitate monitoring of 18S and 25S expression from the plasmid-borne rDNA allele, neutral sequence tags had been inserted into the 5′-terminal regions of the 18S and 25S rRNA genes. Moreover, we introduced nucleotide changes into the rm1 and rm2 motifs of the 18S rRNA gene that reduced the potential of the resulting 18Srm1 (pTH25rm1) and 18Srm2 (pTH25rm2) rRNAs for base pairing with the m1 and m2 motifs of snR30, respectively.
Figure 4.
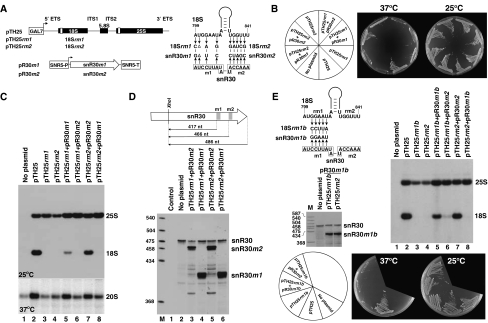
Analysis of yeast strains expressing mutant 18S and snR30 RNAs. (A) Expression constructs used to express wild-type (pTH25) and mutant (pTH25rm1 and pTH25rm2) 18S rRNAs and mutant snR30 (pR30m1 and pR30m2) snoRNAs. Tag sequences in the 18S and 25S rRNA genes are indicated by open boxes (Beltrame and Tollervey, 1992). The GAL7 and SNR5 promoters (SNR5-P) and the SNR5 terminator (SNR5-T) are shown. Nucleotide alterations introduced into 18S (18Srm1 and 18Srm2) or snR30 (snR30m1 and snR30m2) RNAs are shown. The m1 and m2 motifs of snR30 are boxed. (B) Growth properties of yeast NOY504 strains not transformed (no plasmid) or transformed with the indicated expression plasmids on galactose medium at 37 and 25°C. (C) Expression of mutant 18S RNAs. RNAs extracted from NOY504 cells transformed with the indicated plasmids were separated on a 1.2% agarose-formaldehyde gel and blotted onto a nylon membrane. Accumulation of 25S and 18S rRNAs and 20S pre-rRNA was determined by probing the blots with oligonucleotide probes specific for the tag sequences in the ectopically expressed 18S and 25S rRNAs or complementary to the ITS1 region of yeast 35S pre-rRNA (20S pre-RNA). Growth temperatures are indicated. (D) Expression of mutant snR30m1 and snR30m2 snoRNAs. RNAs extracted from yeast NOY504 strains transformed with the indicated plasmids were annealed with an antisense RNA probe complementary to wild-type snR30 and digested with a mixture of RNase A and T1. The protected fragments were separated on a 6% sequencing gel. Structures and sizes of the protected probe RNAs are shown. Positions of the protected RNAs corresponding to the wild-type and the mutant snR30 RNAs are indicated on the right. Lane control, mapping performed with E. coli tRNA. Lane M, size markers. (E) Processing of 18S rRNA carrying an altered rm1 motif can be restored by compensatory base changes in snR30. The nucleotide changes in 18Srm1b and snR30m1b RNAs are shown. Yeast NOY504 strains transformed with the indicated plasmids were grown on galactose medium. Accumulation of mutant snR30 and 18S RNAs was monitored by RNase A/T1 mapping and northern blot hybridization, respectively.
The mutant pTH25rm1 and pTH25rm2 expression constructs, as well as the pTH25 ‘wild-type' control plasmid, were transformed into the yeast NOY504 strain that carries a temperature-sensitive mutation in RNA Pol I (Nogi et al, 1991). At the 37°C non-permissive temperature, the RNA Pol I-mediated transcription of chromosomal rDNAs is blocked in the NOY504 strain. Therefore, in the transformed NOY504-pTH25, NOY504-pTH25rm1 and NOY504-pTH25rm2 strains grown on galactose-containing medium at 37°C, pre-rRNA synthesis occurred only from the Pol II-transcribed plasmid-borne rDNA alleles (Nogi et al, 1991, 1993; Beltrame et al, 1994; Beltrame and Tollervey, 1995). Consistent with this, the non-transformed NOY504 strain (no plasmid) failed to grow on galactose medium at 37°C, but the NOY504-pTH25 strain carrying the control pTH25 expression plasmid was viable under the same conditions (Figure 4B). Northern blot analysis performed with oligonucleotide probes specific for the tag sequences of 18S and 25S rRNAs expressed from the pTH25 construct confirmed that the NOY504-pTH25 strain accumulated both 18S and 25S rRNAs (Figure 4C, lane 2). In contrast, the NOY504 strains harbouring pTH25rm1 or pTH25rm2 failed to grow at 37°C (Figure 4B) and, as revealed by northern blot analysis, they did not accumulate tagged 18S rRNA at the permissive temperature 25°C (Figure 4C, lanes 3 and 4). We concluded that mutations introduced into the rm1 and rm2 motifs fully abolished accumulation of 18S rRNA. Apparently, 35S pre-rRNA synthesis occurred from the mutant pTH25rm1 and pTH25rm2 constructs, as tagged 25S rRNA accumulated in both strains. As compared with the NOY504-pTH25 control strain (lane 2), the 20S precursor of 18S RNA showed a reduced accumulation in both NOY504-pTH25rm1 and NOY504-pTH25rm2 cells (lanes 3 and 4), providing strong support to the notion that the rm1 and rm2 mutations interfered with 18S processing.
