Abstract
Phytochromes are a widespread family of red/far-red responsive photoreceptors first discovered in plants, where they constitute one of the three main classes of photomorphogenesis regulators. All phytochromes utilize covalently attached bilin chromophores that enable photoconversion between red-absorbing (Pr) and far-red-absorbing (Pfr) forms. Phytochromes are thus photoswitchable photosensors; canonical phytochromes have a conserved N-terminal photosensory core and a C-terminal regulatory region which typically includes a histidine-kinase-related domain. The discovery of new bacterial and cyanobacterial members of the phytochrome family within the last decade has greatly aided biochemical and structural characterization of this family, with the first crystal structure of a bacteriophytochrome photosensory core appearing in 2005. This structure and other recent biochemical studies have provided exciting new insights into the structure of phytochrome, the photoconversion process that is central to light sensing, and the mechanism of signal transfer by this important family of photoreceptors.
Keywords: phytochrome, biochemistry, biliprotein, photoreceptor, light signaling, photochemistry
GENERAL INTRODUCTION
Phytochrome was first discovered in plants in 1959 as the photoreceptor that mediates plant growth and development in response to long-wavelength visible light (9). Phytochrome measures the ratio of red light (R) to far-red light (FR), thereby allowing the plant to assess the quantity of photosynthetically active light and trigger shade avoidance responses (89). Phytochromes are found in all flowering plants and cryptophytes, and this important family of developmental regulators constitutes one of the three major classes of photoreceptors in higher plants, with the others being cryptochromes and phototropins (3, 8, 91).
More recently, phytochrome-related proteins have been isolated from other taxa. The first such protein to be discovered was the cyanobacterial chromatic adaptation sensor RcaE (51). Since this initial discovery, R/FR-sensing phytochromes have been discovered in cyanobacteria (Cph1/CphA, Cph2, and CphB/BphP), non-oxygenic bacteria (bacteriophytochromes or BphPs), and even fungi (Fphs), demonstrating that this class of photosensors is not limited to photosynthetic organisms (49). The true extent of the phytochrome family is only now becoming apparent with the advent of genome sequencing. Supplemental Table 1 lists currently known (or suspected) phytochromes and phytochrome-related proteins, and a sequence alignment of more than 120 of these proteins is shown in Supplemental Figure 1.
While microbial phytochromes have proven amenable systems for biochemical and spectroscopic analyses, much remains to be determined about the function of many of these molecules in vivo. The BphPs from Rhodopseudomonas palustris have been shown to regulate the biosynthesis of the photosynthetic apparatus in this organism (32, 33), while BphPs are known to regulate pigment biosynthesis in Deinococcus radiodurans and Rhodospirillum centenum (14, 45). The phytochrome from the filamentous fungus Aspergillus nidulans has recently been implicated in sexual development (5). These functions are thus conceptually analogous to functions of plant Phys: phytochromes regulate the metabolic response of the organism to its light environment.
PHYTOCHROMES ARE BILIPROTEIN PHOTOSWITCHES
Photoconversion: the “Central Dogma” of Phytochromes
An early key in defining the action of phytochrome in plant biology was the observation that both the spectrum of phytochrome preparations and the action spectrum of many plant responses were reversibly altered by illumination (89). Illuminating dark-grown tissues with red light converts phytochrome from the R-absorbing Pr form to the FR-absorbing Pfr form which triggers photomorphogenesis (Figure 1A). This change is reversible, with far-red illumination restoring Pr, and involves both primary photochemistry and subsequent thermal steps. For detailed review of the microscopic steps involved in the phytochrome photocycle, we refer the reader to detailed reviews on this topic (36, 93).
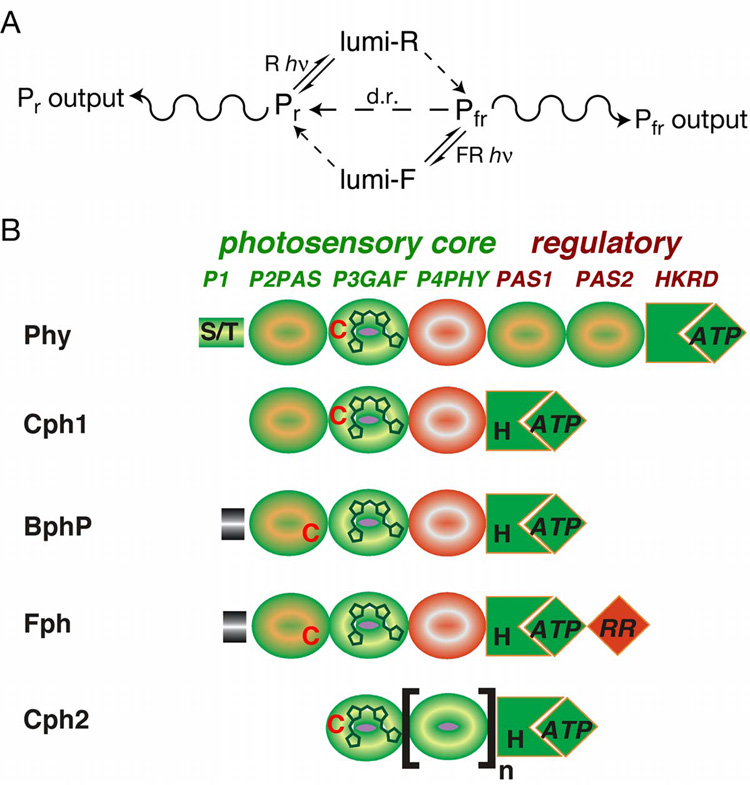
Figure 1. Domain structure and chromophores of phytochromes.
(a) The phytochrome photocycle. Illumination of Pr phytochrome with red light (R) produces lumi-R as the primary photoproduct. This is subsequently converted to Pfr via multiple light-independent steps. Pfr can be converted into Pr either by illumination with far-red light (FR), producing lumi-F and then Pr via subsequent thermal steps, or by an entirely thermal process known as dark reversion (d.r., center). The ratio between Pr and Pfr (and hence between the two physiological outputs) is thus determined by the light environment and by the rate of dark reversion. (b) Domain architecture of the extended phytochrome family. The five classes of phytochromes possess an N-terminal photosensory core region and typically share regulatory output domains related to those found on two component histidine kinases (HKRD). The P3/GAF domain is associated with the bilin chromophore and is highly conserved. All phytochromes except those found in the Cph2 subfamily share P2/PAS domains, while P4/PHY photosensory domains are specific to phytochromes and are thought to have folds similar to GAF domains (69). Plant phytochromes (Phys) possess two additional PAS domains within the regulatory region. Fungal phytochromes (Fphs) have a domain structure similar to those of the cyanobacterial phytochrome 1 (Cph1) and bacteriophytochrome (BphP) families, except for an additional C-terminal response regulator receiver domain (RR) extension and variable N-terminal extensions.
R/FR photoreversibility, the hallmark of plant Phys, is rarely observed in other organisms, because higher fluences or continuous light are typically required to maintain a threshold Pr/Pfr ratio for light responsiveness (91). This reflects a thermal process known as dark reversion, in which Pfr phytochrome is slowly converted to Pr phytochrome in the absence of light. While dark reversion is an intrinsic property of all phytochromes, plant phytochromes have apparently evolved to exhibit slower dark reversion (31). Interestingly, BphPs that possess far-red absorbance maxima in the thermal ground state with photoconversion to Pr-like species have recently been described (32, 48, 100). Whether this spectral inversion proceeds via a reverse of the normal dark reversion pathway or via some other mechanism is not yet clear for any of these bathyBphPs. However, despite these spectral variations, it is clear that the biological outputs from phytochromes reflect the ratio of the Pr and Pfr forms, and that this ratio is determined by the light environment, by the forward and reverse rates of photoconversion, and by the rates for thermal interconversion between these forms.
The Modular Domain Architecture of Phytochromes
The large and steadily growing number of phytochrome sequences now known permit classification into subfamilies (Supplemental Table 1; (50, 69)) and delineation of conserved sequences and domains that are either ubiquitous among phytochromes or conserved in different subfamilies (Figure 1B; Supplemental Figure 1). Plant Phys, Cph1s, and most BphPs share a common architecture consisting of an N-terminal photosensory region with 3 conserved domains (termed P2 or PAS domain, P3 or GAF domain, and P4 or PHY domain) and a C-terminal regulatory histidine kinase or histidine kinase-related domain (HKRD). Plant Phys have an additional N-terminal extension termed P1 known to inhibit dark reversion (102) and two additional regulatory PAS domains recently shown to be important for nuclear localization (13). Fungal Fphs have distinct N-terminal extensions and additional C-terminal response regulator domains (RR/REC) (5, 29). The other class of cyanobacterial phytochromes, the Cph2s, lack the N-terminal P2/PAS domain altogether but have other GAF domains duplicated C-terminal to the P4/PHY domain.
Canonical phytochromes thus consist of a PAS-GAF-PHY N-terminal photosensory module typically combined with a C-terminal HKRD module. PAS and GAF domains are present in other signaling molecules; for example, the photosensory Lov domains of the phototropins are PAS domains (19, 37), while GAF domains have been implicated as regulators of cyclic nucleotide metabolism in organisms as diverse as cyanobacteria and mammals (62, 63). Although there is not yet experimental structural information about the PHY domain, P4/PHY domains typically exhibit low similarity to GAF domains. It has therefore been proposed that this domain also assumes a GAF fold (69). The concatenation of PAS, GAF, and PHY domains attached to HKRD modules typify all classes of phytochromes and phytochrome-related proteins.
