Abstract
B cell differentiation and humoral immune responses are markedly suppressed by the persistent environmental contaminant, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). The suppression of humoral immune responses by TCDD occurs by direct actions on the B cell and involves activation of the aryl hydrocarbon receptor. Transcriptional regulation of paired box gene 5 (Pax5), an important regulator of B cell differentiation, is altered by TCDD in concordance with the suppression of B cell differentiation and humoral immunoglobulin M response. We hypothesized that TCDD treatment leads to dysregulation of Pax5 transcription by interfering with the basic B cell differentiation mechanisms and aimed to determine the effects of TCDD on upstream regulators of Pax5. A critical regulator of B cell differentiation, B lymphocyte–induced maturation protein-1 (Blimp-1) acts as a transcriptional repressor of Pax5. In lipopolysaccharide (LPS)-activated murine B cell lymphoma, CH12.LX, Blimp-1 messenger RNA, and DNA-binding activity within the Pax5 promoter were suppressed by TCDD. Furthermore, LPS activation of CH12.LX cells upregulated DNA-binding activity of activator protein 1 (AP-1) at three responsive element–like motifs within the Blimp-1 promoter. TCDD treatment of LPS-activated CH12.LX cells suppressed AP-1 binding to these motifs between 24 and 72 h, in concordance with the suppression of Blimp-1 by TCDD. A more comprehensive analysis at 72 h demonstrated that the suppression of AP-1 binding within the Blimp-1 promoter by TCDD was concentration dependent. In summary, our findings link the TCDD-mediated suppression of Blimp-1 through AP-1 to the dysregulation of Pax5, which ultimately leads to the suppression of B cell differentiation and humoral immune responses.
Keywords: TCDD, LPS, B cell, Pax5, Blimp-1, AP-1
INTRODUCTION
The environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) is a potent immunosuppressant. In the suppression of primary humoral immune responses, which requires cooperativity between several immune cell types, the B cell has been identified as the primary cellular target for TCDD (Dooley and Holsapple, 1988; Holsapple et al., 1986; Tucker et al., 1986.) Studies performed in murine B cell lymphoma, CH12.LX, have demonstrated that direct addition of TCDD to B cells in culture is sufficient to produce suppression of the immunoglobulin M (IgM) response, corroborating that TCDD directly acts on B cells (Sulentic et al., 1998, 2000). A wide range of biological effects produced by TCDD is mediated through a ligand-activated transcription factor termed the aryl hydrocarbon receptor (AHR). This basic helix loop helix (bHLH) protein resides in the cytosol as part of a multimeric complex comprised of hsp90, ARA9, and additional proteins (Lees and Whitelaw, 2002; Perdew, 1988). TCDD binding activates the AHR, which then dissociates from the cytosolic complex and translocates to the nucleus, where it heterodimerizes with another bHLH protein termed aryl hydrocarbon receptor nuclear translocator (ARNT) (Hankinson, 1995). AHR-ARNT dimers modulate transcription of susceptible genes in a DNA-binding site-specific manner through cis elements containing the core sequence GCGTG motif, termed dioxin response elements (DREs) (Swanson et al., 1993). Inappropriate transcriptional activation or repression of susceptible genes by AHR-DRE interaction has been postulated to mediate the majority of known TCDD toxicities. Likewise, functional AHRs are crucial for the TCDD-mediated immunosuppression, as demonstrated by analysis of TCDD-mediated immunosuppression in mice and murine B cell lines that differ in AHR expression (Kerkvliet et al., 1990; Sulentic et al., 1998; Vorderstrasse et al., 2001).
The exact mechanism whereby TCDD suppresses humoral immune responses remains to be elucidated. A putative link has been established between the expression of the IgM heavy chain gene (IgH) and the AHR-DRE signal transduction pathway. Regulatory hypersensitive domains within the 3′α enhancer region located 40 kb downstream from the α heavy chain locus play an important role in the regulation of IgH expression during terminal B cell differentiation (Singh and Birshtein, 1993; Sulentic et al., 1998). DRE-like motifs have been identified in the hs1,2 and hs4 domains of the 3′α enhancer, and the activity of a reporter gene construct driven by the entire 3′α enhancer was suppressed by TCDD treatment (Sulentic et al., 2000, 2004b). Detailed analysis of the DRE-like motifs from hs1,2 and hs4 revealed AHR involvement in the regulation of hs4 domain but could not solely explain the repression of the IgH gene by TCDD (Sulentic et al., 2004a,b). Recent studies have demonstrated that in addition to suppression of the IgH gene, TCDD treatment resulted in the suppression of two other IgM components, immunoglobulin κ light chain (Igκ) and joining chain (IgJ). Notably, the suppression of IgH, IgJ, and Igκ by TCDD was synchronous and concordant with the abnormally elevated levels of transcriptional repressor Pax5 (Yoo et al., 2004), which is known to regulate the activity of 3′α IgH enhancer, as well as Igκ and IgJ genes (Singh and Birshtein, 1993). Another Pax5-regulated gene, transcription factor x-box protein 1 (XBP-1) (Reimold et al., 1996), controls the expression of a large set of genes that participate in the expansion of the endoplasmic reticulum and in protein trafficking. Through this mechanism, XBP-1 is involved in many secretory responses including immunoglobulin secretion. In previous studies, TCDD treatment of activated B cells suppressed XBP-1 levels in concordance with an impairment of Pax5 downregulation (Yoo et al., 2004). In addition, ectopic expression of Pax5 in lipopolysaccharide (LPS)-activated CH12.LX cells closely mimicked the suppression of the IgM response by TCDD (Schneider et al., 2008). More importantly, the derepression of Pax5 in LPS-activated CH12.LX cells treated with TCDD occurred at the transcriptional level (Yoo et al., 2004), implicating upstream regulatory factors.
In light of the above findings, the objective of this investigation was to examine the upstream events leading to derepression of Pax5 in the presence of TCDD as well as regulation of the Pax5 promoter in the absence and presence of TCDD. Attenuation of Pax5 during terminal B cell differentiation is dominated by the transcriptional repressor B lymphocyte–induced maturation protein-1 (Blimp-1), which acts by binding to its cognate recognition motif, located in the Pax5 promoter (Lin et al., 2002). Although signaling events leading to Blimp-1 induction following B cell activation are poorly understood, they involve the transcription factor activator protein 1 (AP-1) (Ohkubo et al., 2005; Vasanwala et al., 2002), a dimeric complex comprised of jun and fos family members (Karin et al., 1997). AP-1 acts as a transcriptional activator through specific DNA recognition sequences termed 12-O-tetradecanoylphorbol 13-acetate responsive elements (TREs). Notably, the expression of AP-1 component c-jun and AP-1 DNA-binding activity were suppressed by TCDD treatment in LPS-activated CH12.LX cells (Suh et al., 2002). Present studies link the impairment of AP-1 and Blimp-1 by TCDD to the previously reported derepression of Pax5, which ultimately leads to suppression of terminal B cell differentiation and the IgM response. The direct effect of AHR on Pax5 transcriptional control was also investigated in light of the identification of multiple DRE-binding sites within the 3.5 kb of the Pax5 promoter.
MATERIALS AND METHODS
Chemicals.
TCDD, in 100% dimethyl sulfoxide (DMSO), was purchased from AccuStandard (New Haven, CT). DMSO and LPS were purchased from Sigma (St Louis, MO).
Mice.
Virus-free, female B6C3F1mice (6 weeks old) were purchased from Charles River (Portage, MI). On arrival, mice were randomly grouped five per plastic cage with sawdust bedding. Mice had free access to food (Purina-certified laboratory chow) and water at all times. The mouse holding rooms were maintained at 21°C–24°C and 40–60% relative humidity with a 12-h light/dark cycle. All the experimental procedures and conditions were performed according to the guidelines of the All University Committee on Animal Use and Care at Michigan State University (East Lansing, MI).
Cell line.
The CH12.LX B cell line was derived from the murine B cell lymphoma, CH12, which arose in B10.H-2aH-4bP/Wts mice (B10.A x B10.129) and has been previously characterized (Bishop and Haughton, 1986). CH12.LX cells were maintained in RPMI-1640 (Gibco BRL, Grand Island, NY) supplemented with 10% bovine calf serum (Hyclone, Logan, UT), 13.5mM HEPES, 23.8mM sodium bicarbonate, 100 U/ml penicillin, 100 μg/ml streptomycin, 2mM L-glutamine, 0.1mM nonessential amino acids, 1.0mM sodium pyruvate, and 50μM β-mercaptoethanol. Cells (1 × 105/ml) were activated with 5 μg/ml LPS and treated with TCDD and/or 0.01% DMSO for the indicated times.