Processing of mutant 18S rRNAs can be restored by compensatory base changes in snR30
Next, we wanted to demonstrate that abolishment of accumulation of the mutant 18Srm1 and 18Srm2 rRNAs in the NOY504-pTH25rm1 and NOY504-pTH25rm2 strains was a direct consequence of the reduced base-pairing capacity of the altered rm1 and rm2 motifs with the m1 and m2 elements of snR30. To this end, we attempted to restore the processing of 18Srm1 and 18Srm2 rRNAs by expressing mutant snR30m1 and snR30m2 snoRNAs which, in principle, were capable of base pairing with 18Srm1 and 18Srm2, respectively (Figure 4A). The snR30m1 and snR30m2 genes were placed under the control of the SNR5 promoter and the resulting pR30m1 and pR30m2 expression constructs were transformed into the NOY504-pTH25rm1 and NOY504-pTH25rm2 strains. Accumulation of snR30m1 and snR30m2 was confirmed by RNase A/T1 protection that could distinguish between the endogenous wild-type and the ectopically expressed mutant snoRNAs (Figure 4D).
NOY504 cells harbouring the pTH25rm2 and pR30m2 plasmids grew efficiently on galactose medium at 37oC, demonstrating that expression of snR30m2 could suppress the lethal mutations carried by 18Srm2 (Figure 4B). Northern blot analysis revealed that accumulation of the mutant 18Srm2 rRNA and its 20S precursor was re-established in the NOY504-pTH25rm2-pR30m2 strain (Figure 4C, lane 7). As expected, expression of snR30m2 in NOY504-pTH25rm1 cells failed to rescue 18Srm1 expression (lane 6) and cell growth on galactose at 37oC (Figure 4B).
Expression of snR30m1 in the NOY504-pTH25rm1 strain restored accumulation of the 20S precursor of 18Srm1, but only partially rescued expression of the mutant 18Srm1 rRNA (Figure 4C, lane 5). The simplest interpretation of this result was that expression of snR30m1 restored processing of 18Srm1, but the rm1 mutations severely compromised the metabolic stability of the excised 18Srm1 rRNA. To test this hypothesis, we investigated the effects of further rm1 mutations on 18S rRNA processing (Figure 4E). Again, replacement of the wild-type rm1 motif (GGAAU) of pTH25 for CCUUA fully abolished accumulation of the mutant 18Srm1b rRNA in NOY504-pTH25rm1b cells (lane 3). However, co-expression of snR30m1b that carried an altered m1 motif complementary to the mutant rm1b element of 18Srm1b largely restored 18S accumulation (lane 5). It is important to note that the accumulating mutant 18Srm1 and 18Srm1b rRNAs failed to support cell growth on galactose medium at 37°C (Figure 4B and E). This strongly suggests that the rm1 motif of 18S rRNA is important also for ribosome function. In summary, we concluded that processing of mutant yeast 18S rRNAs carrying altered rm1 or rm2 elements can be restored by re-establishing the base-pairing capacity of the m1 and m2 motifs of snR30. Demonstration that the mutant snR30m1, snR30m1b and snR30m2 snoRNAs can support the processing of 18Srm1, 18Srm1b and 18Srm2 rRNAs indicated that the m1 and m2 motifs snR30, apart from interacting with the rm1 and rm2 motifs, have no other essential function in 18S processing.
