The crystal structure of the second PDZ domain from human zonula occludens 2 has been determined at 1.75 Å resolution.
Keywords: PDZ domains, MAGUK family, domain swapping, ligand binding
Abstract
Human zonula occludens 2 (ZO-2) protein is a multi-domain protein that consists of an SH3 domain, a GK domain and three copies of a PDZ domain with slight divergence. The three PDZ domains act as protein-recognition modules that may mediate protein assembly and subunit localization. The crystal structure of the second PDZ domain of ZO-2 (ZO-2 PDZ2) was determined by molecular replacement at 1.75 Å resolution, revealing a dimer in the asymmetric unit. The dimer is stabilized by extensive symmetrical domain-swapping of the β1 and β2 strands. Structural comparison shows that the ZO-2 PDZ2 homodimer may have a similar ligand-binding pattern to the ZO-1 PDZ2–connexin 43 complex.
1. Introduction
Zonula occludens proteins (ZO proteins) are multi-domain proteins that belong to the membrane-associated guanylate kinase (MAGUK) protein family (Anderson et al., 1995 ▶). It has been demonstrated that ZO proteins serve as a platform for the recruitment or assembly of signalling complexes in response to cell–cell adhesion (Angst et al., 2001 ▶; Zahraoui et al., 2000 ▶). They also act as linkers between transmembrane proteins and the actin-based cytoskeleton (Fanning et al., 1998 ▶; Itoh et al., 1997 ▶; Wittchen et al., 1999 ▶). These highly conserved ZO proteins (ZO-1, ZO-2 and ZO-3) share the same domain organization consisting of three PDZ domains, an Src homology 3 (SH3) domain and a guanylate kinase-like (GK) domain (Haskins et al., 1998 ▶).
The three PDZ domains of ZO proteins play a crucial role in membrane-associated scaffolds (Bazzoni et al., 2000 ▶; Glaunsinger et al., 2001 ▶; Hunter et al., 2005 ▶; Kausalya et al., 2004 ▶; Li et al., 2004 ▶; Penes et al., 2005 ▶; Singh et al., 2005 ▶; Traweger et al., 2003 ▶). The PDZ domains have been well characterized as structurally conserved modules consisting of 80–100 amino acids (Sheng & Sala, 2001 ▶). Generally, they are composed of a six-β-strand core flanked by two α-helices. PDZ domains typically bind to short peptide fragments at the carboxy-terminus of their target proteins (Doyle et al., 1996 ▶; Songyang et al., 1997 ▶) in a structurally conserved groove between the β2 strand and the α2 helix (Hung & Sheng, 2002 ▶). In addition, the PDZ domains can form homo- and hetero-oligomers with other PDZ domains (Im, Lee et al., 2003 ▶; Im, Park et al., 2003 ▶).
Interestingly, recent reports have shown that ZO-1 and ZO-2 can form heterodimers and homodimers via the second PDZ domain (Itoh et al., 1999 ▶; Utepbergenov et al., 2006 ▶). The second PDZ domain of ZO-1 and ZO-2 can also interact with connexin 43, a well known gap-junction protein (Giepmans & Moolenaar, 1998 ▶; Singh & Lampe, 2003 ▶; Singh et al., 2005 ▶). However, ZO-2 PDZ2 has a lower affinity for the connexin 43 C-terminus compared with ZO-1 PDZ2 (Singh et al., 2005 ▶; Chen et al., 2008 ▶). For membrane-associated scaffold proteins, dimerization provides an opportunity to cluster the integral membrane proteins and signalling proteins. Deletion experiments (Utepbergenov et al., 2006 ▶) have proved that the dimerization interface of the second PDZ domain is different from the ‘β-finger’ of neuronal nitric oxide synthase-syntrophin (Hillier et al., 1999 ▶) and is likely to create a specific interface for the PDZ domains of Shank (Im, Lee et al., 2003 ▶) and GRIP1 (Im, Park et al., 2003 ▶). In order to obtain a deeper insight into the dimerization interface of the second PDZ domain of ZO proteins, we have solved the crystal structure of recombinant human ZO-2 PDZ2 domain at a resolution of 1.75 Å. The crystal structure reveals that the dimerization of human ZO-2 PDZ2 domain takes place via domain-swapping. Structural comparison with the ZO-1 PDZ2–ligand complex indicates that ZO-2 PDZ2 might have a similar pattern of interaction with the connexin 43 C-terminus. It also shows that structural differences may explain the different affinity for the connexin 43 C-terminus.