Phytochrome Chromophore Structure
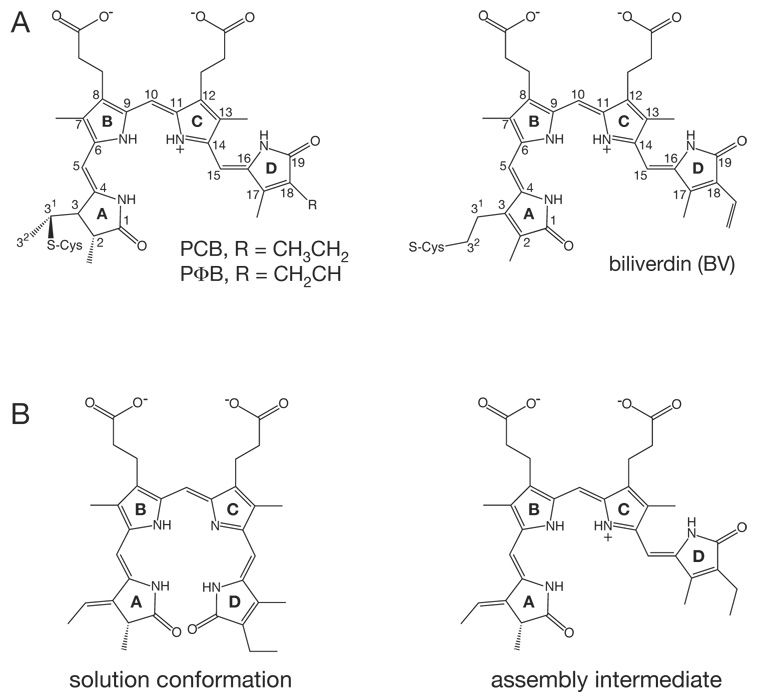
The characteristic absorbance spectra and photoconversion of phytochromes reflect their association with a linear tetrapyrrole bilin chromophore which is normally covalently attached via a thioether linkage (Fig 2A). Photoconversion is known to involve a Z–E isomerization about the C15–C16 double bond of the bilin, as apophytochrome neither photoconverts nor exhibits a typical phytochrome absorbance spectrum (36, 94). The exact nature of this chromophore varies for different subfamilies of phytochromes: plant Phys use phytochromobilin (PΦB, Fig 2A), while Cph1s and Cph2s instead utilize phycocyanobilin (PCB). Both bilins are covalently attached at C31 to a conserved Cys in the P3/GAF domain of the photosensory core (56, 59, 80, 105). In contrast, BphPs and Fphs incorporate biliverdin IXα (BV) chromophores (Fig 2A). In these proteins, the more oxidized BV is attached to a conserved Cys upstream of the P2/PAS domain, apparently via a C32 linkage (58, 103). This linkage appears less stable in BphPs than the C31 linkage to PCB, based on recent evidence for its reversibility (86). Covalent attachment does not appear to be a prerequisite for photoconversion; indeed, a mutant BphP lacking the nucleophilic Cys residue has been shown to function as an enzyme for production of C15–C16 E bilins (60). Covalent attachment likely provides a more stable holoprotein that is better suited to reversible photoswitching. Phytochromes are thus photoswitchable photosensors that utilize bilin chromophores.
Figure 2. Chromophore structure and sssembly.
(a) The structures of the bilin chromophores utilized by known phytochromes are shown. Left, phycocyanobiliin (PCB) and phytochromobilin (PΦB) chromophores share a reduced A ring and differ only at the C18 side chain. These chromophores are utilized by plant and algal Phys and cyanobacterial Cph1s and Cph2s. Right, the BphPs and Fphs instead utilize biliverdin (BV) as chromophore. All chromophores are shown in the C5–Z,syn C10–Z,syn C15–Z,anti configuration adopted in the Pr state (103). (b) Conformations of PCB thought to be present during the assembly reaction with Cph1 are shown (6). The cyclic, porphyrin-like C15–Z,syn species (left) is the most stable in solution at neutral pH and initially binds to apoCph1. After binding, the B/C ring system becomes protonated, driving adoption of a C15–Z,anti conformation (right) which is characterized by enhanced, red-shifted long wavelength absorbance. This species then becomes covalently attached to Cys259 of Cph1 to give the Pr structure shown in (a). BV is bound to a different Cys upstream of the P2/PAS domain of BphPs (58, 103).
THE STRUCTURE AND FUNCTION OF THE PHYTOCHROME PHOTOSENSORY CORE
Phytochromes are Bilin Lyases
Because the bilin precursor of the Cph1 and Cph2 phytochrome chromophores are identical to those used in assembly of the cyanobacterial phycobiliprotein antennae complexes, it was expected that phytochromes would also require bilin lyases for assembly of holoprotein. Surprisingly, plant, bacterial, and fungal apophytochromes are all able to self-ligate to appropriate bilins in vitro in the absence of other proteins or cofactors. This intrinsic bilin lyase activity has been mapped to the P3/GAF domain by truncation analysis, with the P2/PAS and P4/PHY domains important for tuning the spectroscopic porperties of the bound bilin (105). While removal of the P4/PHY domain permits bilin assembly for Phys, Cphs and BphPs, such truncated phytochromes typically exhibit reduced efficiency of photoconversion and enhanced dark reversion (50, 77, 81, 105). The P4/PHY domain thus seems to play an accessory role in reducing both unproductive modes of de-excitation and dark reversion. In contrast, deletion of either the P2/PAS or P3/GAF domains typically yielded unstable or misfolded protein; indeed, AphA, a Cph1 from Anabaena sp. PCC7120, is known to absolutely require a small extension N-terminal to the P2/PAS domain for bilin assembly (108). However, because the cyanobacterial Cph2 phytochromes lack a P2/PAS domain, it was possible to express a Cph2 P3/GAF domain alone and demonstrate formation of a covalent PCB adduct (105). The bilin lyase activity of phytochromes thus wholly resides in the P3/GAF domain.
The assembly reaction of Cph1 with PCB was recently examined by stopped-flow absorbance spectroscopy (6). PCB is shown to rapidly bind noncovalently in a cyclic, porphyrin-like conformation with Z,syn geometry at C5, C10, and C15, which is the most stable configuration in solution (18; Figure 2B). A second intermediate with red-shifted long-wavelength absorbance maximum and enhanced long-wavelength absorbance intensity appears soon after, followed by a blue-shift of the long-wavelength absorbance maximum concomitant with thioether bond formation. The second intermediate exhibits the spectral signature of a bilin in a more extended conformation with all four nitrogens protonated (18, 34, 35). 15N-NMR characterization of the Pr state in Cph1 corroborates this interpretation (97). Recent studies have isolated point mutations in the P3/GAF domain of Cph1 that form covalent PCB or PΦB adducts exhibiting a porphyrin-like conformation or even form covalent adducts with an endogenous porphyrin (21). The extended chromophore configuration is thus not necessary for covalent attachment, and indeed this work raises the intriguing possibility that porphyrins or metalloporphyrins may also modulate phytochrome function.
The Crystal Structure of the DrBphP Photosensory Core
A major breakthrough in understanding the photochemistry and structure of phytochrome was the unveiling of a 2.5 Å crystal structure for the BV-bound P2/PAS and P3/GAF domains of DrBphP, the BphP from Deinococcus radiodurans, in the Pr state (103). This structure confirms that the P2 and P3 domains adopt PAS and GAF folds, as expected (Fig. 3A). Gaps and insertions within these domains in the extended phytochrome family largely fall outside of secondary structure elements, as would be expected for a conserved fold, and both domains exhibit conserved cores (Figure 3A). Consistent with biochemical studies, the BV chromophore is covalently attached to Cys24, apparently by linkage to C32 rather than C31 as in Phys or Cph1 (Figure 2A), and is deeply buried within a conserved pocket in the P3/GAF domain.
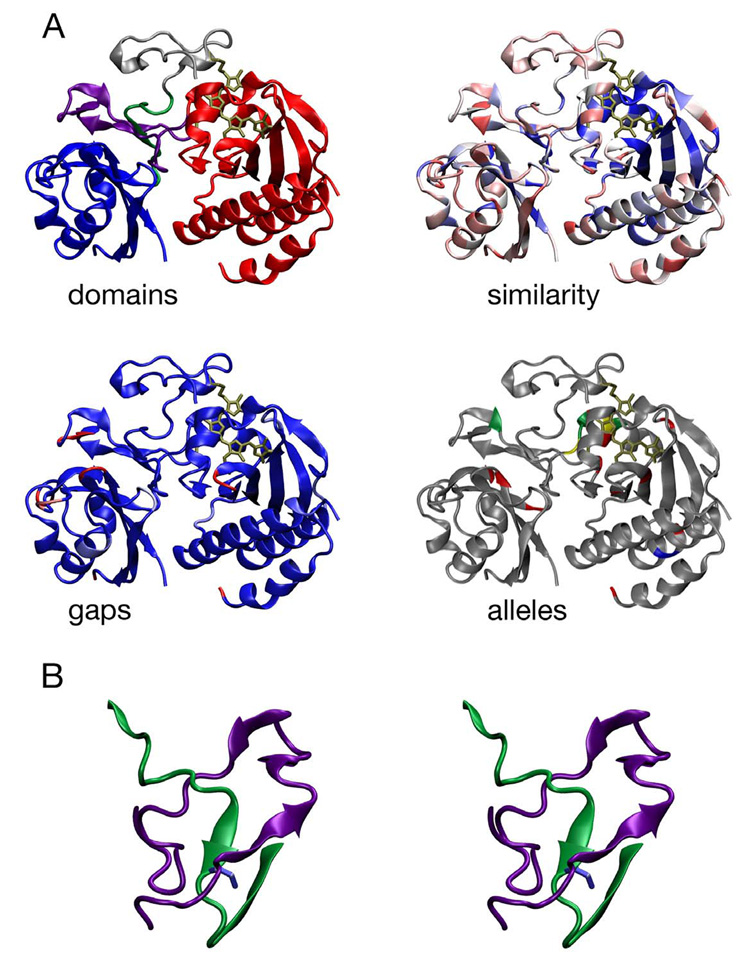
Figure 3. Conservation of the PAS and GAF domains of phytochromes.