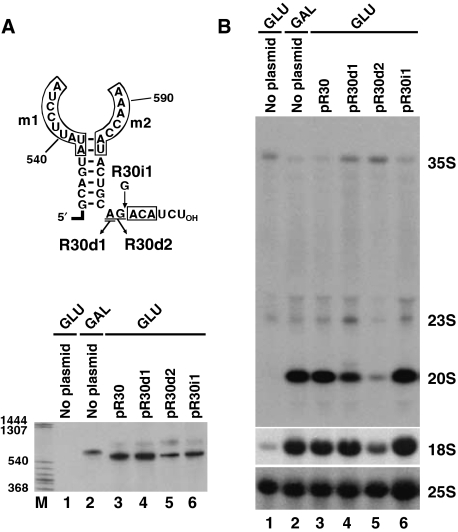
Correct spacing of the ACA box and the m1/m2 rRNA recognition motif is important for efficient 18S processing
Proper function of H/ACA pseudouridylation guide RNPs requires a 14- to 15-nt-long distance between the selected uridine and the H or ACA box of the guide RNA (Ganot et al, 1997a; Bortolin et al, 1999). We noticed that the m1/m2 rRNA recognition element of all vertebrate, yeast and protozoan snR30/U17 snoRNAs is located invariantly seven nucleotides upstream of the ACA box (Figure 3A). To test the functional significance of this structural conservation of snR30/U17 snoRNAs, mutant snR30 RNAs with increased (R30i1) or decreased (R30d1 and R30d2) ACA-m1/m2 spacing were expressed in the GAL∷snR30 strain (Figure 5A). Northern blot analysis was used to confirm accumulation of the mutant snR30 RNAs and to monitor 18S processing in the transformed cells after shifting to glucose-containing medium (Figure 5A and B). Insertion of one nucleotide between the ACA box and 3′-terminal stem of the R30i1 snoRNA had no detectable effect on rRNA processing (Figure 5B, lane 6). In contrast, removal of the A601 residue upstream of the ACA box of R30d1 had no significant effect on 18S accumulation, but reduced the steady-state level of the 20S pre-rRNA and increased accumulation of the 35S and 23S pre-rRNAs, indicating that rRNA processing was slightly compromised in these cells (lane 4). Indeed, placing the ACA box two nucleotides closer to the m1/m2 element of R30d2 by removal of the A601 and G602 tail residues almost fully abolished 20S and 18S accumulation (lane 5), demonstrating that correct spacing of the 18S recognition element and the ACA box of snR30 is essential for efficient pre-rRNA processing.
Figure 5.
Effects of altered ACA box positioning on 18S processing. (A) Structure and expression of mutant snR30 snoRNAs. The 3′-terminal portion of yeast snR30 is shown. The conserved m1, m2 and ACA motifs (boxed), deleted (underlined) and inserted (G) nucleotides are indicated. The pR30, pR30i1, pR30d1 and pR30d2 expression plasmids were transformed into the GAL∷snR30 strain and expression of the mutant snR30 RNAs was monitored by northern blot analysis after grown on glucose for 24 h. Accumulation of snR30 in non-transformed (no plasmid) cells grown either on glucose or galactose was also tested. (B) Processing of yeast pre-rRNA. Accumulation of 18S and 25S rRNA as well as 35S, 23S and 20S pre-rRNAs was monitored by Northern blot analysis.
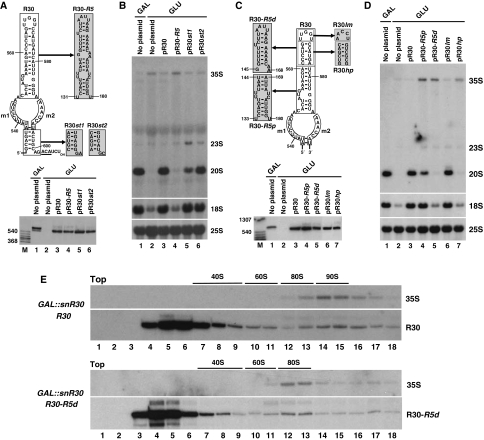
The distal part of the 3′-hairpin of snR30 carries a novel element essential for 18S processing
Earlier, biochemical purification of yeast snR30 snoRNP detected seven snoRNP proteins of 10, 23, 25, 38, 46, 48 and 65 kDa (Lübben et al, 1995). Although four of these proteins apparently corresponded to the Cbf5 (65 kDa), Gar1 (25 kDa), Nhp2 (23 kDa) and Nop10 (10 kDa) H/ACA core proteins, the remaining three putative snoRNP proteins (38, 46 and 48 kDa) seemed to be specific for the snR30 snoRNP. This raises the possibility that in addition to the H and ACA boxes, the snR30 snoRNA contains at least one protein-binding site responsible for the recruitment of snR30-specific snoRNP proteins. We have demonstrated earlier that all RNA elements essential for 18S processing are confined to the 3′-hairpin of yeast snR30 (Atzorn et al, 2004). To examine whether besides the m1 and m2 18S recognition motifs, the 3′-hairpin of snR30 carries additional essential element(s) that could function as protein-docking site(s), we first tested the functional importance of the G532–U536/A597–C600 basal 3′-terminal helix of snR30 (Figure 6A). Switching the orientation of the base pairs (snR30st1) or transition of the purines and pyrimidines (snR30st2) with simultaneous alteration of the A601 and G602 tail nucleotides preceding the ACA box had no effect on the accumulation of the mutant snR30 RNAs in transformed GAL∷snR30 cells (lanes 5 and 6). Moreover, both snR30st1 and snR30st2 supported efficient cell growth (data not shown) and 18S rRNA accumulation on glucose medium (Figure 6B, lanes 5 and 6). Although the GAL∷snR30-snR30st1 cells showed a slightly reduced 20S pre-RNA accumulation (lane 5), we concluded that the basal stem of the snR30 3′-hairpin contains no vital sequence elements for 18S processing.