Electrophoretic mobility shift assay.
To identify the putative TRE and DRE motifs in the promoter region of Pax5 (accession #: AF148961) and Blimp-1 (accession #: AF305534) genes, sequence analysis was performed on the 3.5 kb of the Pax 5 and 2.0 kb of the Blimp-1 region located 5′ to transcription start site using MacVector software (MacVector Inc., San Diego, CA). In addition, position weight matrix with qualifying match score of 0.75 and higher was used to verify the identity of the identified DRE sites (Sun et al., 2004). Ten putative DRE motifs were identified in the Pax5 promoter. Three putative TRE motifs, TRE-267, TRE-1451, and TRE-2001, and two putative DRE motifs, DRE-297 and DRE-329, were identified in the Blimp-1 promoter. Consensus TRE oligonucleotide (Faubert and Kaminski, 2000) containing the core sequence TGACTCA (Novak et al., 1990) and high-affinity DRE oligonucleotide, DRE3, containing the core sequence GCGTG (Denison and Yao, 1991) were used as positive control probes. For AHR electrophoretic mobility shift assay (EMSA), 20 μg nuclear protein was placed in 20 μl AHR-binding buffer (25mM HEPES [pH = 7.5], 1mM EDTA, 2mM dithiotretiol [DTT], 10% glycerol, 110mM KCl). For AP-1 and Blimp-1 EMSA, 11 μg and 20 μg nuclear protein, respectively, were placed in 20 μl of binding buffer (30mM HEPES, 1.5mM MgCL2, 0.3mM EDTA, 100mM NaCl, 10% glycerol, 0.05% Nonidet P-40, 1mM DTT). Each binding buffer contained protease inhibitor cocktail (Roche Boehringer Mannheim, Germany). Samples were then incubated with 1.0 μg poly dI-dC (Roche) at room temperature for 15 min. Double-stranded 32P-labeled probes (Table 1) were added and incubated at room temperature for another 30 min. The binding of protein to DNA was resolved by electrophoresis on a nondenaturing 4.0% polyacrylamide gel. The gel was then dried on 3-mm filter paper (Bio-Rad, Hercules, CA) and autoradiographed.
TABLE 1.
EMSA probes
| Probe name | Top strand 5′–3′ | Bottom strand 5′–3′ |
| TRE CON. | GATCCGGCTGACTCATCAGTA | CTACTGATGAGTCAGCCGGAT |
| TRE-267 | GCTGGTAGGAGTGAATCAGACCGT | ACTGACGGTCTGATTCACTCCTAC |
| TRE-1451 | ACTTCATTGTATGACTAAGTTGGT | TGATACCAACTTAGTCATACAATG |
| TRE-2001 | CATAGTGGTGCTGACTCAGCATCG | TAACCGATGCCTGAGTCAGCACCAC |
| DRE3 | GATCTGGCTCTTCTCACGCAACTCCG | GATCCGGAGTTGCGTGAGAAGAGCCA |
| DRE-297 | CCAGGTGCGGCCACGCCCCCATCGCG | GCCGCGATGGGGGCGTGGCCGCACCT |
| DRE-329 | GCCCTGAACCCCACGCTGCACGGCTG | CCCAGCCGTGCAGCGTGGGGTTCAGG |
| Blimp-1 | TCGGAGAGCGATTCACTTTCCAAAACTGC | GCAGTTTTGGAAAGTGAATCGCTCTCCGA |
Positions of the response elements are in relationship to the ATG site.
Plasmid preparation.
The mouse Pax5 promoter fragment of 3.5 kb size was obtained by PCR using mouse genomic DNA as a template with the following primers—forward primer: 5′-CCTCCCTTGGCTCGAGCCTCCTAGCT-3′ (Xho I restriction site is underlined) and reverse primer: 5′–GTCGGGTAAGCTTTCTAAATCCAT-3′ (Hind III restriction site is underlined). In both primers, small caps show the nucleotides that were modified in order to introduce the restriction sites. The PCR fragment, after XhoI and HindIII restriction reaction, was purified by Qiagen columns and then inserted in to pGL4.10 luciferase reporter vector (Promega, Madison, WI) at the XhoI-HindIII sites to generate a fusion gene plasmid called pPax5-Luc.
Transient transfection and luciferase assay.
CH12.LX cells (5.0 × 106) were transfected with 5 μg of pPax5-Luc, pGL-4.10 vector, or pCYP1A1-Luc (Generous gift from Dr Julie Hall, Hamner Institutes for Health Sciences Research Triangle Park, NC) using the Amaxa Nucleofector (Amaxa Inc., Gaithersburg, MD). Cells plus DNA and 100 μl of 95:5 (solution V: supplement) were mixed and electroporated using program A-20 following manufacturer’s recommendations. After electroporation, CH12.LX cells were allowed to recover for 4 h at 37°C in an atmosphere of 5% CO2 before initiation of treatments. Hepa1c1c7 and NIH3T3 cells were transfected using lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer's recommendations for 24-well transfection protocol. Briefly, DNA and lipofectamine were mixed in 1:3 proportion, 0.8 μg DNA:2.0 μl lipofectamine, after 20 min the cells were transfected overnight and the next day placed into fresh media and treated. In all instances cells were cotransfected with the pTK-Renilla in a proportion of 1:20 as an internal control for transfection efficiency.
At the end of the treatment period, luciferase activity was quantified by lysing the cells using 1X reporter lysis buffer from Promega. Twenty microliters of lysate was used for the kinetic colorimetric detection that was performed in a Falcon 96-well flat bottom opaque plate over a 10-s period every 0.02 s using a KC-4–automated microplate reader with Synergy HTTR injectors (Bio-Tek, Winooski, VT) and the Dual-Luciferase assay kit from Promega. Luciferase values were normalizing to the values of Renilla activity read in the same well.
Real-time PCR.
Total RNA was isolated from naive or LPS-activated cells using a SV Total RNA Isolation kit (Promega). To synthesize complementary DNA (cDNA), 1000 ng total RNA/sample was incubated with 600 ng random primer (Invitrogen) in 10 μl endonuclease-free water at 70°C for 10 min, cooled on ice for 10 min, and reverse transcribed in 20 μl 1× First Strand Synthesis buffer (Invitrogen), containing 0.2mM deoxyribonucleotide triphosphate, 10mM DTT, and 200 U SuperScript II reverse transcriptase (Invitrogen). The reaction mixture was incubated at 42°C for 60 min, and the reaction was stopped by incubation at 75°C for 15 min. Amplification of Blimp-1 cDNA was performed using the following TaqMan primers and probes (Applied Biosystems, Foster City, CA): forward primer: 5′-CTTTGGACTCTTACTCAACTGTACAAGCT-3′, reverse primer: 5′-CAG CTCTGCCAGTCCTTGAAA-3′, probe: 5′-CCCAAGTCTAGCTCCGGCTCCGTG-3′. The PCR cycling conditions were as follows: initial denaturation and enzyme activation for 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. For real-time PCR detection, the levels of target messenger RNA (mRNA) were calculated by the comparative CT method using 18S ribosomal subunit expression as a loading control (Applied Biosystems). Detection of PCR transcripts for real-time PCR was performed on PE Applied Biosystems PRISM 7900HT Sequence Detection System.
In vitro activation of mouse splenocytes with LPS or sRBC.
Single-cell suspensions of splenocytes from naive mice were adjusted to 5 × 106 cells/ml for LPS activation and 1 × 107 cells/ml for sheep erythrocytes (sRBC) activation in RPMI 1640 (Invitrogen) supplemented with 10% bovine colf serum (Hyclone, Utah), 100 U penicillin/ml, 100 μg streptomycin/ml, and 50μM 2-mercaptoethanol. Spleen cells were transferred to 24-well culture plates in 1 ml aliquots with four wells per treatment group. TCDD (10nM) for kinetic studies and (3, 10, or 30nM) for concentration response studies and/or vehicle (0.01% DMSO) were added directly to each well in 5 μl aliquots. The splenocytes were sensitized with 10 μg/ml LPS and cultured for 3 days or 1 × 109 sRBC for 5 days in a pressurized chamber at 5.0 psi containing 10% O2, 7% CO2, and 83% N2 gas mixture with continuous rocking at 37°C. At the end of the culture period, the cells were harvested and RNA isolated.