2. Materials and methods
2.1. Protein purification
ZO-2 PDZ2 domain (residues 306–385) was amplified by polymerase chain reaction technology using the appropriate primers and then inserted between the NdeI site and the XhoI site of pET22b (Novagen), which adds a C-terminal hexahistidine tag to the cloned gene. The recombinant plasmid was transformed into Escherichia coli BL21 (DE3) cells (Novagen) for large-scale protein production. The cells were cultured at 310 K in LB medium containing ampicillin (50 µg ml−1) until the OD600 reached 0.6–0.8. The cells were induced by adding 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After induction, the cells were grown at 289 K for 24 h and harvested by centrifugation at 8000g for 8 min at 281 K. The cell pellet was suspended in lysis buffer (20 mM Tris–HCl pH 8.0 containing 500 mM NaCl and 1 mM PMSF) and then lysed by sonication on ice. The lysate was centrifuged at 22 600g at 277 K for 20 min twice. The supernatant solution was dialyzed against binding buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl) three times for 2 h each. The dialyzed solution was centrifuged at 22 600g at 277 K for 20 min and then loaded onto a HisTrap column (GE Healthcare) equilibrated with binding buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl). The column was washed with ten column volumes of binding buffer and the column was then eluted with a linear gradient of imidazole from 20 to 175 mM using an ÄKTA Purifier instrument (GE Healthcare) in ten column volumes to remove impurities from the column. Finally, ZO-2 PDZ2 was eluted with elution buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl and 250 mM imidazole) in two column volumes. ZO-2 PDZ2 was highly expressed as a soluble protein in E. coli strain BL21 (DE3) cells and the majority of the target protein appeared in the eluted fraction. The fractions containing ZO-2 PDZ2 were pooled and desalted by ultrafiltration using Ultracel Amicon YM10 (Millipore). The protein was then concentrated to a final concentration of 10 mg ml−1 and stored in crystallization buffer (5 mM Tris–HCl pH 8.0). The protein concentration was determined by the Bradford method (Bio-Rad protein assay; Bradford, 1976 ▶) prior to crystallization trials; the His6 tag at the C-terminus was not removed. The purity was checked by SDS–PAGE and estimated to be over 95%.
2.2. Crystallization and data collection
Crystallization experiments were performed at constant temperature (283 K) using the hanging-drop vapour-diffusion method. The initial crystallization conditions were screened using Crystal Screens I and II (Hampton Research). Equal volumes (1 µl) of protein solution and crystallization reagent were mixed and the resulting drops were equilibrated against 350 µl crystallization reagent in the well. Similar procedures were used in the optimization process. Two conditions (10% PEG 6000, 100 mM HEPES pH 7.5, 5% MPD and 12% PEG 8000, 100 mM Tris–HCl pH 8.3) produced ZO-2 PDZ2-domain crystals belonging to space groups P322 and P1, respectively. Preliminary diffraction data were collected using an imaging plate (MAR Research dtb345), a rotating-anode X-ray source (Cu Kα; Rigaku Micro007) and a crystal-to-detector distance of 220 mm. Diffraction data were integrated and scaled using AUTOMAR (v.1.4; MAR Research). The processing statistics for the data from the two different crystal forms are summarized in Table 1 ▶.