Figure 6.
Identification of a novel snR30 element required for 18S processing. (A) Structure and expression of snR30 RNAs carrying altered 3′-terminal hairpins. Sequences used to replace wild-type nucleotides in the pR30 expression construct are highlighted in shaded boxes. The resulting pR30st1, pR30st2 and pR30-R5 expression plasmids were transformed into yeast GAL∷snR30 cells and expression of the mutant snR30 RNAs was monitored by northern blot analysis. (B) Northern blot analysis of pre-rRNA processing in GAL∷snR30 cells transformed with the indicated expression plasmids. (C) Structure and expression of mutant snR30-R5d, snR30-R5p, snR30hp and snR30lm RNAs in GAL∷snR30 cells. (D) Processing of pre-rRNA in GAL∷snR30 cells transformed with the indicated plasmids. (E) Sedimentation of R30-R5d snoRNA in sucrose gradient. Extracts of GAL∷snR30 cells expressing R30-R5d or R30 snoRNAs were fractionated in a 4.5–45% sucrose gradient. RNA was isolated from each of 18 fractions and analysed by northern blotting with R30- and 35S-specific oligonucleotide probes. Positions of 40S, 60S and 80S ribosomal particles are indicated.
To test the functional importance of the upper part of the 3′-hairpin of snR30, the C548–A588 sequences were replaced for the equivalent region of the yeast snR5 pseudouridylation guide snoRNA (Figure 6A). The chimaeric snR30-R5 snoRNA efficiently accumulated in GAL∷snR30 cells (Figure 6A, lane 4), but it failed to support cell growth (data not shown) and 18S processing on glucose (Figure 6B, lane 4), demonstrating that upper part of the 3′-hairpin of snR30 contains crucial element(s) for 18S processing.
To delimit more precisely the snoRNA sequences essential for 18S processing, the distal and proximal segments of the upper part of the 3′-hairpin of snR30 were replaced for the equivalent sequences of snR5 (Figure 6C). Although both R30-R5p and R30-R5d chimaeric snoRNAs accumulated in GAL∷snR30 cells (lanes 4 and 5), only R30-R5p supported cell growth (data not shown) and, with a slightly reduced efficiency, 18S processing (Figure 6D, lane 4). Cells expressing R30-R5d were not viable on glucose (data not shown) and failed to accumulate 18S rRNA (lane 5), indicating that the distal part of the 3′-hairpin of snR30 contains elements indispensable for 18S processing. Alteration of the terminal loop sequence in R30lm RNA failed to influence cell growth (data not shown) and 18S processing (Figure 6C and D, lane 6). In contrast, expression of the mutant R30hp snoRNA in which the C561–G566/C572–A577 distal region of the wild-type 3′-hairpin of snR30 was altered failed to support cell growth (data not shown) and 18S accumulation (Figure 6C and D, lane 7). Further corroborating the functional importance of these sequences, we found that alteration of either the C561–G566 or the C572–A577 sequence alone inhibited 18S processing (data not shown). Thus, we concluded that the 3′-terminal hairpin of snR30 carries an additional snoRNA element that is essential for 18S rRNA processing.
To test whether the newly identified essential sequences of snR30 are important for interaction with pre-ribosomal particles containing the 35S pre-RNA, extracts prepared from GAL∷snR30 cells expressing the mutant R30-R5d or the wild-type R30 snoRNA were fractionated by velocity sedimentation in sucrose gradients (Figure 6E). Northern blot analysis confirmed that the control R30 snoRNA efficiently associated with large 90S pre-ribosomes containing the 35S pre-rRNA (upper panel, lanes 13–17). As compared with the wild-type 90S pre-ribosomes, the 35S rRNA-containing pre-ribosomal particles derived from the mutant R30-R5d cells migrated in the gradient with reduced sedimentation velocity (lower panel, lanes 12 and 13). As a fraction of R30-R5d co-sedimented with the aberrant 35S-containing pre-ribosomal particles (lanes 12 and 13), we concluded that R30-R5d can interact with 35S pre-rRNA, but it fails to support the assembly of wild-type 90S pre-ribosomes. Importantly, the R30-R5d snoRNP monoparticles sedimented significantly slower in sucrose gradient than did the wild-type R30 snoRNPs (lower panel, lanes 3–6 versus upper panel lanes 4–7), suggesting that the R30-R5d snoRNP lacks R30-specific snoRNP protein(s). These results support the idea that instead of interacting with pre-rRNA sequences, the newly identified, functionally essential C561–G566/C572–A577 sequences in the 3′-hairpin of snR30 provide the docking site(s) for putative snR30-specific snoRNP proteins, which are essential for 90S pre-ribosome assembly and 18S rRNA processing.