The 2.5 Å crystal structure of DrBphP P2/PAS and P3/GAF domains (PDB code 1ZTU: (103)) is shown with bound BV chromophore covalently attached to Cys24 (bronze) colored by domains (top left), similarity (top right), gaps (bottom left), and known alleles of plant Phys (bottom right). The DrBphP structure colored by domains (top left) uses the following color scheme: PAS, blue; GAF, red; N-terminal knot interface, green; GAF insert knot interface, purple; N-terminus, grey. The DrBphP structure colored by similarity (top right) uses a normalized BLOSUM62 matrix (38) and the alignment of 122 phytochromes presented in Supplemental Figure 1. A continuous color scale is used, ranging from dark blue (100% conserved) to bright red (variable). The DrBphP structure colored by length of gaps (bottom left) uses the alignment in Supplemental Figure 1. A continuous color scale ranges from light blue (no gaps) to bright red (gaps ≥ 5 amino acids long), with a gap defined as a position where any phytochrome has insertions relative to DrBphP. The DrBphP structure colored by the location of alleles in plant phytochromes (bottom right) shows alleles that have been reported within the PAS/GAF domains of DrBphP against a grey background (see Supplemental Table 2). Loss-of-function alleles are colored red, gain-of-function alleles are colored blue, positions with multiple phenotypes are colored yellow, and silent alleles are colored green. Figure 3 and Figure 4 were prepared using VMD (40), Tachyon (96), STRIDE (28) and homolmapper (N. C. R. and J. C. L., unpublished results). (b) Stereoview of the conserved trefoil knot at the interface between the PAS and GAF domains by residues 27–43 (green, upstream of the PAS domain and the first beta strand of the PAS domain) and 227–257 (purple). Ile35 (blue) is at the center of the knot.
Unexpectedly, the interface between the PAS and GAF domains is formed by a deep trefoil knot (Figure 3B). Such knots have only been recognized relatively recently, so known examples are relatively few (76, 101). Phytochrome biosynthesis thus holds the promise of providing new insight into knot formation. The phytochrome knot is formed from sequence lying between Cys24 and the P2/PAS domain proper, which passes through a “lasso” formed by P3/GAF sequence between the fourth and fifth strands of the central GAF β sheet (103). The trefoil knot is centered on the conserved Ile35, which lies within the N-terminal sequence element required for bilin assembly of AphA (108), and also contains Arg254, a conserved residue directly interacting with the B-ring carboxylate of the biliverdin chromophore. It is therefore highly likely that this architecture will be conserved among other phytochromes. Additional N-terminal extensions such as the P1 region in plant phytochrome or the large N-terminal extensions of Fphs must either be largely unstructured or else could be proteolytically removed to facilitate this folding process. The potential regulatory role of such extensions is an interesting topic for future investigation.
Mutations in the PAS and GAF domains of plant phytochromes that result in altered function in vivo (Supplemental Table 2) have been mapped onto the DrBphP structure (Figure 3A). While several loss-of-function mutations cluster about the chromophore-binding pocket as expected, others occur in residues at the interface between the PAS domain and the trefoil knot, such as Gly118, Ser134, and Ile208 in Arabidopsis PhyB (Supplemental Table 2). Such mutations might well affect the proper folding of these domains. Both loss-of-function and gain-of-function alleles have been isolated in the “back-side” helices of the GAF domain, which lie on the other side of the central β sheet from the chromophore-binding pocket (e.g., mutation of Arabidopsis PhyA Glu229 and pea PhyB Val238). It is thus conceivable that these helices play a role in signal transduction via light-mediated regulation of either intramolecular interactions (with the P4/PHY domain, C-terminal regulatory domains, or plant Phy P1 sequence) or direct intermolecular interactions with downstream signaling components.
The BV chromophore of DrBphP is unequivocally bound in the C5–Z,syn C10–Z,syn C15–Z,anti configuration in the Pr state (Figure 4), ending a controversy which has lasted for some time. The chromophore is deeply buried in the GAF domain, and both the carboxylate side chains and the tetrapyrrole ring system are excluded from bulk solvent. The B-ring carboxylate forms a tight, bidentate association with Arg254, while the C ring carboxylate is associated with His260, Ser272, and Ser274 (Figure 4A).
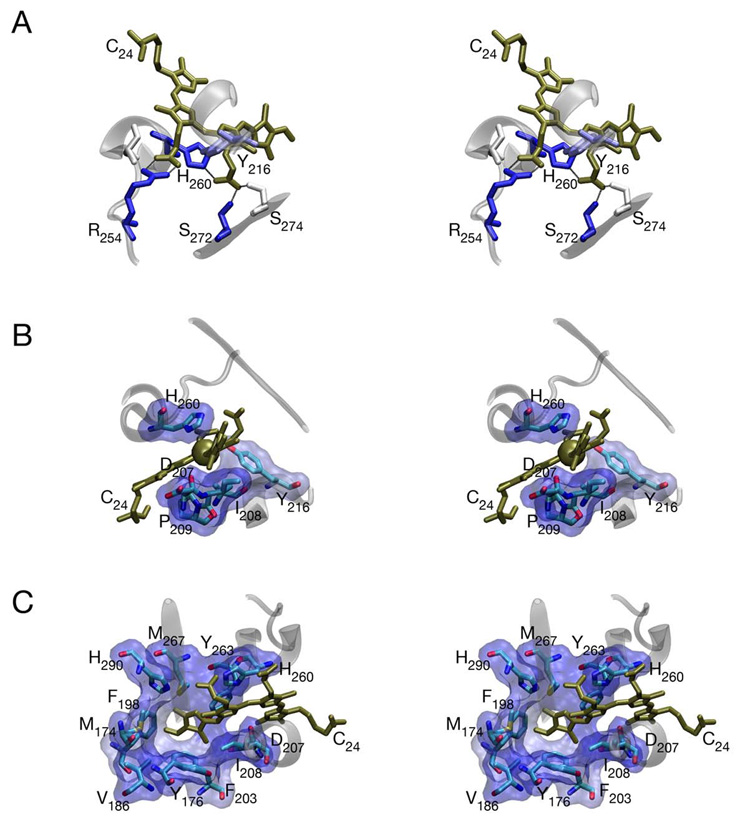
Figure 4. Chromophore-protein interactions in DrBphP are conserved.
(a) Interaction of the buried carboxylate side chains of BV (bronze) with DrBphP (103). The B ring carboxylate interacts with the conserved Arg254, which is part of the trefoil knot, while the C ring carboxylate interacts with conserved Ser272 and His260. All protein residues within 3.5 Å of the carboxylate oxygens are shown colored by similarity as in Figure 2. Secondary structure elements are shown in transparent grey for residues 214–218, 254–262, and 271–275 for reference. (b) Environment of the C10 bridging carbon (bronze sphere). This atom is held in place by the B and C rings of biliverdin along with the conserved Asp207, Ile208, Tyr216, His260, and Pro209 (shown as sticks colored by atom type and as solvent-accessible surface, with the surface colored by similarity as in Figure 2). (c) Environment of the D ring. Residues within 5 Å of the chromophore D ring and/or C15 methine bridge are shown as sticks and surface as in part (b).
The chromophore ring system is on one side packed onto the highly conserved motif formed by Asp207, Ile208, and Pro209 and on the other closely apposed to His260. Assuming that this structure reflects a protonated BV species, as seems likely based on spectral characterization of the crystals, the positive charge that is delocalized across the B- and C-ring NH moieties is sandwiched between the backbone carbonyl oxygen of Asp207 and the side chain of His260. The charge of the ring system is thus closely associated with the partial negative charges of the His260 δ1 nitrogen and the backbone carbonyl oxygen of Asp207, which together should suffice to stabilize the charged, protonated BV ring system.
Although DrBphP was crystallized without the PHY domain, this crystal structure nevertheless provides unique insight into the photoconversion process. The BV A-ring is sandwiched between secondary structure elements of the GAF domain and is covalently attached to Cys24. The C10 methine bridge is tightly packed by the B- and C-rings of chromophore as well as by the conserved Asp207, Ile208, Pro209, Ala212, Tyr216, and His260 (Figure 4B). C10 is thus held tightly in place, so photochemistry cannot occur about this position, explaining the lack of photoconversion and the intense fluorescence observed upon assembly of apophytochrome with a bilin containing a saturated C15 bridge (71). In contrast, the D-ring is in a looser environment that would sterically permit more ready rotation. The D-ring pocket is also lined with highly conserved residues (Figure 4C), at least one of which is known to be critical for photochemistry (20, 21). This structure thus provides strong evidence that the conversion of Pr to Pfr proceeds with rotation of the D-ring and only the D-ring of the chromophore.