Isolation of nuclear AHR protein.
CH12.LX cells were incubated with 0.01% DMSO or 30nM TCDD in DMSO for 1 h at 37°C. Cells were harvested by centrifugation at 300 × g for 10 min, washed once with 1× PBS and incubated on ice in lysis buffer (10mM HEPES, 3mM MgCl2) for 15 min, and centrifuged as above. All buffers below contained protease inhibitor cocktail (Roche). One milliliter of MDH buffer (3mM MgCl2, 1mM DTT, 25mM HEPES) was added to the cell pellet and homogenized with tight-fitting pestle. Nuclei were pelleted by centrifugation at 1000 × g for 5 min, washed twice with MDHK buffer (3mM MgCl2, 1mM DTT, 25mM HEPES, 100mM KCl) and then resuspended in 100 μl of HEDGK buffer (25mM HEPES, 1mM EDTA, 1mM DTT, 10% glycerol, 400mM KCl), incubated on ice with agitation for 40 min, and centrifuged at 14,000 × g for 15 min. The supernatant was aliquoted and stored at −80°C before use in the EMSA. Protein concentrations were determined using the bicinchonic acid protein determination assay (Sigma).
Isolation of nuclear AP-1 and Blimp-1.
CH12.LX cells treated with LPS, TCDD, or both were harvested at the indicated times by centrifugation at 300 × g for 10 min, washed once with 1× PBS, and lysed in lysis buffer (10mM HEPES, 1.5mM MgCl2). Nuclei were pelleted by centrifugation at 6700 × g for 10 min, and the pellet was lysed in a hypertonic buffer (30mM HEPES, 1.5mM MgCl2, 450mM NaCl, 0.3mM EDTA, and 10% glycerol), which contained 1mM DTT and protease inhibitors cocktail (Roche), for 15 min on ice. Samples were then centrifuged at 17,500 × g for 15 min, and the supernatant was retained. Protein determinations were performed using the bicinchoninic acid assay (Sigma).
Statistical analysis of data.
Mean ± SE was calculated for each treatment group of a given PCR experiment. Statistical differences between groups were determined by a two-way ANOVA followed by Bonferroni post hoc test.
RESULTS
Pax5 Promoter Reporter Analysis
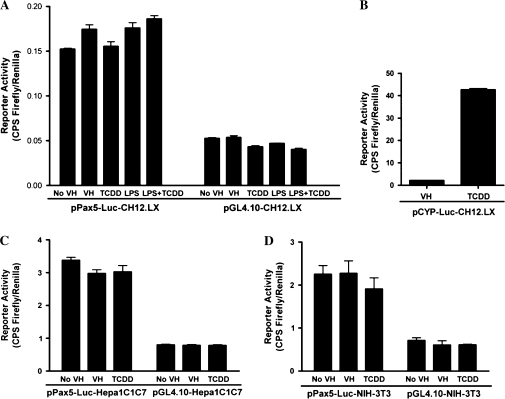
In light of our recent findings demonstrating the dysregulation of Pax5 at the level of mRNA, protein and DNA-binding activity by TCDD in LPS-activated B cells, which was followed by marked suppression of the IgM antibody response (Yoo et al., 2004), studies were initiated to examine whether AHR directly regulates the Pax5 promoter. A motif analysis of the 3.5-kb 5′ from the transcriptional start site identified multiple (10 in total) putative DRE-like sites. To assess whether AHR activation can directly regulate the Pax5 promoter, a reporter gene approach was employed. A reporter containing 3529 bp of the mouse Pax5 promoter (pPax5-Luc) and the 10 putative DRE was prepared for preliminary transient transfection experiments. Electroporation of pPax5-Luc into CH12.LX cells followed by a 4-h recovery period and then LPS treatment for various times including 8, 24, and 48 h was performed in the absence or presence of TCDD. In spite of constitutive background promoter activity, as evidenced by luciferase activity above the vector control at 8 h, which was anticipated since nonactivated CH12.LX cells express significant levels of Pax5, treatment of CH12.LX cells for 8 h with LPS, TCDD, or LPS plus TCDD, following a 4-h recovery period postelectroporation produced no additional effects on the promoter (Fig. 1A). Similar results were obtained at 24 h after LPS treatment of CH12.LX cells with the exception that the constitutive activity of pPax5-Luc was significantly diminished as evidenced by decreased luciferase activity and which was similar to vector-only (pGL4.10) transfected cells (data not shown). These later results suggest that the transiently transfected CH12.LX cells rapidly expel pPax5-Luc plasmid. Consistent with this premise, virtually no luciferase activity was detected at 48 h after LPS treatment of transfected CH12.LX cells (data not shown). To confirm that electroporation of CH12.LX cells did not render the cells unresponsive to treatment; CH12.LX cells were also transiently transfected with a reporter plasmid containing 1.4 kb of the human CYP promoter cloned into a pGL3.1 vector from Promega called pCYP1A1-Luc followed by TCDD treatment for 8 h. The CH12.LX cells were highly responsive to TCDD treatment as evidenced by induction of pCYP1A1-Luc (Fig. 1B). In addition, cell viability employing electroporation of CH12.LX routinely yielded cell viability above 60%. To assess whether the AHR directly modulates the Pax5 promoter, pPax5-Luc was transiently transfected into Hepa1c1c7 cells and NIH3T3 cells, both of which express high levels of AHR. Compared to CH12.LX cells, Hepa1c1c7 cells and NIH3T3 cells transfected with the vector control (i.e., pGL4.10), and pPax5-Luc demonstrated significantly greater constitutive activity. Since both Hepa1c1c7 and NIH3T3 cells are significantly more amenable to transfection, the higher background activity is, at least in part, due to an increase in transfection efficiency. As with the CH12.LX cells, no differences in luciferase activity were observed in pPax5-Luc–transfected cells between the naive, vehicle, and TCDD (30nM) treatment groups (Figs. 1C and D). These results show that even in cells that express high levels of AHR such as Hepa1c1c7, Pax5 promoter activity was not induced in spite of the putative DRE-like sites within −3529 bp of the Pax5 promoter.
FIG. 1.
TCDD dysregulation of the Pax5 promoter is not mediated directly by the AHR. CH12.LX, Hepa1c1c7, and NIH-3T3 cells were transfected with different promoter-driven luciferase reporters, pPax5-Luc–containing 3529 bp of Pax5 promoter–containing multiple DRE sites, or pGL4.10 vector control or pCYP-Luc and cotransfected with pTK-Renilla vector. Then cells were treated with 30nM TCDD or LPS (10 μg/ml) in the presence of TCDD (30nM) and/or vehicle (0.01% DMSO), harvested 8 h after treatment and analyzed for luciferase expression. Firefly luciferase reporter activity is expressed relative to pTK-Renilla luciferase. (A) Activity of pPax5 and pGL4.10 reporters in CH12.LX cells 8 h after treatment with no vehicle (VH), TCDD, LPS, and LPS + TCDD. (B) pCYP-Luc reporter activity in CH12.LX cells after 8 h treatment with VH and TCDD. (C) Activity of pPax5-Luc and pGL4.10 reporters in Hepa1c1c7 cells. (D) Activity of pPax5-Luc and pGL4.10 reporters in NIH-3T3 cells. Results from triplicate determinations are represented as the mean ± SE. Results are representative of more than three separate experiments.