Discussion
The yeast snR30 box H/ACA snoRNA has long been known to have an essential function in 18S rRNA processing, but the molecular mechanism supporting its function remained unknown (Bally et al, 1988; Morrissey and Tollervey, 1993; Atzorn et al, 2004). In this study, by using a novel in vivo crosslinking RNA affinity selection approach and yeast molecular genetics, we demonstrated that during pre-rRNA processing, two short internal sequence motifs of the 18S rRNA, called rm1 and rm2, transiently base-pair with the previously identified, functionally essential m1 and m2 sequences of snR30. We showed that formation of the newly discovered m1/rm1 and m2/rm2 Watson–Crick interactions of snR30 and 18S rRNA is required for 18S production and cell viability. The m1 and m2 motifs of snR30/U17 snoRNAs and the complementary rm1 and rm2 sequences of 18S/17S rRNAs are conserved in all vertebrates, budding and fission yeasts and in the unicellular protozoan T. thermophila. Even more tellingly, the m1/rm1 base-pairing interaction of snR30 and 18S rRNA in T. brucei is preserved by compensatory base changes. Thus, we can predict with great certainty that the snR30/U17 snoRNA has an evolutionarily conserved function in pre-rRNA processing.
Similarly to the antisense elements of H/ACA pseudouridylation guide RNAs, the m1 and m2 18S recognition motifs of snR30 are located on the opposite strands of an internal pseudouridylation loop-like structure of the 3′-terminal hairpin of the snoRNA. However, contrary to this obvious resemblance, there are fundamental structural differences between the interaction of pseudouridylation guide RNAs formed with their target sequences and the proposed association of snR30 with 18S rRNA. The antisense elements of H/ACA guide RNAs are located in the upper (distal) part of the pseudouridylation loop (Figure 1A), whereas the m1 and m2 elements of snR30/U17 snoRNAs occupy the lower (proximal) part of an internal loop of the 3′-hairpin (Figure 7). The pseudouridylation guide elements bind to contiguous rRNA sequences, whereas the m1 and m2 elements interact with two distantly located sequence motifs that are folded together by an evolutionarily conserved stem-loop structure of 18S/17S rRNAs (Alkemar and Nygard, 2006). Consequently, snR30/U17 forms a complex three-dimensional local structure with 18S/17S sequences that is novel to box H/ACA RNAs and according to our knowledge no similar interaction has been reported for other cellular RNAs either (Figure 3C).
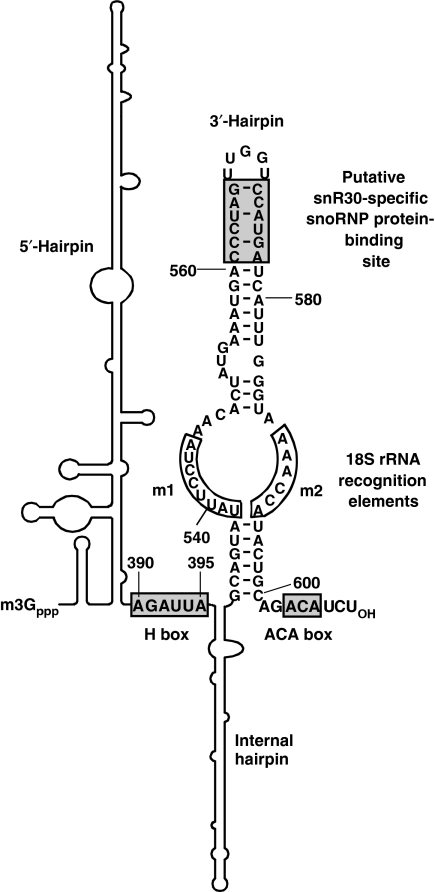
Figure 7.
Functionally essential elements of yeast snR30. The 5′-terminal and internal hairpins (shown schematically) lack functionally important elements (Atzorn et al, 2004). Sequences binding either box H/ACA or snR30-specific proteins are in shaded boxes. The m1 and m2 18S recognition sequences are in open boxes.
Determination of the crystal structure of archaeal H/ACA pseudouridylation guide RNP revealed a great structural flexibility for the pseudouridylation loop of substrate-free H/ACA RNA (Li and Ye, 2006). Although the ACA box sequences, the lower and upper stem regions of the single-hairpin archaeal H/ACA RNA, are tightly fastened to the composite surface of the Cbf5, L7ae and Nop10 core H/ACA RNP proteins, the pseudouridylation loop nucleotides show very few intermolecular contacts with RNP proteins. This structural plasticity of the entire pseudouridylation loop indicates that both the distal and proximal loop nucleotides are available for base-pairing interactions and provides the structural basis for the interaction of the m1 and m2 proximal loop sequences of snR30 with pre-rRNA sequences. Identification of a novel target recognition strategy for the ‘pseudouridylation loop' of snR30 further emphasizes the functional diversity of H/ACA RNAs and in the future, it may facilitate understanding of the molecular role of additional ‘orphan' H/ACA RNAs.