THE PHYTOCHROME PHOTOCYCLE AND DARK REVERSION
The “Forward” Reaction: Pr to Pfr Phototransformation
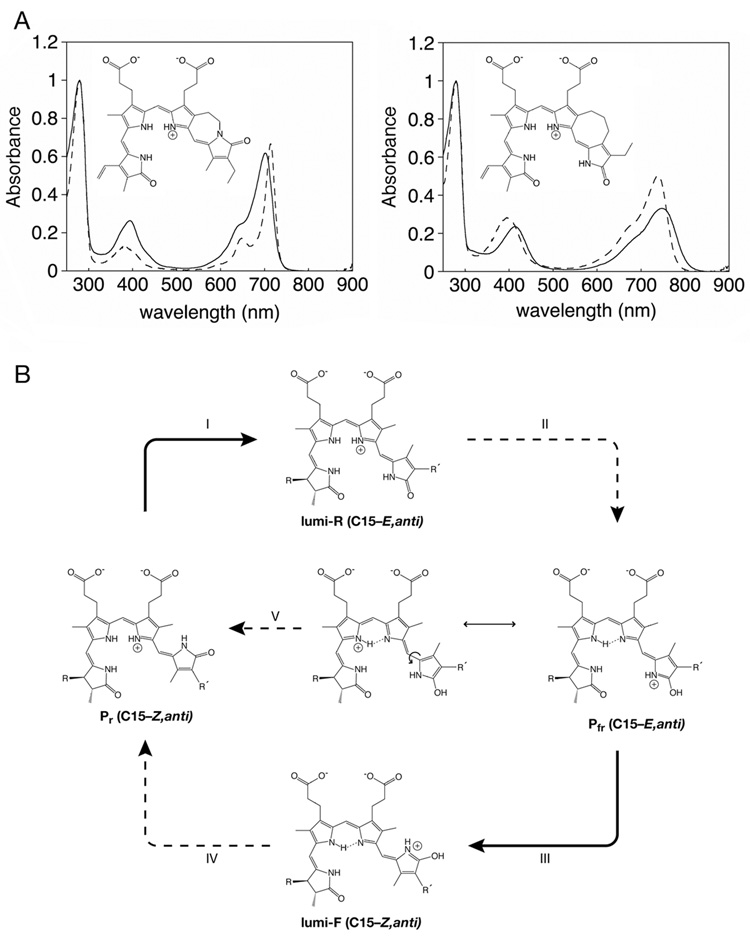
The DrBphP crystal structure is in the Pr state, so the exact structure of Pfr must await future investigation. However, in combination with other data, this structure provides a new basis for more directed speculation about the photochemical pathway than was possible without any experimental structure. Several Resonance Raman studies have led to proposal of a C15–E,anti geometry for the Pfr chromophore, although without any consensus as to the structure of the Pr chromophore (1, 22, 54, 66, 67, 70). Examination of both Phys and Cph1 by FT-IR spectroscopy provides evidence that the primary photoproduct formed upon irradiation of Pr, lumi-R, has a Pfr-like configuration (23–25). The subsequent dark reactions leading to Pfr have been proposed to involve a further rotation about C15 to generate a C15–E,syn conformation (2), but the crystal structure of DrBphP indicates that this conformer would be sterically disfavored (103). Moreover, a recent study utilizing synthetic bilins that are unable to rotate about C15 (44) demonstrated that only the C15–E,anti BV analog yielded a BphP adduct with properties similar to those of Pfr (Figure 5A). This approach also correctly identified the C15–Z,anti conformation of the Pr chromophore (44). Taken together with the crystal structure, these data provide good evidence that conversion of Pr to Pfr is best described by a single photochemical isomerization of the chromophore about the C15–C16 double bond, with both the lumi-R primary photoroduct and Pfr adopting a C15–E,anti conformation (Figure 5B).
Figure 5. Chemical delineation of the phytochrome photocycle.
(a) Structures and spectra for synthetic, sterically locked bilins (44) assembled with the bacteriophytochrome Agp1 from Agrobacterium tumefaciens. (left) Spectra for the C15–Z,anti locked bilin (dashed) and Pr biliverdin (solid) adducts. (b) Spectra for the C15–E,anti locked bilin (dashed) and Pfr biliverdin (solid) adducts. Spectra in (a) and (b) are courtesy of Drs. Tilman Lamparter and Katsuhiko Inomata. (c) Proposed photocycle for phytochromes utilizing PCB or PΦB. The Pr conformation is assigned based on the crystal structure of DrBphP, the locked bilin data presented in part (a), and the known stereochemistry of the 3 stereocenters in these molecules. Illumination with red light triggers photoisomerization about the C15–C16 double bond (I) to give the C15–E,anti primary photoproduct lumi-R, which is subsequently converted to Pfr in several light-independent steps (II). As discussed in the text, the proposed Pfr is hypothetical but would account for the observed instability of the Pfr chromopeptide, the red-shifted Pfr absorbance maximum, and the observed Pfr dark reversion. Illumination of Pfr with far-red light (III) triggers the reverse photoisomerization to yield the C15–Z,anti lumi-F primary photoproduct, which is subsequently converted to Pr in a series of light-independent steps (IV). Dark reversion would proceed through the Pfr resonance form with single-bond character about C15–C16, which would readily undergo thermal rotation about this bond and then convert to Pr(V).
Unlike Pr, the Pfr state is not stable in solution and can only be observed within its native protein matrix (88). The substantial red-shift of Pr relative to Pfr (Figure 5A) indicates either a much more extended conformation, which is not the case for the proposed C15–E,anti configuration, or greater electron density on the D-ring that extends the effective length of the conjugated system and red-shifts the resulting spectrum. Such electron density could readily result from the hypothetical Pfr structure shown in Figure 5B, which would arise from two proton transfers between the chromophore and the protein. The greater electron density on the D ring is consistent with the observed red shift of Pfr. The O-protonated Pfr species proposed in Figure 5B would explain the instability of the Pfr chromophore upon denaturation (87), because it is not significantly populated in solution (18). A recent NMR study of Cph1 presented evidence that all four nitrogens were protonated in the Pfr state (97), but it is unclear whether these technically challenging experiments would have been able to distinguish between such a model and one in which one proton is shared between the B- and C-ring nitrogens, particularly in light of the apparently weak NMR intensities seen for deprotonated bilin nitrogens (18).
The proposed Pfr structure in Figure 5B would be stabilized in the chromophore-binding pocket through the action of a proton acceptor (taking a proton from the B/C-ring NH moieties) and a proton donor (transferring a proton to the D-ring carbonyl oxygen). Recently, the pKa of the chromophore ring system in holoprotein has been estimated at ~9.5, suggesting that conserved Tyr or Cys residues could be viable proton donors in addition to conserved His, Asp, or Glu residues. It should now be possible to test this model and others by mutagenizing candidate proton donors and acceptors based on the DrBphP crystal structure.
In this proposed model, the photoconversion of Pr to Pfr proceeds via initial photoisomerization of the C15–C16 double bond followed by proton transfers and conformational changes of the protein matrix. The Pfr state is known to be much less fluorescent than the Pr state, and the proposed structure in Figure 5B provides a possible explanation for this: excited chromophore molecules that do not undergo photochemistry could readily undergo proton transfer either via tunneling between the B- and C-rings or at the D-ring carbonyl, leading to spectrally silent de-excitation.
The “Reverse” Reaction: Pfr to Pr Phototransformation
The conversion of Pfr to Pr is known to proceed via a distinct pathway from that of conversion from Pr to Pfr (36). For at least one phytochrome, recent FT-IR data provide evidence that the lumi-F primary photoproduct adopts a Pr-like configuration, which would imply a C15–Z,anti configuration, but this may not be universal (24). While the Pfr structure proposed in Figure 5B is hypothetical and is presented here as a conceptual aid, one can see that the reverse reaction from this species would indeed proceed via a different pathway with a different primary photoproduct. Subsequent thermal relaxation of lumi-F to Pr would entail a different pathway of proton transfers and protein conformational changes, with the residue donating a proton to the D-ring in the Pfr state again becoming protonated and the proton acceptor returning the proton to the B/C-ring system. Additional structural information about the Pfr state will be needed before a more informed description of the photochemical reverse reaction can be attained.
Dark Reversion
The Pfr state is also thermally unstable in most phytochromes, with restoration of the Pr state over time in a process known as dark reversion. It has long been known that multiple factors are capable of modulating the rate of dark reversion, such as changes in pH, ionic strength, reducing agents or metal ion concentration (27). By definition, this process cannot be triggered by spectral techniques, so it is much less amenable to study than photoconversion. The proposed Pfr structure suggests a mechanism for dark reversion via an alternate resonance form with single bond character about C15–C16 that could therefore thermally rotate to the C15–Z,anti configuration of Pr (Figure 5B, center). Reversion of this intermediate to Pr via a series of steps reminiscent of the photochemical process can then be envisaged. Although dark reversion is not yet well characterized, it makes an important contribution to the balance between Pr and Pfr and hence to determining the output state of a given phytochrome. Indeed, evidence that dark reversion of plant Phys is fluence rate dependent (39), can be reduced by interaction with other proteins (99) or enhanced by missense mutations (16) suggests that regulation of dark reversion may play a significant role in Phy signal output.
PHYTOCHROME AFTER DARK: FROM PHOTOCHEMISTRY TO SIGNALING
In view of the diversity of regulatory domains associated with a conserved bilin-binding GAF domain of phytochromes, the molecular mechanisms of signal output are expected to vary widely. Phytochromes with histidine kinase(-related) regulatory domains are the most widespread - an observation that strongly suggests that plant phytochromes evolved from a two component sensor precursor with a tetrapyrrole binding pocket (69). Indeed, prokaryotic phytochromes of the Cph1 and BphP families are predominantly ATP-dependent histidine kinases that mediate phosphotransfer to aspartate residues of their cognate response regulators, which are often encoded within the same operon (50). Despite the nature of the output domain, it is well accepted that phytochrome signaling involves light-mediated changes in interactions between photosensory and regulatory domains that are best understood for plant phytochromes (79).