TCDD Suppresses Protein-DNA–binding Activity of Blimp-1 within the Pax5 Promoter
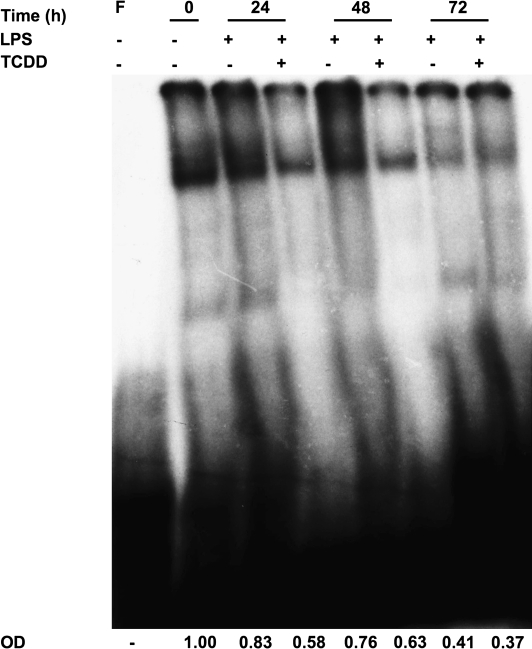
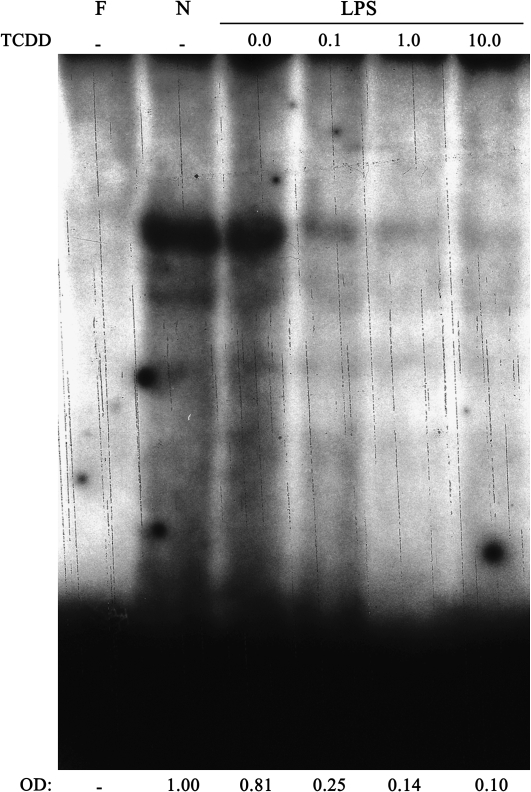
Based on the absence of direct AHR-mediated regulation of the Pax5 promoter, the role of the major Pax5 regulator and transcriptional repressor, Blimp-1, was evaluated in B cells in the absence and presence of TCDD. The repression of Pax5 expression by Blimp-1 is mediated through a Blimp-1 recognition motif located within the Pax5 promoter (Lin et al., 2002). Hence, EMSA was performed to assess Blimp-1 DNA-binding activity within the Pax5 promoter under conditions of LPS activation in the absence and presence of TCDD (Fig. 2). During the 72-h time course, TCDD treatment produced a marked decrease in Blimp-1 DNA-binding activity at 24 and 48 h when compared to the respective time-matched controls (i.e., LPS in the absence of TCDD). At 72 h, Blimp-1 DNA-binding activity was minimal in both the vehicle- and TCDD-treated cells. Overall, the TCDD-mediated reduction in Blimp-1 DNA-binding activity was concordant with previously reported derepression of Pax5 mRNA levels by TCDD (Schneider et al., 2008; Yoo et al., 2004).
FIG. 2.
Blimp-1 DNA-binding activity within the Pax5 promoter is reduced by TCDD in LPS-activated CH12.LX cells. CH12.LX cells were activated with LPS (5 μg/ml) in the presence of TCDD (10nM) and/or vehicle (0.01% DMSO) and harvested at the indicated times after LPS activation. Nuclear protein (20 μg per lane) and the 32P-labeled double-stranded oligonucleotides containing the Blimp-1 DNA-binding motif were incubated for 30 min and resolved on a 4% nondenaturing PAGE gel, dried on 3-mm filter paper and analyzed by autoradiography. Binding of Blimp-1 to the 32P-labeled Blimp-1 DNA-binding motif was quantified by densitometry. The adjusted volumes (OD × area) for all samples are expressed as fold-change from time 0. Results are representative of at least two separate experiments. F denotes free probe.
TCDD Alters Blimp-1 mRNA Levels in LPS-activated CH12.LX Cells
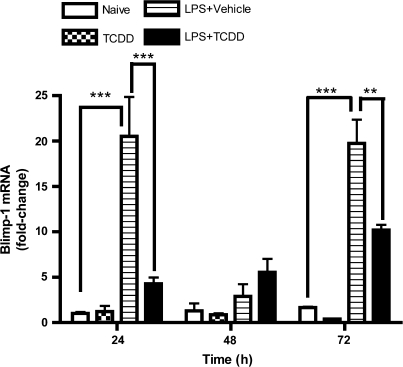
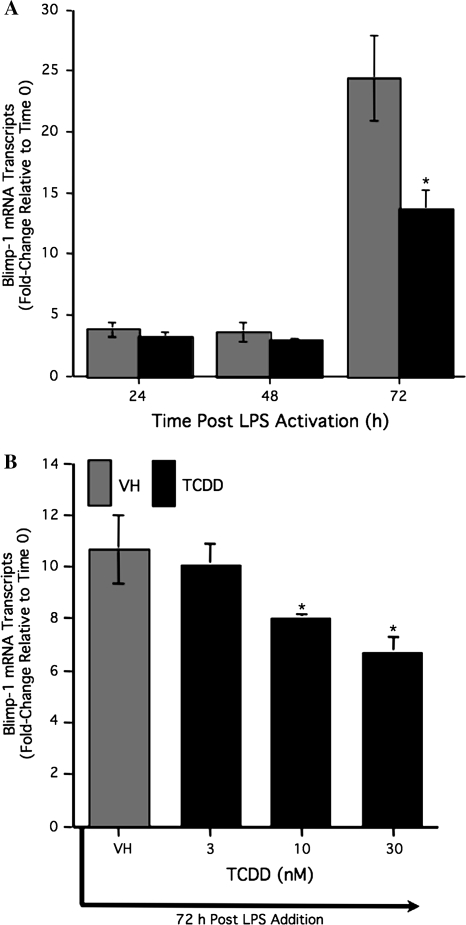
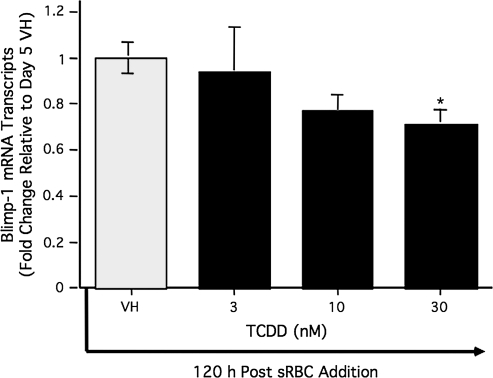
To further investigate the mechanism by which TCDD attenuated LPS-induced Blimp-1 DNA-binding activity within the Pax5 promoter, we examined the effect of TCDD on Blimp-1 regulation in LPS-activated CH12.LX cells. Due to the fact that Blimp-1 is primarily regulated at the transcriptional level (Tunyaplin et al., 2000), Blimp-1 mRNA levels were monitored over a 72-h period in the LPS-activated CH12.LX cells (Fig. 3). As would be predicted, Blimp-1 mRNA levels were strongly induced by LPS activation at 24 and 72 h, when compared to the time-matched naive (i.e., untreated) controls. In contrast, TCDD treatment markedly attenuated Blimp-1 mRNA levels at 24 and 72 h in LPS-activated CH12.LX cells (Fig. 3). Although unclear why, Blimp-1 mRNA levels were strikingly low in LPS as well as in LPS- and TCDD-cotreated cells at 48 h, when compared to their respective treatment groups at 24 and 72 h. Repeatedly, a biphasic profile of Blimp-1 mRNA levels was observed after LPS activation. Similar experiments were performed using LPS-activated mouse splenocytes. By comparison, peak induction of Blimp-1 mRNA levels in splenocytes was observed 72 h after LPS activation (Fig. 4A). As observed in CH12.LX cells, TCDD treatment significantly attenuated LPS-mediated induction of Blimp-1 mRNA levels in primary cells in a time- and concentration-dependent manner (Fig. 4B). To explore whether the TCDD-mediated attenuation of Blimp-1 mRNA levels was unique to LPS-induced B cell activation, the T cell–dependent antigen, sRBC, was employed to activate B cells. Similar to LPS, sRBC-activated splenocytes demonstrated a concentration-dependent decrease in Blimp-1 mRNA level, as measured on day 5, the peak day of anti-sRBC IgM antibody–forming response, which was markedly attenuated by TCDD treatment (Fig. 5). It is notable that the latter B cell activation studies (i.e., LPS- and sRBC-induced activation) were conducted with a heterogeneous preparation of cells from the spleen and therefore we cannot rule out the possibility that other cell types may have contributed to the overall levels of Blimp-1 that were detected in these experiments.
FIG. 3.