Besides snR30, the nucleolytic processing of eukaryotic rRNAs also requires the U3, U8, U14 and U22 box C/D snoRNAs. However, these snoRNAs do not seem to be directly involved in nucleolytic pre-rRNA cleavage reactions. They are believed to function as RNA chaperones that transiently base-pair with pre-rRNA to safeguard its correct folding (Beltrame and Tollervey, 1995; Liang and Fournier, 1995; Hughes, 1996; Peculis, 1997; Sharma and Tollervey, 1999; Borovjagin and Gerbi, 2000, 2004). In principle, snR30 may also facilitate the proper temporal folding of the maturing pre-rRNA, as its binding prevents the formation of the internal stem loop that includes the rm1 and rm2 sequences in the mature 18S rRNA (Alkemar and Nygard, 2006). However, we demonstrated that in addition to the m1 and m2 18S rRNA recognition elements, snR30 carries another functionally essential element located in the C561–G566/C572–A577 distal region of its 3′-hairpin (Figure 7). The C561–G566/C572–A577 sequences lack obvious complementarity to pre-rRNA sequences and they are dispensable for interaction with 35S pre-rRNA (Figure 6E). Thus, we favour the idea that the distal part of the 3′-hairpin of snR30, instead of selecting pre-rRNA sequences, functions as a snoRNP protein-binding site. The terminal stem-loop regions of H/ACA RNA hairpins frequently carry docking sites for RNP proteins that target H/ACA scaRNPs to the Cajal body or control processing of the human telomerase RNA (Richard et al, 2003; Jády et al, 2004; Theimer et al, 2007). Besides the four H/ACA core proteins, the yeast snR30 snoRNP has been reported to contain at least three snR30-specific snoRNP proteins of about 38, 46 and 48 kDa (Lübben et al, 1995). We propose that binding of putative snoRNP proteins to the 3′-hairpin of snR30 is essential for the assembly of functional 90S pre-ribosomal particle and for processing of 18S rRNA (Figure 6E). The snR30 snoRNA, but not other H/ACA and C/D snoRNAs, is released from the maturing pre-rRNA by the Rok1 RNA helicase, suggesting that Rok1 either specifically recognizes the snR30 snoRNP or it is an integral component of this particle (Bohnsack et al, 2008). Identification and functional characterization of the putative snR30-specific snoRNP proteins, which likely interact with the C561–G566/C572–A577 region of the 3′-hairpin of snR30, will be an exciting task for the future.
Thus far, all H/ACA RNAs assigned to a cellular function have been shown to function as guide RNAs that carry specific target recognition antisense elements to select substrate nucleic acids, such as rRNAs (snoRNAs), snRNAs (scaRNAs) or telomeric DNA (telomerase RNA). Other parts of the H/ACA guide RNAs tether the Cbf5/dyskerin pseudouridine synthase or the telomerase reverse transcriptase to the selected target nucleic acid. In this study, demonstration that the m1 and m2 sequences of snR30 function as 18S recognition elements, which probably target essential snR30-associated processing factors to pre-rRNA, indicates that not only pseudouridylation guide RNAs and telomerase RNA but also snR30 functions as a guide RNA.
Materials and methods
General procedures and yeast strains
Unless stated otherwise, standard laboratory procedures were used to manipulate DNA, RNA and oligodeoxynucleotides. Yeast strain NOY504 (MATα rrn4∷LEU2 ade2-101 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100) has been provided by Dr M Nomura (University of California, Irvine, USA) (Nogi et al, 1991). Construction of the GAL∷snR30 strain (MATa ade2 leu2-3 ilv1 met8 lys2 Trp1Δ his3Δ URA3-GAL10∷snR30) has been reported (Atzorn et al, 2004). Yeast S. cerevisiae cells were cultured according to standard protocols (Sherman, 1991).
Plasmid construction
Construction of the pR30 expression plasmid has been described (Atzorn et al, 2004). To generate the pR30m1, pR30m1b, pR30m2, pR30d1, pR30d2, pR30i1, pR30-R5, pR30st1, pR30st2, pR30-R5d, pR30-R5p, pR30hp and pR30lm expression plasmids, nucleotide changes have been introduced into the snR30-coding region of pR30 by using a two-step PCR approach, appropriate oligodeoxynucleotide primers and pR30 as a template. To obtain pMS2-R30, a synthetic DNA fragment containing three copies of a binding motif for MS2 coat protein was inserted into the KpnI site of pR30 (Zhou et al, 2002). The pTH25 yeast rRNA expression construct was kindly provided by Dr D Tollervey (University of Edinburgh, UK). To obtain pY18, the SacI–XhoI fragment of pTH25 was subcloned into pBluescript (Stratagene). To generate pTH25m1, pTH25m1b and pTH25m2, the 670 bp SacII–SacI internal 18S fragment encompassing the rm1 and rm2 motifs was excised from pTH25 and inserted into pBluescript. After introduction of nucleotide alterations into the rm1 and rm2 sequences by PCR mutagenesis, the mutant SacII–SacI fragments were used to replace the same fragment of pTH25. The identity of all constructs was verified by sequence analyses.