Table 1. Data-collection and structure-refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | ||
| Space group | P322 | P1 |
| Unit-cell parameters (Å, °) | a = 66.03, b = 66.03, c = 46.72, α = β = 90, γ = 120 | a = 30.17, b = 41.32, c = 41.29, α = 80.05, β = 68.63, γ = 68.52 |
| Molecules per ASU | 1 | 2 |
| Resolution range (Å) | 20–2.20 | 20–1.75 (1.81–1.75) |
| Completeness (%) | 97.9 (94.9) | 91.5 (89.3) |
| I/σ(I) | 10.4 (1.6) | 9.3 (3.3) |
| Rmerge† (%) | 4.65 (28.11) | 4.92 (14.81) |
| Structure refinement | ||
| Resolution (Å) | 20–1.75 (1.81–1.75) | |
| Rcryst/Rfree‡ (%) | 20.6 (33.0)/23.9 (34.0) | |
| No. of protein atoms | 1241 | |
| No. of waters | 203 | |
| R.m.s.d. from ideal values | ||
| Bond lengths (Å) | 0.015 | |
| Bond angles (°) | 1.545 | |
| Average B factor (Å2) | 27.3 | |
| Ramachandran plot (%) | ||
| Most favoured regions (%) | 93.6 | |
| Allowed regions (%) | 6.4 |
R
merge = 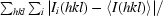
 , where I
i(hkl) is the intensity of the measured reflection and 〈I(hkl)〉 is the mean intensity of all symmetry-related reflections.
, where I
i(hkl) is the intensity of the measured reflection and 〈I(hkl)〉 is the mean intensity of all symmetry-related reflections.
R
cryst = 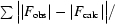
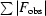 , where F
obs and F
calc are observed and calculated structure factors. R
free =
, where F
obs and F
calc are observed and calculated structure factors. R
free = 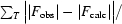
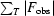 , where T is a test data set of about 10% of the total reflections that were randomly chosen and set aside prior to refinement.
, where T is a test data set of about 10% of the total reflections that were randomly chosen and set aside prior to refinement.
2.3. Structure determination and refinement
The crystal structure of ZO-2 PDZ2 domain with space group P322 was determined by molecular replacement with MOLREP (Vagin & Teplyakov, 1997 ▶) using the 13th PDZ domain of human MPDZ (PDB code 2fne; Elkins et al., 2007 ▶) as a search model. The initial model was input into the program Coot (Emsley & Cowtan, 2004 ▶) for manual revision and model extension was then performed using RESOLVE (Terwilliger, 2000 ▶). After the majority of amino acids had been added, the model with space group P322 was utilized as the search model for the crystal with space group P1 using the MOLREP program (Vagin & Teplyakov, 1997 ▶). Model refinement involved manual adjustment in Coot (Emsley & Cowtan, 2004 ▶) and rigid and restrained refinement in REFMAC (Murshudov et al., 1997 ▶). The final R cryst and R free factors were 20.6% and 23.9%, respectively. The stereochemical quality of the final model of the P1 space-group form was analyzed by PROCHECK (Holm & Sander, 1993 ▶) and the final refinement statistics and geometry are listed in Table 1 ▶.