Blimp-1 mRNA levels are suppressed by TCDD in LPS-activated CH12.LX cells. CH12.LX cells were activated with LPS (5 μg/ml) in the presence of TCDD (10nM) and/or vehicle (0.01% DMSO), harvested at the indicated times after LPS activation and analyzed for the levels of Blimp-1 mRNA. Samples were normalized per 18S ribosomal subunit amplification, which served as loading control. The fold-change in Blimp-1 mRNA levels relative to naive sample at 24 h was arbitrarily given the value of 1, as represented on the y-axis. Results represent the mean ± SE of triplicate determinations in each treatment group from at least three separate experiments. Statistical significance was determined using a two-way ANOVA and Bonferroni post hoc test. Statistical significance is denoted as **p < 0.01, ***p < 0.001.
FIG. 4.
Suppression of Blimp-1 mRNA levels by TCDD treatment in splenocytes after LPS activation. (A) Freshly isolated splenocytes from B6C3F1 were treated with vehicle TCDD (10nM) and/or vehicle (VH, 0.01% DMSO) and activated with LPS (10 μg/ml). Splenocytes were recovered at 24, 48, and 72 h postactivation. (B) Splenocytes were activated with LPS (10 μg/ml), treated with TCDD (0, 3, 10, and 30nM) and/or VH (0.01% DMSO) and isolated at 72 h. The mRNA was isolated from splenocytes, and Blimp-1 mRNA levels were quantified by real-time PCR. Results from quadruplicate determinations are represented as the mean ± SE. Results are representative from one of two separate experiments. Statistical significance is denoted as *p < 0.05.
FIG. 5.
Suppression of Blimp-1 mRNA levels by TCDD treatment in splenocytes after sRBC activation. Freshly isolated splenocytes from B6C3F1 were treated with TCDD (0, 3, 10, and 30nM) and/or vehicle and isolated at 120 h. The mRNA was isolated from splenocytes, and Blimp-1 mRNA levels were quantified by real-time PCR. Results from quadruplicate determinations are represented as the mean ± SE. Results are representative from one of two separate experiments. Statistical significance is denoted as *p < 0.05.
Putative DRE Sites Identified within the Blimp-1 Promoter
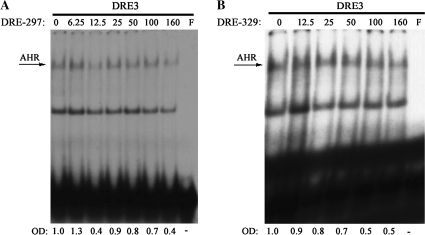
A detailed analysis of the first 2000 bp of Blimp-1 promoter led to the identification of two putative DRE motifs, based on sequence similarity with the consensus core DRE (GCGTG) and the composition of regions flanking the core (Yao and Denison, 1992). These motifs are located −297 and −329 bp upstream from the Blimp-1 transcription start site and are designated here DRE-297 and DRE-329, respectively (Table 1). A radiolabeled double-stranded oligonucleotide probe containing the DRE3 sequence from the murine CYP1A1 promoter (Denison and Yao, 1991) was used as a positive control for assessing AHR DNA-binding activity in EMSA analyses. Using nuclear proteins isolated from TCDD-treated CH12.LX cells, no TCDD-inducible specific DNA-binding activity was detected to either DRE-297 or DRE-329 (data not shown). Coincubation of the previously characterized DRE3 motif with increasing amounts of cold DRE-297 or DRE-329 reduced gel retardation by the radiolabeled DRE3 in a molar excess-dependent manner (Figs. 6A and B). However, 50% reduction in gel retardation by radiolabeled DRE3 required at least 100-fold excess of cold DRE-297 or DRE-329 competitor. This finding is in contrast with a competition EMSA performed for radiolabeled and cold DRE3 probes, where 12.5-fold molar excess of cold DRE3 was sufficient to completely abrogate the radiolabeled DRE3 gel shift band (data not shown), suggesting negligible or no specific protein binding to DRE-297 or DRE-329.
FIG. 6.
Characterization of inducible protein binding to two putative DRE sites in the Blimp-1 promoter by a competition EMSA. CH12.LX cells were incubated with TCDD (30nM) for 1 h at 37°C, and nuclear protein extracts were isolated. Nuclear protein (20 μg per lane) and the 32P-labeled DRE3-binding nucleotide were incubated with the indicated molar excess of (A) cold DRE-297 or (B) cold DRE-329 for 30 min and resolved on a 4% nondenaturing PAGE gel, dried on 3-mm filter paper and analyzed by autoradiography. TCDD-inducible protein binding to the 32P-labeled DRE3 was quantified by densitometry. The adjusted volumes (OD × area) for all samples are expressed as fold-change from TCDD-inducible protein binding in the absence of cold competitor. Results are representative of three separate experiments. F denotes free probe.
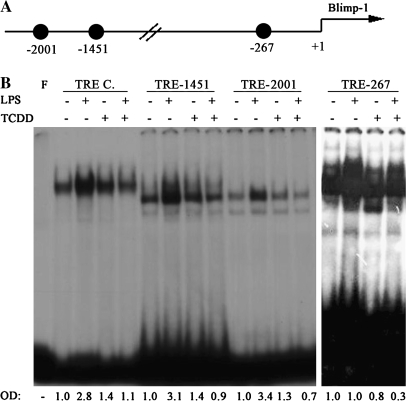
Characterization of TRE-like Motifs in the Blimp-1 Promoter
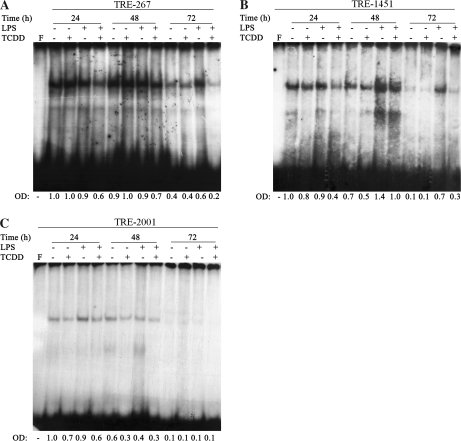
Since no TCDD-inducible specific protein binding was detected to DRE-297 or DRE-329, marked attenuation of Blimp-1 mRNA levels were produced by TCDD treatment of LPS-activated CH12.LX cells as well as LPS- or sRBC-activated splenocytes, and the role of additional transcriptional regulators of Blimp-1 was investigated. Recently, AP-1 has been identified as a critical positive regulator of Blimp-1 transcription (Ohkubo et al., 2005). We identified three TRE-like motifs within the first 2000 bp of Blimp-1 promoter. These motifs are located −267, −1451, and −2001 bp upstream from the Blimp-1 transcription start site and are designated here TRE-267, TRE-1451, and TRE-2001, respectively (Fig. 8A). Notably, TRE-2001 corresponds to the AP-1–binding motif identified by Ohkubo et al. (2005). EMSAs were performed using radiolabeled probes possessing TRE-267, TRE-1451, and TRE-2001 (Fig. 7B). A consensus TRE motif (Faubert and Kaminski, 2000) containing the core TGACTCA (Novak et al., 1990) was used as a positive, as well as comparative, control for AP-1 DNA-binding activity. Gel retardation band indicative of background AP-1 DNA-binding activity was detected in naive cells incubated with the TRE consensus, TRE-267, TRE-1451, and TRE-2001 probes (Fig. 8B). The AP-1 DNA-binding activity was strongly induced 2 h after LPS treatment to the consensus TRE, TRE-1451, and TRE-2001 probes, but not to TRE-267 probe. TCDD treatment of LPS-activated cells resulted in reduced protein binding to all four probes when compared to LPS alone (Fig. 7B). Interestingly, TCDD treatment of nonactivated CH12.LX cells for 2 h resulted in a very modest induction of protein binding to probes TRE-1451 and TRE-2001, but not TRE-267, when compared to the time-matched naive control. The induction of AP-1 DNA-binding activity by TCDD alone was short lived and was not observed at 24, 48, and 72 h (Fig. 8).
FIG. 8.
TCDD alters the kinetics of AP-1 binding in Blimp-1 promoter. CH12.LX cells were activated with LPS (5 μg/ml) in the presence of TCDD (10nM) and/or vehicle (0.01% DMSO) and harvested at the indicated times after LPS activation. Nuclear protein (20 μg per lane) and each 32P-labeled dsDNA probes containing the TRE-like motifs were incubated for 30 min and resolved on a 4% nondenaturing PAGE gel, dried on 3-mm filter paper and analyzed by autoradiography. Protein binding to the 32P-labeled (A) TRE-267, (B) TRE-1451, or (C) TRE-2001 motifs in Blimp-1 promoter was quantified by densitometry. The adjusted volumes (OD × area) for all samples are expressed as fold-change from naive cells at 24 h. Results are representative of three separate experiments. F denotes free probe.