In vivo crosslinking
Yeast GAL∷snR30 cells carrying the pMS2-R30 expression plasmid were grown in YPD liquid medium containing 2% sucrose at 30°C until OD600 0.8. Cells were collected and resuspended in 0.1 volume of spheroplasting buffer containing 50 mM K-phosphate, pH 7.4, 0.9 M sorbitol, 10 mM DTT and 0.5 mg/ml Zymolyase (Seikagaku Corp.) and incubated at 30°C until spheroplasted. The spheroplast suspension was transferred onto ice, supplemented with 100 μg/ml AMT psoralen (4′-aminomethyl-4,5′,8-trimethylpsoralen) (HRI Associates Inc.), incubated in the dark for 15 min and irradiated with UV light (365 nm, ∼25 mW/cm2) for 15 min. Total RNA was extracted from the irradiated spheroplasts. For affinity selection of M2-R30 RNA, about 20 μg of purified recombinant MS2-MBP protein was bound to 20 μl of amylose resin (New England BioLabs) in NET2 buffer (200 mM NaCl, 0.05% Nonidet P-40 and 20 mM Tris–HCl pH 7.4) as described (Zhou et al, 2002). The beads were washed three times with 500 μl of NET2 and about 100 μg of cellular RNA was added in 50 μl of NET2. The beads were incubated on ice for 1 h, washed twice with 500 μl of cold low-salt buffer (10 mM NaCl, 0.5 mM EDTA, 2.0 mM DTT and 20 mM Tris–HCl pH 7.4). The associated MS2-R30 RNA was eluted with 50 μl of low-salt buffer supplemented with 20 mM maltose, extracted with phenol–chloroform and collected by ethanol precipitation. For partial hydrolysis, RNA was dissolved in a mixture of 3 μl of water and 12 μl of deionized formamide containing 0.4 mM MgCl2 and incubated in boiling water for 3–5 min. The fragmented RNA was collected by ethanol precipitation and annealed to an in vitro synthesized internally labelled RNA complementary to snR30 by incubation in 10 μl of S1 hybridization buffer (40 mM PIPES, pH 6.7, 400 mM NaCl, 1 mM EDTA and 80% formamide) at 50°C for 12 h. The annealing mixture was directly used for probing Southern membranes carrying restriction-digested yeast rDNA.
Sucrose gradient velocity sedimentation and RNA analysis
After treatment with 50 μg/ml of cycloheximide, cells were collected and lysed in 20 mM Tris–HCl, pH 7.4, 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 1% protease inhibitor (Roche), 0.1 U/μl RNasin (Promega) and 50 μg/ml cycloheximide. Lysates were fractionated on 4.5–45% sucrose gradients containing 20 mM Tris–HCl, pH 7.4, 50 mM KCl and 10 mM MgCl2. Centrifugation was performed at 39 000 r.p.m. at 4°C for 150 min in a SW40Ti rotor. Gradients were fractionated and RNA was isolated by phenol–chloroform extraction. Northern blot analysis and RNase A/T1 mapping of rRNAs and snR30 RNAs have been described (Atzorn et al, 2004). Yeast endogenous 18S (TTGTGTCTGGACCTGGTGAG) and 25S (CTCACGACGGTCTAAACCC) rRNAs and internally tagged 18S (CGAGGATCCAGGCTTT) and 25S (GTACACTCGAGAGCTTCA) rRNAs were visualized by terminally labelled oligodeoxynucleotide probes (Beltrame and Tollervey, 1992).
Acknowledgments
We are grateful to D Tollervey and M Nomura for providing us with the pTH25 expression vector and the yeast strain NOY504, respectively. We thank A Henras for advice. VA and EF-L were supported by la Fondation pour la Recherche Médicale. This study was supported by la Ligue Nationale contre le Cancer, la Fondation pour la Recherche Médicale and l'Agence Nationale de la Recherche.