Molecular mechanisms of prokaryotic phytochrome signaling
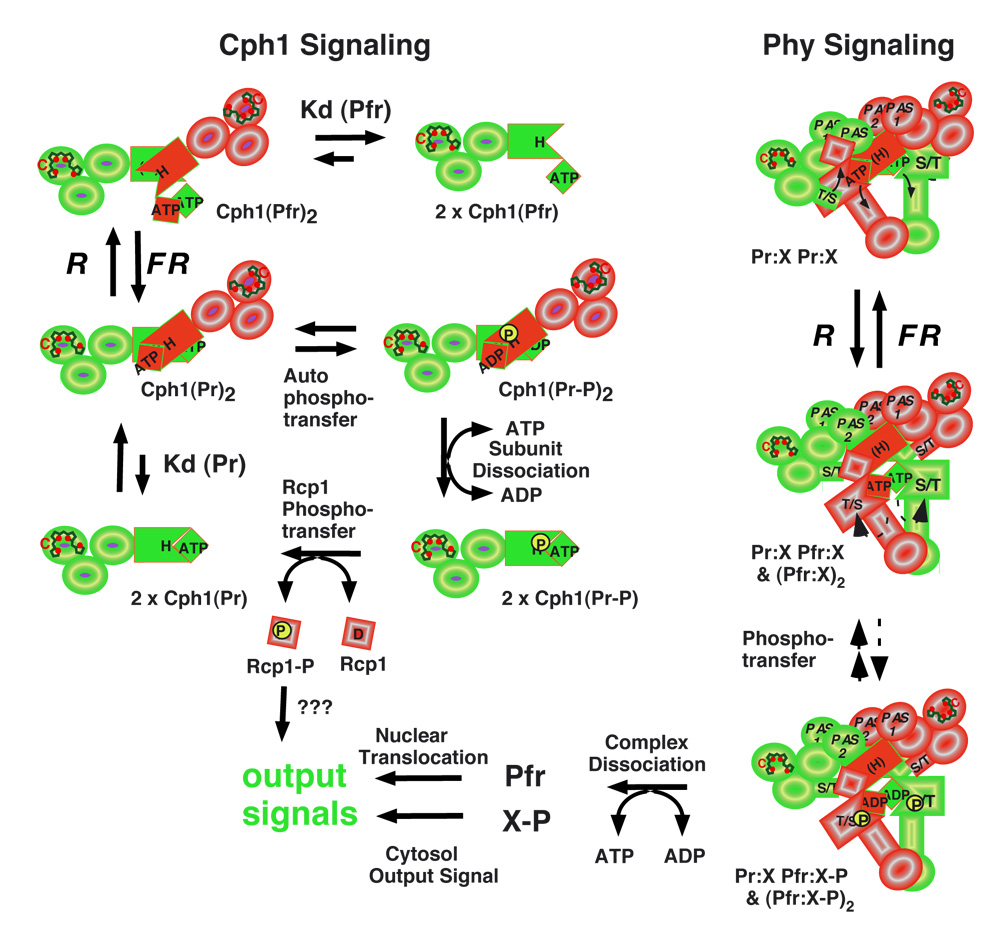
Signal transfer by prokaryotic phytochromes most frequently utilizes the two component signaling paradigm, i.e. ligand-dependent histidine kinase activation and phosphotransfer to a response regulator that directly regulates transcription or motility (82). Phosphotransfer is both bilin- and light-modulated for Cph1s and BphPs, a result consistent with the regulation of photoreceptor homodimerization and substrate interaction dynamics by these input signals (69, 78). Bilin binding stimulates kinase activity for Cph1s, while red light inhibits both autophosphorylation and response regulator phosphorylation in a mechanistic interpretation depicted in Figure 6A (17, 43, 59, 107). Although some BphPs show photoregulation similar to Cph1 (33, 43, 61), other BphPs exhibit a reversal in polarity with Pfr being more active than Pr (4, 48), while others show no effect of light on autophosphorylation (100). Unfortunately, the structural basis of this diversity in biochemical signal output is not readily revealed by comparison of the protein sequences; we expect that compensatory changes in both the photosensory and regulatory domains will be responsible.
Figure 6. Hypothetical signaling mechanisms for prokaryote and eukaryote phytochromes.
Homodimerization of the prokaryote phytochrome (Cph1) is dynamic and light-dependent (upper left) since autophosphorylation is favored by the formation of homodimers in the Pr form (Cph1(Pr)2) and inhibited by conversion to Pfr (Cph1(Pfr)2) which dissociates to an inactive monomer ((Cph1(Pfr)). Exchange of bound ADP with ATP, a process that promotes dissociation of the phosphorylated Pr dimer (Cph1(Pr-P)2) by inhibiting reassociation of the phosphorylated Pr monomer (Cph1(Pr-P)), stimulates histidine to aspartate phosphotransfer to Cph1's substrate Rcp1. The dephosphorylated Pr monomer (Cph1(Pr)) reassociates to form the active homodimer (Cph1(Pr)2). Eukaryote phytochromes (Phys) are obligate homodimers that are associated with a cytosolic anchoring protein X in an ATP-dependent protein complex (upper right). Photoconversion yields a Pr-Pfr heterodimer/Pfr-Pfr homodimer mixture (Pr:X Pfr:X & (Pfr:X)2) which results in activation of the Ser/Thr kinase activity and the stimulation of phosphotransfer to anchoring protein X. The exchange of bound ADP with ATP favors dissociation of the Pfr:X complexes, enabling free Pfr to move to the nucleus and phosphorylated X to mediate a cytosolic output signal.
Molecular mechanisms of plant phytochrome signaling
Our understanding of plant phytochrome signaling has benefited from extensive genetic, biochemical and cell biological investigations going back many years (7, 10, 11, 30, 42, 68, 73, 83, 84, 90, 95). For this reason, the following discussion is limited to recent data that most directly impinge upon the molecular basis of phytochrome signaling. As depicted in Figure 1B, the structure of plant phytochromes (Phys) has been remarkedly preserved throughout evolution (69). In contrast with Cph1s and BphPs, plant phytochromes are obligate dimers consisting of two ~120 kDa subunits with both regulatory PAS and HKRD subdomains contributing to the high affinity subunit-subunit interaction (47). Small x-ray scattering and EM analysis indicates that the phytochrome holoprotein has similar overall dimensions to mammalian immunoglobulin Gs (46, 74, 75). Encoded by small gene families in angiosperms (64), phytochromes fall into two classes - those that are light-labile (phyAs) and those that are light-stable (phyB-F). While phyAs are mostly homodimeric, recent studies reveal that light-stable phytochromes are also found as tightly bound heterodimers (92). Based on this structural property, it is clear that light-regulated subunit-subunit dissociation cannot be the signaling mechanism used by plant phytochromes.
The recent discovery that the P1-P3 photosensory core domains of plant phytochrome are fully sufficient for phytochrome signaling as long as the truncated polypeptide is targeted to the plant cell nucleus as a homodimeric holoprotein was another paradigm-shifting observation in the field of phytochrome research (65, 77). This work, along with a plethora of other studies using GFP-labeled phytochromes (reviewed in (68, 72, 73)) and cytoplasm-anchored phytochrome (41), indicate that phytochrome signaling requires dynamic cytoplasm-to-nuclear relocalization following its photoactivation. Since the nuclear localization signal (NLS) has been localized to the C-terminal PAS domains (13), the regulatory domains must play a dual role in phytochrome signaling - to maintain the homodimer and to target the photoreceptor to the nucleus. The evidence that plant phytochromes are serine/threonine kinases suggests that ATP-binding and/or protein phosphorylation mediated by the regulatory domains also play a role in light signaling (106).
Phytochrome phosphorylation has been shown to contribute to desensitization of the light signal (52, 53). However, the hypothesis that light-regulated protein phosphorylation is the trigger enabling the photoreceptor to 'uncouple' itself from a cytoplasmic anchor thereby exposing the NLS, remains a viable one (90). In this signaling model depicted in Figure 6B, photoconversion of Pr to Pfr with red light effects a conformational change that facilitates phosphotransfer to a bound anchoring molecule (X). The Pfr(ADP):X-P complex dissociates upon ATP-ADP exchange, enabling exposure of the PAS-localized NLS and Pfr migration to the nucleus where the N-terminal photosensory domain can interact with regulatory transcription factors (42). The altered activity of the putative anchoring molecule(s) X-P is envisaged to initiate a cytoplasmic output signal. Little is presently known about the cytoplasmic signaling pathway, candidate phytochrome-interacting cytoplasmic substrates have been identified (90). Once in the nucleus, Pfr accumulates in sub-nuclear foci or speckles whose appearance are correlated with the output signal (12). Speckles are thought to represent sites of transcription factor degradation, although other hypotheses have been proposed (11). Speckle formation requires the intact C-terminus, i.e. both PAS and HKRD domains, suggesting this region plays an additional signaling role in the nucleus (77). Pfr autophosphorylation and/or subsequent dark reversion are envisaged to complete the phytochrome signaling cycle, whereupon free phosphorylated forms of phytochrome are degraded (phyA) or recycled (PhyB-E).
Through isolation of missense alleles of phytochromes, genetic approaches have provided valuable insight into the molecular basis of phytochrome signaling. Such mutant alleles can be categorized into two classes: hyposensitive (loss-of-function) and hypersensitive (gain-of-function) alleles (Supplemental Table 2). While a large majority of the loss-of-function mutations fall within the regulatory PAS domains (85), loss-of-function mutations also occur throughout the photosensory region. Where tested, the molecular bases for loss-of-function phenotypes include increased dark reversion, reduced nuclear targeting and altered sub-nuclear localization. Known gain-of-function mutations are rare (55)(15) (104), with some falling in the photosensory core (Figure 3A). These mutations could enhance the translocation of Pfr to the nucleus or inhibit nuclear turnover of phytochrome (104). The accumulation of additional mutant alleles, together with x-ray crystallographic analysis, will be a powerful combination to assess the molecular basis of phytochrome signaling in the future.
Phytochrome signaling mechanisms are still evolving
There are a number of BphPs that lack HKRDs altogether, and other catalytic/regulatory domains have been inserted in their place during evolution (32, 50). This type of exchange appears to have occurred many times in the past, but the probability that the new phytochrome chimera remained functional is small since few such phytochromes exist (outside the cyanobacteria). Domain exchange is likely responsible for the emergence of the plant phytochrome lineage since their regulatory PAS and HKRD modules appear evolutionarily distinct from HKRDs found on the extant prokaryotic phytochromes (64). Domain exchange has occurred more recently in primitive plants to yield the neochromes, which are functional chimeras of a plant phytochrome photosensory P1-P4 domain and a blue-light sensing phototropin (98). It is clear that the most extensive phytochrome evolution has taken place in the cyanobacteria, which probably reflects the abundance of multiple bilin ligands and the need of these photosynthetic bacteria to adapt to light environments that are enriched in blue, green or red wavelengths (57).