FIG. 7.
Three TRE-like motifs were identified in the Blimp-1 promoter. (A) Location of the three TRE-like sites identified in Blimp-1 promoter. (B) CH12.LX cells were activated with LPS (5 μg/ml) in the presence of TCDD (10nM) and/or vehicle (0.01% DMSO) and harvested 2 h after LPS activation. Nuclear protein (20 μg per lane) and the 32P-labeled dsDNA probes containing the TRE consensus motif (TRE C.) or putative TRE-like motifs from Blimp-1 promoter (TRE-267, TRE-1451, TRE-2001) were incubated for 30 min and resolved on a 4% nondenaturing PAGE gel, dried on 3-mm filter paper and analyzed by autoradiography. Protein binding to the 32P-labeled TRE motifs was quantified by densitometry. The adjusted volumes (OD × area) for all samples are expressed as fold-change from naive control. Results are representative of three separate experiments. F denotes free probe.
TCDD Alters AP-1 DNA-binding Activity within the Blimp-1 Promoter
Since LPS and TCDD cotreatment reduced AP-1–binding activity to TRE-267, TRE-1451, and TRE-2001 probes at 2 h, as compared to LPS alone, we examined the profile of AP-1 DNA binding to these TREs between 24 and 72 h after LPS activation. During the 72-h time course, a similar profile of DNA-binding activity was observed for TRE-267, TRE-1451, and TRE-2001 probes (Figs. 8A–C). In LPS-activated cells, the optical density of the gel shift bands was similar to or above the levels observed in the time-matched untreated controls. Conversely, in LPS-activated and TCDD-treated cells, protein binding to TRE-267, TRE-1451, and TRE-2001 probes was reduced when compared with CH12.LX cells activated with LPS in the absence of TCDD as well as the untreated time-matched control group.
To further examine whether a causal relationship existed between TCDD treatment and the suppression of AP-1 DNA-binding activity in the Blimp-1 promoter, LPS-activated CH12.LX cells were treated with TCDD over a broad concentration range (0.1–10nM) employing TRE-267 (Fig. 9). The 32P-labelled TRE-267–containing probe was incubated with nuclear CH12.LX extracts collected 72 h after LPS activation in presence of increasing concentrations of TCDD. As compared to LPS-activated nontreated cells, TCDD treatment (0.1–10nM) decreased the protein-binding activity to TRE-267 in a concentration-dependent manner. Similar results were observed by EMSA, employing the TRE-1451 and TRE-2001 probes (data not shown), indicating a concentration-dependent reduction in AP-1 DNA-binding activity within the Blimp-1 promoter in TCDD-treated CH12.LX cells as well as providing additional evidence that the suppression of AP-1 DNA-binding activity within the Blimp-1 promoter is mediated by TCDD.
FIG. 9.
The suppression of AP-1 binding in the Blimp-1 promoter is dependent on TCDD concentration. CH12.LX cells were activated with LPS (5 μg/ml) in the presence of the indicated concentrations of TCDD (0, 0.1, 1.0 and 10 nM) and/or vehicle (0.01% DMSO) and harvested 72 h after LPS activation. Nuclear protein (20 μg per lane) and the 32P-labeled dsDNA probes containing the TRE-267 motif were incubated for 30 min and resolved on a 4% nondenaturing PAGE gel, dried on 3-mm filter paper and analyzed by autoradiography. Protein binding to the 32P-labeled TRE-267 motif was quantified by densitometry. The adjusted volumes (OD ×area) for all samples are expressed as fold-change from time 0. Results are representative of at least two separate experiments. F denotes free probe, N denotes naive (i.e., untreated) cells.
DISCUSSION
The objective of the current studies was to further investigate the signaling events leading to the dysregulation of the B cell differentiation program by TCDD. We have previously shown that TCDD markedly impaired the downregulation of the transcriptional repressor, Pax5, at the levels of mRNA-, protein-, and DNA-binding activity in activated B cells (Schneider et al., 2008; Yoo et al., 2004). In concordance with abnormally elevated levels of Pax5, the regulation of downstream target genes that are differentially expressed in plasma cells, in comparison to mature resting B cells, and known to be regulated by Pax5, was also altered. Specifically, IgH, Igκ, IgJ, and XBP-1, all of which are strongly upregulated in plasma cells, were repressed by TCDD treatment, while major histocompatibility class II, which is downregulated as B cells differentiate into plasma cells, remained highly expressed (Schneider et al., 2008; Yoo et al., 2004). Based on the level of Pax5 dysregulation produced by TCDD together with the critical role this transcriptional repressor plays in B cell differentiation, we further examined the molecular mechanisms for this phenomenon including the putative role of the AHR on Pax5 transcriptional regulation as well as signaling events that precede Pax5 repression during B cell differentiation.
In spite of extensive investigation by numerous laboratories, surprisingly little is known concerning the molecular mechanisms by which TCDD alters immune function and specifically how it impairs B cell differentiating into plasma cells, beyond the requisite involvement of the AHR. Since AHR is a ligand-activated transcription factor, a significant effort has been expended on identifying putative target genes whose regulation may be altered directly at the transcriptional level via DREs found within critical regulatory regions. Prior investigations identified functional DREs within the IgH chain 3′α enhancer which contributes, in part, to the decrease in IgH chain expression (Sulentic et al., 2004a,b); however, it cannot account for the concomitant decrease in Igκ and IgJ chain, hence the focus on the upstream transcriptional repressor, Pax5. A motif analysis of the Pax5 promoter identified 10 putative DREs within the first 3.5 kb 5′ of the transcriptional start site. Reporter assays employing the pPax5-Luc reporter, which contained all 10 putative DREs, in transient transfected CH12.LX cells suggested no direct AHR regulation as LPS, TCDD, or LPS plus TCDD treatment of pPax5-Luc–transfected cells showed no effect on reporter activity 8 h after treatment. It is noteworthy that CH12.LX cells exhibit relatively high Pax5 mRNA levels, and likewise, background pPax5-Luc activity was readily observed in untreated CH12LX cells (i.e., above empty vector control) initially after transfection. However, the background pPax5-Luc activity was found to rapidly diminish with time due to the rapid loss of pPax5-Luc. The expulsion of the pPax5-Luc plasmid precluded examination of the TCDD effects on promoter activity at later times (i.e., 24–72 h) on CH12.LX cells. We have previously observed similar expulsion of plasmids by CH12.LX within 24 h after transfection, which was of similar size as the reporter employed in the present studies (Schneider et al., 2008). The absence of modulation by LPS and/or TCDD of pPax5-Luc activity in CH12.LX is unlikely due to either poor cell viability or unresponsiveness due to electroporation, as viability was above 60% and CH12.LX cells transfected with pCYP1A1-Luc; a plasmid under the control of DREs was readily induced by TCDD treatment. Arguably, CYP1A1 is more rapidly induced than Pax5, nevertheless, the control experiment demonstrates that the CH12.LX cells were transfected and viable. The more likely reason is that Pax5 is readily downregulated by LPS treatment between 24 and 48 h, time points for which a reporter assay approach proved to be technically limiting in CH12.LX cells. In light of the challenges surrounding the pPax5-Luc reporter studies in CH12.LX cells, transient transfection studies were also conducted in Hepa1c1c7 and NIH3T3 cells as these cell lines are easily transfected and allowed for an assessment of TCDD-mediated effects on Pax5 promoter activity outside the context of its native B cell environment in order to investigate putative direct regulation by the AHR. In the case of both Hepa1c1c7 and NIH3T3 cells, TCDD treatment did not affect background pPax5-Luc activity in spite of the fact that both cell lines express high levels of AHR, suggesting the absence of direct AHR modulation of the Pax5 promoter. Collectively, these studies suggest that deregulation of Pax5 by TCDD in activated B cells involves other upstream regulators rather than direct AHR regulation of the Pax5 promoter.