References
- Alkemar G, Nygard O (2006) Probing the secondary structure of expansion segment ES6 in 18S ribosomal RNA. Biochemistry 45: 8067–8078 [DOI] [PubMed] [Google Scholar]
- Atzorn V, Fragapane P, Kiss T (2004) U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol Cell Biol 24: 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834 [DOI] [PubMed] [Google Scholar]
- Bally M, Hughes J, Cesareni G (1988) SnR30: a new, essential small nuclear RNA from Saccharomyces cerevisiae. Nucleic Acids Res 16: 5291–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Hury A, Liang XH, Michaeli S (2005) Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J Biol Chem 280: 34558–34568 [DOI] [PubMed] [Google Scholar]
- Beltrame M, Henry Y, Tollervey D (1994) Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res 22: 5139–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D (1992) Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J 11: 1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D (1995) Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J 14: 4350–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Kos M, Tollervey D (2008) Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep 9: 1230–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Borovjagin AV, Gerbi SA (2000) The spacing between functional Cis-elements of U3 snoRNA is critical for rRNA processing. J Mol Biol 300: 57–74 [DOI] [PubMed] [Google Scholar]
- Borovjagin AV, Gerbi SA (2004) Xenopus U3 snoRNA docks on pre-rRNA through a novel base-pairing interaction. RNA 10: 942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin ML, Ganot P, Kiss T (1999) Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J 18: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino GD, Gamper HB, Isaacs ST, Hearst JE (1985) Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu Rev Biochem 54: 1151–1193 [DOI] [PubMed] [Google Scholar]
- Collins K, Mitchell JR (2002) Telomerase in the human organism. Oncogene 21: 564–579 [DOI] [PubMed] [Google Scholar]
- Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T (2002) Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J 21: 2746–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Tollervey D (2002) Making ribosomes. Curr Opin Cell Biol 14: 313–318 [DOI] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T (1997a) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T (1997b) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 11: 941–956 [DOI] [PubMed] [Google Scholar]
- Gu AD, Zhou H, Yu CH, Qu LH (2005) A novel experimental approach for systematic identification of box H/ACA snoRNAs from eukaryotes. Nucleic Acids Res 33: e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR (1993) Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res 21: 3051–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Rivas FV, Murchison EP, Steitz JA (2006) The expanding universe of noncoding RNAs. Cold Spring Harb Symp Quant Biol LXXI: 551–564 [DOI] [PubMed] [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J 13: 2452–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JM (1996) Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol 259: 645–654 [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J 20: 2943–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Bertrand E, Kiss T (2004) Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 164: 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T (2003) Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal bodies following import from the cytoplasm. EMBO J 22: 1878–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss AM, Jády BE, Bertrand E, Kiss T (2004) Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol 24: 5797–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148 [DOI] [PubMed] [Google Scholar]
- Koonin EV (1996) Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res 24: 2411–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D (1998) The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev 12: 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye K (2006) Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443: 302–307 [DOI] [PubMed] [Google Scholar]
- Li SG, Zhou H, Luo YP, Zhang P, Qu LH (2005) Identification and functional analysis of 20 Box H/ACA small nucleolar RNAs (snoRNAs) from Schizosaccharomyces pombe. J Biol Chem 280: 16446–16455 [DOI] [PubMed] [Google Scholar]
- Liang WQ, Fournier MJ (1995) U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev 9: 2433–2443 [DOI] [PubMed] [Google Scholar]
- Lübben B, Fabrizio P, Kastner B, Lührmann R (1995) Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J Biol Chem 270: 11549–11554 [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP (2007) Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220 [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV (2005) Small regulatory RNAs in mammals. Hum Mol Genet 14 (Spec no. 1): R121–R132 [DOI] [PubMed] [Google Scholar]
- Meier UT (2005) The many facets of H/ACA ribonucleoproteins. Chromosoma 114: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JP, Tollervey D (1993) Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol 13: 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573 [DOI] [PubMed] [Google Scholar]
- Nogi Y, Yano R, Dodd J, Carles C, Nomura M (1993) Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol Cell Biol 13: 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y, Yano R, Nomura M (1991) Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci USA 88: 3962–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis BA (1997) The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol Cell Biol 17: 3702–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G (2007) The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res 35: 1452–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T (2003) A common sequence motif determines the Cajal body-specific localisation of box H/ACA scaRNAs. EMBO J 22: 4283–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Barberan-Soler S, Lowe TM (2006) A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA 12: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Tollervey D (1999) Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol 19: 6012–6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F (1991) Getting started with yeast. Methods Enzymol 194: 3–23 [DOI] [PubMed] [Google Scholar]
- Terns MP, Terns R (2006) Noncoding RNAs of the H/ACA family. Cold Spring Harb Symp Quant Biol LXXI: 395–405 [DOI] [PubMed] [Google Scholar]
- Theimer CA, Jády BE, Chim N, Richard P, Breece KE, Kiss T, Feigon J (2007) Structural and functional characterization of human telomerase RNA processing and Cajal body localization signals. Mol Cell 27: 869–881 [DOI] [PubMed] [Google Scholar]
- Vitali P, Royo H, Seitz H, Bachellerie JP, Hüttenhofer A, Cavaillé J (2003) Identification of 13 novel human modification guide RNAs. Nucleic Acids Res 31: 6543–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J (1999) Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol 19: 7461–7472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R (2002) Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185 [DOI] [PubMed] [Google Scholar]