SUMMARY LIST
Phytochromes photoconvert between Pr and Pfr states, and the ratio of these states determines the signaling state of phytochrome.
Phytochromes have a modular domain architecture with a conserved N-terminal photosensory core and a C-terminal regulatory region.
Phytochromes utilize bilin chromophores that photoisomerize during the conversion between Pr and Pfr.
The conserved N-terminal photosensor of phytochromes can be fused to a variety of regulatory domains which can act in bacterial two-component pathways or in more complex pathways in plants.
Recent structural breakthroughs and biochemical results have defined the Pr state of the chromophore and provide new insight into the structure of the Pfr state.
FUTURE DIRECTIONs
Understanding the structural changes associated with photoisomerization.
Understanding how those changes alter the function of the regulatory domains to trigger signaling.
Understanding the biological functions of phytochromes in nonphotosynthetic microbes and of divergent, phytochrome-related molecules in plants and cyanobacteria.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Drs. Katrina Forest and Richard Vierstra for supplying the coordinates for the DrBphP structure prior to publication. The authors also thank Drs. Katsuhiko Inomata and Tilman Lamparter for providing the raw spectral data for Agp1. Our work was supported by grants from the National Institutes of Health (GM068552) and from the National Science Foundation Center for Biophotonics Science and Technology PHY-0120999.
Glossary
GLOSSARY
- bilin
a linear tetrapyrrole, metabolically derived from heme.
- dark reversion
conversion of Pfr to Pr in a light-independent thermal process; also can be used to describe the Pr to Pfr thermal conversion for the bathyBphPs.
- P2/PAS
the PAS domain in the phytochrome photosensory core.
- P3/GAF
the GAF domain in the phytochrome photosensory core.
- P4/PHY
the PHY domain in the phytochrome photosensory core, thought to be a phytochrome-specific GAF domain.
- photoconversion
photochemical conversion of Pr phytochrome to Pfr and back.
- Pr
red-light-absorbing phytochrome state.
- Pfr
far-red-light-absorbing phytochrome state.
ACRONYMS LIST
- bathyBphP
atypical BphP that exhibits Pfr-like spectrum in the thermal ground state, with photoconversion to a species with a Pr-like spectrum.
- BphP
member of the bacteriophoytochrome subfamily.
- BV
biliverdin IXα
- Cph1, Cph2
phytochrome subfamilies named after cyanobacterial phytochromes 1 and 2, exemplars of these subfamilies from Synechocystis sp. PCC 6803.
- EM
electron microscopy.
- Fph
member of the fungal phytochrome subfamily.
- FR
far-red light.
- FT-IR
Fourier transform infrared spectroscopy.
- GAF
domain acronym derived from vertebrate cGMP-specific phosphodiesterases, cyanobacterial adenylate cyclases, and formate hydrogen lyase transcription activator FhlA.
- HKRD
histidine kinase related domain.
- PAS
domain acronym derived from period clock (PER) protein, aromatic hydrocarbon receptor nuclear translocator (ARNT), and single minded (SIM).
- PCB
phycocyanobilin.
- PΦB
phytochromobilin.
- Phy
member of the plant phytochrome subfamily.
- R
red light.
Footnotes
SIDE BAR
Phytochromes as Sensors of Oxygen-Dependent Heme Catabolism. The bilin chromophores incorporated by all phytochromes are synthesized from heme in two steps. First, a heme oxygenase converts heme into BV, which is directly incorporated as the chromophore of BphP and Fph phytochromes. In plants and cyanobacteria, however, BV is further reduced to yield PΦB in higher plants and PCB in cyanobacteria and green algae. Conversion of BV to PΦB is carried out by HY-2 in the chloroplast, while reduction of BV to yield PCB is instead carried out by PcyA. Both HY-2 and PcyA belong to a conserved family of ferredoxin-dependent bilin reductases.
The kinase activity and regulatory signaling state of many phytochromes are regulated not only by light but by the presence or absence of chromophore. The synthesis of chromophore is itself dependent on the heme metabolism of the cell, because chromophore will only be produced sparingly if cells are starved for heme or oxygen. Hence, phytochrome signaling is sensitive to heme metabolism and oxygen levels. Phytochromes therefore integrate both the light environment and the metabolic state of the cell to affect a single signaling readout. Bilin metabolism has recently been reviewed (26).
RELATED CHAPTERS (Vol. 57, 2006)
Complementary Chromatic Adaptation (David Kehoe)
Integrated Signaling Pathways (Klaus Harter)
Tetrapyrrole Synthesis (Ayumi Tanaka)
LITERATURE CITED
- 1.Andel F, Lagarias JC, Mathies RA. Resonance Raman analysis of chromophore structure in the lumi-R photoproduct of phytochrome. Biochem. 1996;35:15997–16008. doi: 10.1021/bi962175k. [DOI] [PubMed] [Google Scholar]
- 2.Andel F, Murphy JT, Haas JA, McDowell MT, van der Hoef I, et al. Probing the photoreaction mechanism of phytochrome through analysis of resonance Raman vibrational spectra of recombinant analogues. Biochem. 2000;39:2667–2676. doi: 10.1021/bi991688z. [DOI] [PubMed] [Google Scholar]
- 3.Batschauer A, editor. Photoreceptors and Light Signaling. Vols. 3. Cambridge, UK: Royal Society of Chemistry; 2003. 388 pp. [Google Scholar]
- 4.Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature. 2001;414:776–779. doi: 10.1038/414776a. [DOI] [PubMed] [Google Scholar]
- 5.Blumenstein A, Vienken K, Tasler R, Purschwitz J, Veith D, et al. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 2005;15:1833–1838. doi: 10.1016/j.cub.2005.08.061. [DOI] [PubMed] [Google Scholar]
- 6. Borucki B, Otto H, Rottwinkel G, Hughes J, Heyn MP, Lamparter T. Mechanism of Cph1 phytochrome assembly from stopped-flow kinetics and circular dichroism. Biochem. 2003;42:13684–13697. doi: 10.1021/bi035511n. This study examines the assembly reaction of Cph1 with PCB in real time using stopped-flow techniques.
- 7.Briggs WR, Rice HV. Phytochrome: Chemical and physical properties and mechanism of action. Annual Review of Plant Physiology. 1972;23:293–334. [Google Scholar]
- 8.Briggs WR, Spudich JA, editors. Handbook of Photosensory Receptors. Weinheim: Wiley VCH; 2005. 473 pp. [Google Scholar]
- 9.Butler WL, Norris KH, Seigelman HW, Hendricks SB. Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc. Natl. Acad. Sci. USA. 1959;45:1703–1708. doi: 10.1073/pnas.45.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casal JJ, Luccioni LG, Oliverio KA, Boccalandro HE. Light, phytochrome signalling and photomorphogenesis in Arabidopsis [Review] Photochemical & Photobiological Sciences. 2003;2:625–636. doi: 10.1039/b300094j. [DOI] [PubMed] [Google Scholar]
- 11.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Ann. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 12.Chen M, Schwabb R, Chory J. Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc. Natl. Acad. Sci. USA. 2003;100:14493–14498. doi: 10.1073/pnas.1935989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen M, Tao Y, Lim J, Shaw A, Chory J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 2005;15:637–642. doi: 10.1016/j.cub.2005.02.028. This work demonstrates that the PAS repeat domains of plant Phys contain a cryptic NLS that is key for light-mediated signaling.
- 14.Davis SJ, Vener AV, Vierstra RD. Bacteriophytochromes: Phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science. 1999;286:2517–2520. doi: 10.1126/science.286.5449.2517. [DOI] [PubMed] [Google Scholar]
- 15.Dieterle M, Bauer D, Buche C, Krenz M, Schäfer E, Kretsch T. A new type of mutation in phytochrome A causes enhanced light sensitivity and alters the degradation and subcellular partitioning of the photoreceptor. Plant J. 2005;41:146–161. doi: 10.1111/j.1365-313X.2004.02286.x. [DOI] [PubMed] [Google Scholar]
- 16.Elich TD, Chory J. Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell. 1997;9:2271–2280. doi: 10.1105/tpc.9.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteban B, Carrascal M, Abian J, Lamparter T. Light-induced conformational changes of cyanobacterial phytochrome cph1 probed by limited proteolysis and autophosphorylation. Biochem. 2005;44:450–461. doi: 10.1021/bi0484365. [DOI] [PubMed] [Google Scholar]
- 18. Falk H. The Chemistry of Linear Oligopyrroles and Bile Pigments. Vienna: Springer-Verlag; 1989. 621 pp. This classic work covers the chemistry of bilins and provides invaluable information on how bilin chromophores behave in solution.