In the absence of evidence supporting direct transcriptional regulation of Pax5 by AHR, specific emphasis was placed on the plasma cell commitment factor Blimp-1, a major repressor of Pax5 (Angelin-Duclos et al., 2000; Lin et al., 2002). Indeed, Blimp-1 DNA-binding activity within the Pax5 promoter was suppressed by TCDD with the most pronounced effects observed at 24 and 48 h when compared to the time-matched LPS-activated control. The repression of Blimp-1 DNA-binding activity by TCDD corresponded to the previously reported abnormally elevated Pax5 mRNA-, protein-, and DNA-binding activity in TCDD-treated LPS-activated CH12.LX cells as well as in mouse splenocytes (Schneider et al., 2008; Yoo et al., 2004) and implicate the involvement of Blimp-1 in the TCDD-induced attenuation of Pax5 repression. Importantly, Blimp-1 is primarily regulated at the transcriptional levels and is strongly induced through toll-like receptor 4 (TLR4)–initiated signaling; however, whether TLR4 is essential for Blimp-1 induction has remained controversial (Savitsky and Calame, 2006; Tunyaplin et al., 2000). Concordantly, Blimp-1 mRNA levels were induced by LPS activation of CH12.LX cells as compared with the time-matched naive control at 24 and 72 h, but not 48 h. The absence of Blimp-1 mRNA induction at 48 h was consistently observed throughout this series of experiments. Although the explanation for this effect is unknown, a similar biphasic pattern of Blimp-1 mRNA levels has been reported in the BCL-1 cell line where after the initial wave of Blimp-1 expression, Blimp-1 mRNA levels subsided and then increased again (Turner et al., 1994). A recent report revealed that Pax5 can act as a reciprocal repressor for Blimp-1 (Mora-Lopez et al., 2007), which may contribute to the downregulation in Blimp-1 levels in LPS-activated CH12.LX cells at 48 h. Importantly, the suppression of Blimp-1 mRNA levels by TCDD was robust at 24 and 72 h, demonstrating the ability of TCDD to interfere with the LPS-mediated induction of the Blimp-1 gene. TCDD-mediated effects on Blimp-1 mRNA levels at 24 and 72 h were concordant with Blimp-1 DNA-binding activity observed within the Pax5 promoter. Attenuation of Blimp-1 mRNA by TCDD was also observed in LPS-activated splenocytes and in response to B cell activation with the T cell–dependent antigen, sRBC, demonstrating that the deregulation of Blimp-1 by TCDD is a generalized effect on B cells and is not unique to CH12.LX cells or LPS-induced B cell activation. Moreover, the decrease in Blimp-1 mRNA levels likely account for the decrease in Blimp-1 DNA-binding activity by TCDD.
Although the ability of AHR to repress transcription through inhibitory DRE motifs has been previously reported for a number of estrogen-regulated genes (Safe et al., 1998, 2000), no specific TCDD-inducible protein binding was detected to two DRE-like motifs, DRE-297 or DRE-329, identified within the most proximal 2000 bp of the Blimp-1 promoter. Alternatively, in the human Blimp-1 promoter, two consensus TRE motifs have been shown to participate in the regulation of Blimp-1 expression (Vasanwala et al., 2002). Somewhat similarly, in the mouse Blimp-1 promoter, whose sequence differs from the human Blimp-1 promoter in the region where the TRE-like motifs have been identified, published results confirmed that AP-1 binding to TRE-2001 contributed to the induction of Blimp-1 in terminally differentiating B cells (Ohkubo et al., 2005). In the current investigation, EMSA assays revealed that AP-1 binding to a consensus TRE motif (Faubert and Kaminski, 2000; Novak et al., 1990) and to TRE-1451 and TRE-2001 (but not TRE-267) was induced at 2 h after LPS-activation which was suppressed in the presence of TCDD. A modest transient induction of protein binding to TRE-1451 and TRE-2001 probes was detected in nonactivated TCDD-treated CH12.LX cells. This effect is in agreement with the previously reported results obtained in cultured murine hepatoma cells (Puga et al., 1992), suggesting that the induction of AP-1 DNA binding by TCDD may occur independently of cell type. In contrast with LPS activation alone, TCDD treatment of LPS-activated CH12.LX cells produced marked suppression of AP-1 binding to TRE-267, TRE-1451, and TRE-2001 at 24, 48, and 72 h at concentrations that were previously demonstrated to suppress the IgM response in the LPS-activated CH12.LX cells (Sulentic et al., 2000; Yoo et al., 2004).
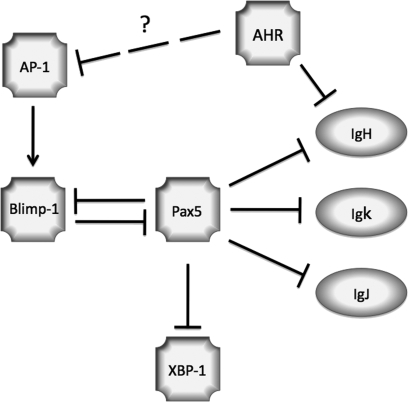
In extension of previous studies demonstrating TCDD-mediated suppression of c-jun expression and AP-1 DNA-binding activity in LPS-activated CH12.LX cells (Suh et al., 2002), we have identified a putative link between the repression of AP-1 DNA-binding activity and the impairment of a critical step in the B cell differentiation program (i.e., repression of Pax5 by Blimp-1) following TCDD treatment (Fig. 10). In contrast to the direct AHR-DRE interactions we identified as contributing, at least in part, to the dysregulation of IgH 3′α enhancer by TCDD (Sulentic et al., 2004a,b), we found no evidence of DRE-mediated involvement in the TCDD-induced alterations in Pax5 or Blimp-1 regulation. Furthermore, the repression of Blimp-1 in activated CH12.LX cells in the presence of TCDD appeared to be associated with diminished AP-1 DNA-binding activity within the Blimp-1 promoter. Interestingly, our results reported here and elsewhere (Schneider et al., 2008) demonstrate that TCDD alters numerous steps in the well-orchestrated molecular program of B cell differentiation, in concordance with the suppression of the IgM response. The observed attenuation by TCDD of Blimp-1 mRNA levels in response to several different B cell activation stimuli, in a cell line as well as primary cells, and impaired Blimp-1 DNA-binding activity in the Pax5 promoter are especially important in light of the fact that Blimp-1 is one of the few regulatory factors known, whose activation is required for terminal B cell differentiation (Kallies and Nutt, 2007). Taken together, these studies strongly implicate impaired regulation of Blimp-1 and AP-1 in the molecular mechanism responsible for suppression of the B cell differentiation program by TCDD.
FIG. 10.
Signaling model summarizing the effect of TCDD on the interactions between critical regulators of B cell differentiation and immunoglobulin gene expression. Transcription factors are depicted by  , positive regulation by
, positive regulation by  , and negative regulation by
, and negative regulation by  .
.
FUNDING
National Institutes of Health (R01ES5220, P42ES04911, T32ES07255).
Acknowledgments
We thank the members of Dr Timothy Zacharewski's laboratory for sharing the position weight matrix software for the identification of putative DRE-like sites in the Blimp-1 promoter (Sun et al. 2004), Dr Julie Hall for the pCYP1A1-Luc reporter, and Dr Barbara Kaplan for critical review of this manuscript. We also thank Mrs Kimberly Hambleton for assisting in the formatting and electronic submission of this manuscript.