- 19.Fedorov R, Schlichting I, Hartmann E, Domratcheva T, Fuhrmann M, Hegemann P. Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii. Biophysical Journal. 2003;84:2474–2482. doi: 10.1016/S0006-3495(03)75052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer AJ, Lagarias JC. Harnessing phytochrome's glowing potential. Proc. Natl. Acad. Sci. USA. 2004;101:17334–17339. doi: 10.1073/pnas.0407645101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer AJ, Rockwell NC, Jang AY, Ernst LA, Waggoner AS, et al. Multiple roles of a conserved GAF domain tyrosine residue in cyanobacterial and plant phytochromes. Biochem. 2005;44 doi: 10.1021/bi051633z. online October 25, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fodor SPA, Lagarias JC, Mathies RA. Resonance Raman analysis of the Pr and Pfr forms of phytochrome. Biochem. 1990;29:11141–11146. doi: 10.1021/bi00502a018. [DOI] [PubMed] [Google Scholar]
- 23.Foerstendorf H, Benda C, Gärtner W, Storf M, Scheer H, Siebert F. FTIR studies of phytochrome photoreactions reveal the C=O bands of the chromophore: consequences for its protonation states, conformation, and protein interaction. Biochem. 2001;40:14952–14959. doi: 10.1021/bi0156916. [DOI] [PubMed] [Google Scholar]
- 24.Foerstendorf H, Lamparter T, Hughes J, Gärtner W, Siebert F. The photoreactions of recombinant phytochrome from the cyanobacterium Synechocystis: A low-temperature UV-Vis and FT-IR spectroscopic study. Photochem. Photobiol. 2000;71:655–661. doi: 10.1562/0031-8655(2000)071<0655:tporpf>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Foerstendorf H, Mummert E, Schäfer E, Scheer H, Siebert F. Fourier-Transform Infrared Spectroscopy of Phytochrome - Difference Spectra of the Intermediates of the Photoreactions. Biochem. 1996;35:10793–10799. doi: 10.1021/bi960960r. [DOI] [PubMed] [Google Scholar]
- 26.Frankenberg NF, Lagarias JC. Biosynthesis and biological function of bilins. In: Kadish KM, Smith KM, Guilard R, editors. The Porphyrin Handbook. Chlorophylls and Bilins: Biosynthesis Structure and Degradation. New York: Academic Press; 2003. pp. 211–235. [Google Scholar]
- 27.Franklin B. Biosynthesis and dark transformations of phytochrome. In: Mitrakos K, Shropshire W, editors. Phytochrome. New York: Academic Press; 1972. pp. 195–225. [Google Scholar]
- 28.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 29.Froehlich AC, Noh B, Vierstra RD, Loros J, Dunlap JC. Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryotic Cell. 2005. in press. [DOI] [PMC free article] [PubMed]
- 30.Furuya M. Phytochromes - Their molecular species, gene families, and functions. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1993;44:617–645. [Google Scholar]
- 31.Furuya M, Schäfer E. Photoperception and signalling of induction reactions by different phytochromes. Trends in Plant Science. 1996;1:301–307. [Google Scholar]
- 32.Giraud E, Fardoux J, Fourier N, Hannibal L, Genty B, et al. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature. 2002;417:202–205. doi: 10.1038/417202a. [DOI] [PubMed] [Google Scholar]
- 33.Giraud E, Zappa S, Vuillet L, Adriano JM, Hannibal L, et al. A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. J. Biol. Chem. 2005;280:32389–32397. doi: 10.1074/jbc.M506890200. [DOI] [PubMed] [Google Scholar]
- 34.Goller AH, Strehlow D, Hermann G. Conformational flexibility of phycocyanobilin: An AM1 semiempirical study. Chemphyschem. 2001;2:665–671. doi: 10.1002/1439-7641(20011119)2:11<665::AID-CPHC665>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 35.Goller AH, Strehlow D, Hermann G. The excited-state chemistry of phycocyanobilin: a semiempirical study. Chemphyschem. 2005;6:1259–1268. doi: 10.1002/cphc.200400667. [DOI] [PubMed] [Google Scholar]
- 36.Gärtner W, Braslavsky SE. The phytochromes: Spectroscopy and function. In: Batschauer A, editor. Photoreceptors and Light Signalling. Cambridge, UK: Royal Society of Chemistry; 2003. pp. 136–180. [Google Scholar]
- 37.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 38.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hennig L, Schäfer E. Both subunits of the dimeric plant photoreceptor phytochrome require chromophore for stability of the far-red light-absorbing form. J. Biol. Chem. 2001;276:7913–7918. doi: 10.1074/jbc.M009793200. [DOI] [PubMed] [Google Scholar]
- 40.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of Molecular Graphics. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 41.Huq E, Al-Sady B, Quail PH. Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 2003;35:660–664. doi: 10.1046/j.1365-313x.2003.01836.x. [DOI] [PubMed] [Google Scholar]
- 42.Huq E, Quail PH. Phytochrome Signaling. In: Briggs WR, Spudich JA, editors. Handbook of Photosensory Receptors. Weinheim: Wiley VCH; 2005. pp. 151–170. [Google Scholar]
- 43.Hübschmann T, Jorissen HJ, Börner T, Gärtner W, Tandeau de Marsac N. Phosphorylation of proteins in the light-dependent signalling pathway of a filamentous cyanobacterium. Eur. J. Biochem. 2001;268:3383–3389. doi: 10.1046/j.1432-1327.2001.02229.x. [DOI] [PubMed] [Google Scholar]
- 44. Inomata K, Hammam MAS, Kinoshita H, Murata Y, Khawn H, et al. Sterically locked synthetic bilin derivatives and phytochrome Agp1 from Agrobacterium tumefaciens form photoinsensitive Pr- and Pfr-like adducts. J. Biol. Chem. 2005;280:24491–24497. doi: 10.1074/jbc.M504710200. This study uses novel synthetic bilins unable to rotate about the C15 methine bridge to delineate the geometry of Pr and Pfr.
- 45.Jiang ZY, Swem LR, Rushing BG, Devanathan S, Tollin G, Bauer CE. Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science. 1999;285:406–409. doi: 10.1126/science.285.5426.406. [DOI] [PubMed] [Google Scholar]
- 46.Jones A, Erickson H. Domain Structure of Phytochrome From Avena sativa Visualized by Electron Microscopy. Photochem. Photobiol. 1989;49:479–483. doi: 10.1111/j.1751-1097.1989.tb09198.x. [DOI] [PubMed] [Google Scholar]
- 47.Jones AM, Edgerton MD. The Anatomy of Phytochrome, a Unique Photoreceptor in Plants. Sem. Cell Biol. 1994;5:295–302. doi: 10.1006/scel.1994.1036. [DOI] [PubMed] [Google Scholar]
- 48.Karniol B, Vierstra RD. The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. USA. 2003;100:2807–2812. doi: 10.1073/pnas.0437914100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karniol B, Vierstra RD. Structure, function, and evolution of microbial phytochromes. In: Schäfer E, Nagy F, editors. Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms. 3rd Edition. Dordrecht, The Netherlands: Springer; 2005. in press. [Google Scholar]
- 50.Karniol B, Wagner JR, Walker JM, Vierstra RD. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem. J. 2005;392:103–116. doi: 10.1042/BJ20050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kehoe DM, Grossman AR. Similarity of a Chromatic Adaptation Sensor to Phytochrome and Ethylene Receptors. Science. 1996;273:1409–1412. doi: 10.1126/science.273.5280.1409. This paper presents the first description of a phytochrome from a bacterial system.
- 52.Kim JI, Park JE, Zarate X, Song PS. Phytochrome phosphorylation in plant light signaling. Photochemical & Photobiological Sciences. 2005;4:681–687. doi: 10.1039/b417912a. [DOI] [PubMed] [Google Scholar]
- 53.Kim JI, Shen Y, Han YJ, Park JE, Kirchenbauer D, et al. Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell. 2004;16:2629–2640. doi: 10.1105/tpc.104.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kneip C, Schlamann W, Braslavsky SE, Hildebrandt P, Schaffner K. Resonance Raman spectroscopic study of the tryptic 39-kDa fragment of phytochrome. FEBS Lett. 2000;482:252–256. doi: 10.1016/s0014-5793(00)02069-x. [DOI] [PubMed] [Google Scholar]
- 55.Kretsch T, Poppe C, Schäfer E. A new type of mutation in the plant photoreceptor phytochrome B causes loss of photoreversibility and an extremely enhanced light sensitivity. Plant J. 2000;22:177–186. doi: 10.1046/j.1365-313x.2000.00715.x. [DOI] [PubMed] [Google Scholar]
- 56.Lagarias JC, Rapoport H. Chromopeptides from phytochrome. The structure and linkage of the Pr form of the phytochrome chromophore. J. Am. Chem. Soc. 1980;102:4821–4828. [Google Scholar]
- 57.Lamparter T. Evolution of cyanobacterial and plant phytochromes. FEBS Lett. 2004;573:1–5. doi: 10.1016/j.febslet.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 58. Lamparter T, Carrascal M, Michael N, Martinez E, Rottwinkel G, Abian J. The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of Agrobacterium phytochrome Agp1. Biochem. 2004;43:3659–3669. doi: 10.1021/bi035693l. This paper demonstrates the nature of covalent attachment for phytochromes that utilize BV as chromophore.
- 59.Lamparter T, Esteban B, Hughes J. Phytochrome Cph1 from the cyanobacterium Synechocystis PCC6803 - Purification, assembly, and quaternary structure. Eur. J. Biochem. 2001;268:4720–4730. doi: 10.1046/j.1432-1327.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- 60.Lamparter T, Michael N. Agrobacterium phytochrome as an enzyme for the production of ZZE bilins. Biochem. 2005;44:8461–8469. doi: 10.1021/bi047510g. [DOI] [PubMed] [Google Scholar]
- 61.Lamparter T, Michael N, Mittmann F, Esteban B. Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proc. Natl. Acad. Sci. USA. 2002;99:11628–11633. doi: 10.1073/pnas.152263999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martinez SE, Bruder S, Schultz A, Zheng N, Schultz JE, et al. Crystal structure of the tandem GAF domains from a cyanobacterial adenylyl cyclase: modes of ligand binding and dimerization. Proc. Natl. Acad. Sci. USA. 2005;102:3082–3087. doi: 10.1073/pnas.0409913102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez SE, Wu AY, Glavas NA, Tang XB, Turley S, et al. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc. Natl. Acad. Sci. USA. 2002;99:13260–13265. doi: 10.1073/pnas.192374899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mathews S, Sharrock RA. Phytochrome gene diversity. Plant Cell Environ. 1997;20:666–671. [Google Scholar]
- 65.Matsushita T, Mochizuki N, Nagatani A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature. 2003;424:571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- 66.Matysik J, Hildebrandt P, Schlamann W, Braslavsky SE, Schaffner K. Fourier-Transform Resonance Raman Spectroscopy Of Intermediates Of the Phytochrome Photocycle. Biochem. 1995;34:10497–10507. doi: 10.1021/bi00033a023. [DOI] [PubMed] [Google Scholar]
- 67.Mizutani Y, Tokutomi S, Kitagawa T. Resonance Raman Spectra Of The Intermediates In Phototransformation Of Large Phytochrome - Deprotonation Of The Chromophore In The Bleached Intermediate. Biochem. 1994;33:153–158. doi: 10.1021/bi00167a020. [DOI] [PubMed] [Google Scholar]
- 68.Moller SG, Ingles PJ, Whitelam GC. The cell biology of phytochrome signalling. New Phytologist. 2002;154:553–590. doi: 10.1046/j.1469-8137.2002.00419.x. [DOI] [PubMed] [Google Scholar]
- 69.Montgomery BL, Lagarias JC. Phytochrome ancestry. Sensors of bilins and light. Trends in Plant Science. 2002;7:357–366. doi: 10.1016/s1360-1385(02)02304-x. [DOI] [PubMed] [Google Scholar]
- 70.Mroginski MA, Murgida DH, von Stetten D, Kneip C, Mark F, Hildebrandt P. Determination of the chromophore structures in the photoinduced reaction cycle of phytochrome. J. Am. Chem. Soc. 2004;126:16734–16735. doi: 10.1021/ja043959l. [DOI] [PubMed] [Google Scholar]
- 71.Murphy JT, Lagarias JC. The Phytofluors: A new class of fluorescent protein probes. Curr. Biol. 1997;7:870–876. doi: 10.1016/s0960-9822(06)00375-7. [DOI] [PubMed] [Google Scholar]
- 72.Nagatani A. Light-regulated nuclear localization of phytochromes. Current Opinion in Plant Biology. 2004;7:708–711. doi: 10.1016/j.pbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 73.Nagy F, Schäfer E. Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol. 2002;53:329–355. doi: 10.1146/annurev.arplant.53.100301.135302. [DOI] [PubMed] [Google Scholar]
- 74.Nakasako M, Iwata T, Inoue K, Tokutomi S. Light-induced global structural changes in phytochrome A regulating photomorphogenesis in plants. FEBS Journal. 2005;272:603–612. doi: 10.1111/j.1742-4658.2004.04508.x. [DOI] [PubMed] [Google Scholar]
- 75.Nakasako M, Wada M, Tokutomi S, Yamamoto KT, Sakai J, et al. Quaternary Structure Of Pea Phytochrome-I Dimer Studied With Small-Angle X-Ray Scattering And Rotary-Shadowing Electron Microscop. Photochem. Photobiol. 1990;52:3–12. [Google Scholar]
- 76.Nureki O, Shirouzu M, Hashimoto K, Ishitani R, Terada T, et al. An enzyme with a deep trefoil knot for the active-site architecture. Acta Crystallogr D Biol Crystallogr. 2002;58:1129–1137. doi: 10.1107/s0907444902006601. [DOI] [PubMed] [Google Scholar]
- 77.Oka Y, Matsushita T, Mochizuki N, Suzuki T, Tokutomi S, Nagatani A. Functional analysis of a 450-amino Acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell. 2004;16:2104–2116. doi: 10.1105/tpc.104.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Otto H, Lamparter T, Borucki B, Hughes J, Heyn MP. Dimerization and inter-chromophore distance of Cph1 phytochrome from Synechocystis, as monitored by fluorescence homo and hetero energy transfer. Biochem. 2003;42:5885–5895. doi: 10.1021/bi026946y. [DOI] [PubMed] [Google Scholar]
- 79.Park CM, Bhoo SH, Song PS. Inter-domain crosstalk in the phytochrome molecules. Sem. Cell Dev. Biol. 2000;11:449–456. doi: 10.1006/scdb.2000.0200. [DOI] [PubMed] [Google Scholar]
- 80.Park CM, Kim JI, Yang SS, Kang JG, Kang JH, et al. A second photochromic bacteriophytochrome from Synechocystis sp PCC 6803: Spectral analysis and down-regulation by light. Biochem. 2000;39:10840–10847. doi: 10.1021/bi992831r. [DOI] [PubMed] [Google Scholar]
- 81.Park CM, Shim JY, Yang SS, Kang JG, Kim JI, et al. Chromophore-apoprotein interactions in Synechocystis sp PCC6803 phytochrome Cph1. Biochem. 2000;39:6349–6356. doi: 10.1021/bi992916s. [DOI] [PubMed] [Google Scholar]
- 82.Parkinson JS. Signal Transduction Schemes Of Bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 83.Pratt LH. Phytochrome: The protein moiety. Annual Review of Plant Physiology. 1982;33:557–582. [Google Scholar]
- 84.Quail PH. Phytochrome - A light-activated molecular switch that regulates plant gene expression. Ann. Rev. Genet. 1991;25:389–409. doi: 10.1146/annurev.ge.25.120191.002133. [DOI] [PubMed] [Google Scholar]
- 85.Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: Photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 86.Quest B, Gärtner W. Chromophore selectivity in bacterial phytochromes: dissecting the process of chromophore attachment. Eur. J. Biochem. 2004;271:1117–1126. doi: 10.1111/j.1432-1033.2004.04015.x. [DOI] [PubMed] [Google Scholar]
- 87.Rüdiger W, Brandlmeier T, Blos I, Gossauer A, Weller J-P. Isolation of the phytochrome chromophore. The cleavage reaction with hydrogen bromide. Z. Naturforsch. 1980;35c:763–769. [Google Scholar]
- 88.Rüdiger W, Thümmler F, Cmiel E, Schneider S. Chromophore structure of the physiologically active form (Pfr) of phytochrome. Proc. Natl. Acad. Sci. USA. 1983;80:6244–6248. doi: 10.1073/pnas.80.20.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sage LC. Pigment of the Imagination: A History of Phytochrome Research. San Diego: Academic Press, Inc.; 1992. 562 pp. xx. This interesting popular work provides a unique perspective on the early years of phytochrome research.
- 90.Schepens I, Duek P, Fankhauser C. Phytochrome-mediated light signalling in Arabidopsis. Current Opinion in Plant Biology. 2004;7:564–569. doi: 10.1016/j.pbi.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 91.Schäfer E, Nagy F, editors. Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms. 3rd Edition. Dordrecht, The Netherlands: Springer; 2005. 667 pp. [Google Scholar]
- 92.Sharrock RA, Clack T. Heterodimerization of type II phytochromes in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2004;101:11500–11505. doi: 10.1073/pnas.0404286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sineshchekov AV, Koppel LA, Sineshchekov VA, Mokronosov AT. Dynamics Of Phytochrome Content And Photoactivity In Germinating Pea And Bean Seeds. Soviet Plant Physiology. 1989;36:165–172. [Google Scholar]
- 94.Sineshchekov VA. Photobiophysics and Photobiochemistry Of the Heterogeneous Phytochrome System. Biochimica Et Biophysica Acta-Bioenergetics. 1995;1228:125–164. [Google Scholar]
- 95.Smith H. Physiological and Ecological Function Within the Phytochrome Family. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1995;46:289–315. [Google Scholar]
- 96.Stone J. Masters thesis. University of Missouri-Rolla; 1998. An Efficient Library for Parallel Ray Tracing and Animation. [Google Scholar]
- 97.Strauss HM, Hughes J, Schmieder P. Heteronuclear solution-state NMR studies of the chromophore in cyanobacterial phytochrome Cph1. Biochem. 2005;44:8244–8250. doi: 10.1021/bi050457r. [DOI] [PubMed] [Google Scholar]
- 98.Suetsugu N, Mittmann F, Wagner G, Hughes J, Wada M. A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proc. Natl. Acad. Sci. USA. 2005;102:13705–13709. doi: 10.1073/pnas.0504734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Baurle I, et al. Interaction of the Response Regulator ARR4 with Phytochrome B in Modulating Red Light Signaling. Science. 2001;294:1108–1111. doi: 10.1126/science.1065022. [DOI] [PubMed] [Google Scholar]
- 100.Tasler R, Moises T, Frankenberg-Dinkel N. Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. FEBS Journal. 2005;272:1927–1936. doi: 10.1111/j.1742-4658.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- 101.Taylor WR. A deeply knotted protein structure and how it might fold. Nature. 2000;406:916–919. doi: 10.1038/35022623. [DOI] [PubMed] [Google Scholar]
- 102.Vierstra RD. Illuminating Phytochrome Functions. Plant Physiol. 1993;103:679–684. doi: 10.1104/pp.103.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore binding domain of phytochrome. Nature. 2005. in press. This paper presents a pivotal breakthrough: the first crystal structure of a phytochrome photosensory core.
- 104.Weller JL, Batge SL, Smith JJ, Kerckhoffs LH, Sineshchekov VA, et al. A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiol. 2004;135:2186–2195. doi: 10.1104/pp.103.036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wu SH, Lagarias JC. Defining the bilin lyase domain: Lessons from the extended phytochrome superfamily. Biochem. 2000;39:13487–13495. doi: 10.1021/bi001123z. This paper describes a series of truncation experiments demonstrating that the P3/GAF domain is sufficient for covalent attachment of chromophore.
- 106.Yeh K-C, Lagarias JC. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yeh K-C, Wu S-H, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. This paper demonstrates that the prokaryotic phytochrome Cph1 functions as a light-regulated protein kinase.
- 108.Zhao KH, Ran Y, Li M, Sun YN, Zhou M, et al. Photochromic biliproteins from the cyanobacterium Anabaena sp. PCC 7120: lyase activities, chromophore exchange, and photochromism in phytochrome AphA. Biochem. 2004;43:11576–11588. doi: 10.1021/bi0491548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.