References
- Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J. Immunol. 2000;165:5462–5471. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- Bishop GA, Haughton G. Induced differentiation of a transformed clone of Ly-1+ B cells by clonal T cells and antigen. Proc. Natl Acad. Sci. U.S.A. 1986;83:7410–7414. doi: 10.1073/pnas.83.19.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MS, Yao EF. Characterization of the interaction of transformed rat hepatic cytosolic Ah receptor with a dioxin responsive transcriptional enhancer. Arch. Biochem. Biophys. 1991;284:158–166. doi: 10.1016/0003-9861(91)90278-q. [DOI] [PubMed] [Google Scholar]
- Dooley RK, Holsapple MP. Elucidation of the cellular targets responsible for 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-induced suppression of the antibody response: I. The role of the B lymphocyte. Immunopharmacology. 1988;16:167–180. doi: 10.1016/0162-3109(88)90005-7. [DOI] [PubMed] [Google Scholar]
- Faubert BL, Kaminski NE. AP-1 activity is negatively regulated by cannabinol through inhibition of its protein components, c-fos and c-jun. J. Leuk. Biol. 2000;67:259–266. doi: 10.1002/jlb.67.2.259. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Holsapple MP, Dooley RK, McNerney PJ, McCay JA. Direct suppression of antibody responses by chlorinated dibenzodioxins in cultured spleen cells from (C57BL/6 x C3H)F1 and DBA/2 mice. Immunopharmacology. 1986;12:175–186. doi: 10.1016/0162-3109(86)90001-9. [DOI] [PubMed] [Google Scholar]
- Kallies A, Nutt SL. Terminal differentiation of lymphocytes depends on Blimp-1. Curr. Opin. Immunol. 2007;19:156–162. doi: 10.1016/j.coi.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr. Opin. Cell. Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- Kerkvliet NI, Steppan LB, Brauner JA, Deyo JA, Henderson MC, Tomar RS, Buhler DR. Influence of the Ah locus on the humoral immunotoxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin: Evidence for Ah-receptor-dependent and Ah-receptor-independent mechanisms of immunosuppression. Toxicol. Appl. Pharmacol. 1990;105:26–36. doi: 10.1016/0041-008x(90)90356-y. [DOI] [PubMed] [Google Scholar]
- Lees MJ, Whitelaw ML. Effect of ARA9 on dioxin receptor mediated transcription. Toxicology. 2002;181-182:143–146. doi: 10.1016/s0300-483x(02)00270-6. [DOI] [PubMed] [Google Scholar]
- Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol. Cell. Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Lopez F, Reales E, Brieva JA, Campos-Caro A. Human BSAP and BLIMP1 conform an autoregulatory feedback loop. Blood. 2007;110:3150–3157. doi: 10.1182/blood-2007-05-092262. [DOI] [PubMed] [Google Scholar]
- Neurath MF, Strober W, Wakatsuki Y. The murine Ig 3’ alpha enhancer is a target site with repressor function for the B cell lineage-specific transcription factor BSAP (NF-HB, S alpha-BP) J. Immunol. 1994;153:730–742. [PubMed] [Google Scholar]
- Novak TJ, White PM, Rothenberg EV. Regulatory anatomy of the murine interleukin-2 gene. Nucleic Acids Res. 1990;18:4523–4533. doi: 10.1093/nar/18.15.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo Y, Arima M, Arguni E, Okada S, Yamashita K, Asari S, Obata S, Sakamoto A, Hatano M, O-Wang J, et al. A role for c-fos/activator protein 1 in B lymphocyte terminal differentiation. J. Immunol. 2005;174:7703–7710. doi: 10.4049/jimmunol.174.12.7703. [DOI] [PubMed] [Google Scholar]
- Perdew GH. Association of the Ah receptor with the 90-kDa heat shock protein. J. Biol. Chem. 1988;263:13802–13805. [PubMed] [Google Scholar]
- Puga A, Nebert DW, Carrier F. Dioxin induces expression of c-fos and c-jun proto-oncogenes and a large increase in transcription factor AP-1. DNA Cell Biol. 1992;11:269–281. doi: 10.1089/dna.1992.11.269. [DOI] [PubMed] [Google Scholar]
- Reimold AM, Ponath PD, Li YS, Hardy RR, David CS, Strominger JL, Glimcher LH. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J. Exp. Med. 1996;183:393–401. doi: 10.1084/jem.183.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkenberger JL, Wallin JJ, Johnson KW, Koshland ME. An interleukin-2 signal relieves BSAP (Pax5)-mediated repression of the immunoglobulin J chain gene. Immunity. 1996;5:377–386. doi: 10.1016/s1074-7613(00)80263-0. [DOI] [PubMed] [Google Scholar]
- Safe S, Wang F, Porter W, Duan R, McDougal A. Ah receptor agonists as endocrine disruptors: Antiestrogenic activity and mechanisms. Toxicol. Lett. 1998;102–103:343–347. doi: 10.1016/s0378-4274(98)00331-2. [DOI] [PubMed] [Google Scholar]
- Safe S, Wormke M, Samudio I. Mechanisms of inhibitory aryl hydrocarbon receptor-estrogen receptor crosstalk in human breast cancer cells. J. Mammary Gland Biol. Neoplasia. 2000;5:295–306. doi: 10.1023/a:1009550912337. [DOI] [PubMed] [Google Scholar]
- Savitsky D, Calame K. B-1 B lymphocytes require Blimp-1 for immunoglobulin secretion. J. Exp. Med. 2006;203:2305–2314. doi: 10.1084/jem.20060411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Manzan MA, Crawford RB, Chen W, Kaminski NE. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD)-mediated impairment of B cell differentiation involves dysregulation of paired box 5 (Pax5) isoform, Pax5a. J. Pharmacol. Exp. Ther. 2008;326:463–474. doi: 10.1124/jpet.108.139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Peng A, Schlissel MS. In vivo occupancy of the kappa light chain enhancers in primary pro- and pre-B cells: A model for kappa locus activation. Immunity. 1997;6:131–143. doi: 10.1016/s1074-7613(00)80420-3. [DOI] [PubMed] [Google Scholar]
- Singh M, Birshtein BK. NF-HB (BSAP) is a repressor of the murine immunoglobulin heavy-chain 3’ alpha enhancer at early stages of B-cell differentiation. Mol. Cell. Biol. 1993;13:3611–3622. doi: 10.1128/mcb.13.6.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Jeon YJ, Kim HM, Kang JS, Kaminski NE, Yang KH. Aryl hydrocarbon receptor-dependent inhibition of AP-1 activity by 2,3,7,8-tetrachlorodibenzo-p-dioxin in activated B cells. Toxicol. Appl. Pharmacol. 2002;181:116–123. doi: 10.1006/taap.2002.9403. [DOI] [PubMed] [Google Scholar]
- Sulentic CE, Holsapple MP, Kaminski NE. Aryl hydrocarbon receptor-dependent suppression by 2,3,7,8-tetrachlorodibenzo-p-dioxin of IgM secretion in activated B cells. Mol. Pharmacol. 1998;53:623–629. [PubMed] [Google Scholar]
- Sulentic CE, Holsapple MP, Kaminski NE. Putative link between transcriptional regulation of IgM expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin and the aryl hydrocarbon receptor/dioxin-responsive enhancer signaling pathway. J. Pharmacol. Exp. Ther. 2000;295:705–716. [PubMed] [Google Scholar]
- Sulentic CE, Kang JS, Na YJ, Kaminski NE. Interactions at a dioxin responsive element (DRE) and an overlapping kappaB site within the hs4 domain of the 3′alpha immunoglobulin heavy chain enhancer. Toxicology. 2004a;200:235–246. doi: 10.1016/j.tox.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Sulentic CE, Zhang W, Na YJ, Kaminski NE. 2,3,7,8-tetrachlorodibenzo-p-dioxin, an exogenous modulator of the 3′alpha immunoglobulin heavy chain enhancer in the CH12.LX mouse cell line. J. Pharmacol. Exp. Ther. 2004b;309:71–78. doi: 10.1124/jpet.103.059493. [DOI] [PubMed] [Google Scholar]
- Sun YV, Boverhof DR, Burgoon LD, Fielden MR, Zacharewski TR. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 2004;32:4512–4523. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson HI, Tullis K, Denison MS. Binding of transformed Ah receptor complex to a dioxin responsive transcriptional enhancer: Evidence for two distinct heteromeric DNA-binding forms. Biochemistry. 1993;32:12841–12849. doi: 10.1021/bi00210a037. [DOI] [PubMed] [Google Scholar]
- Tucker AN, Vore SJ, Luster MI. Suppression of B cell differentiation by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Mol. Pharmacol. 1986;29:372–377. [PubMed] [Google Scholar]
- Tunyaplin C, Shapiro MA, Calame KL. Characterization of the B lymphocyte-induced maturation protein-1 (Blimp-1) gene, mRNA isoforms and basal promoter. Nucleic Acids Res. 2000;28:4846–4855. doi: 10.1093/nar/28.24.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: A mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J. Immunol. 2002;169:1922–1929. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- Vorderstrasse BA, Steppan LB, Silverstone AE, Kerkvliet NI. Aryl hydrocarbon receptor-deficient mice generate normal immune responses to model antigens and are resistant to TCDD-induced immune suppression. Toxicol. Appl. Pharmacol. 2001;171:157–164. doi: 10.1006/taap.2000.9122. [DOI] [PubMed] [Google Scholar]
- Yao EF, Denison MS. DNA sequence determinants for binding of transformed Ah receptor to a dioxin-responsive enhancer. Biochemistry. 1992;31:5060–5067. doi: 10.1021/bi00136a019. [DOI] [PubMed] [Google Scholar]
- Yoo BS, Boverhof DR, Shnaider D, Crawford RB, Zacharewski TR, Kaminski NE. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) alters the regulation of Pax5 in lipopolysaccharide-activated B cells. Toxicol. Sci. 2004;77:272–279. doi: 10.1093/toxsci/kfh013. [DOI] [PubMed] [Google Scholar]