3. Results and discussion
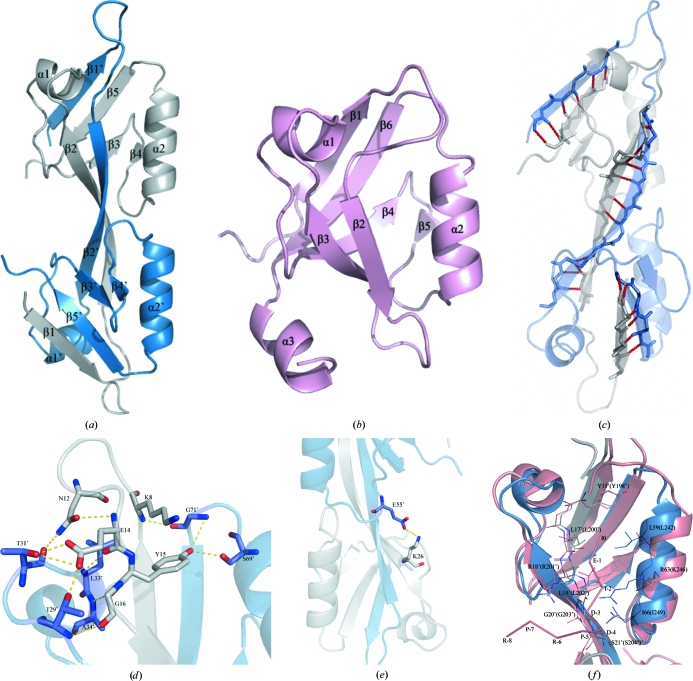
The ZO-2 PDZ2 domain produces a homodimer consisting of two identical molecules (molecules A and B). The secondary-structure elements of molecule B are indicated with a prime (Fig. 1 ▶ a). Both molecules are composed of two α-helices (α1 and α2) and five β-strands (β1–β5). Comparison with the prototype PDZ domain (Cabral et al., 1996 ▶; Fig. 1 ▶ b) reveals that the structural resemblance at the tertiary level is broken in the N-terminal β1 and β2 strands. In the prototype PDZ fold, the β2 strand is followed by a loop which induces the formation of an antiparallel arrangement with the β3 strand. In the corresponding region in the ZO-2 PDZ2 domain, the lack of a loop connecting the β2 and β3 strands leads to their replacement by an exceptionally long β2 strand, which makes the β1 and β2 strands protrude from the core PDZ fold. This protrusion contributes to a PDZ fold that is apparently incomplete. The incomplete PDZ fold from one molecule interacts with the β1, β2 and β5 strands from the other molecule. These three β-strands (residues 2–7, 17–28 and 72–77) in one monomer forms 24 pairs of main-chain hydrogen bonds and hydrophobic contacts with the NCS-related strands in the neighbouring monomer in an antiparallel fashion (Fig. 1 ▶ c). Hydrogen-bond interactions at the interface are shown in Figs. 1 ▶(c), 1 ▶(d) and 1 ▶(e). The total buried area spans 37.4% of the total monomeric surface area. [These data were obtained using the Protein–Protein Interaction Server (Jones & Thornton, 1996 ▶).] Size-exclusion chromatography and analytical ultracentrifugation also indicate that the ZO-2 PDZ2 domain is a dimer in solution (Wu et al., 2007 ▶). This high-resolution crystal structure of ZO-2 PDZ2 has the same topology as the previously reported NMR structure (PDB code 2osg; Wu et al., 2007 ▶). Comparison between the crystal structure and the solution structure of ZO-2 PDZ2 was performed in order to investigate the differences between the two structures. The results showed that the main differences are found in the loop regions and may arise from crystal packing.
Figure 1.
(a) A ribbon diagram of the domain-swapped dimeric structure. The secondary-structure elements are numbered in order of appearance in the primary structure. One monomer is represented in blue and the other monomer is in grey. (b) The fold of the third PDZ domain from the human DLG protein (PDB code 1pdr; Cabral et al., 1996 ▶), defining the topology of the prototype PDZ domain. (c) Cartoon representation of the interaction between the β1, β2 and β5 strands of the homodimer. The residues that interact via direct hydrogen bonds are shown in stick reprsentation. There are six hydrogen bonds between strands β1 and β5′, 12 hydrogen bonds between strands β2 and β2′ and six hydrogen bonds between strands β1′ and β5. The hydrogen bonds are labelled in red. (d, e) The interactions in the homodimer. Only relevant side chains are shown in this figure for clarity. The broken lines represent hydrogen bonds. (f) Superposition of the putative ligand-binding site of ZO-2 PDZ2 domain (blue and grey) and the corresponding residues that interact with ligands in the structure of the ZO-1 PDZ domain (salmon). The ligand of ZO-1 PDZ2 is also shown (salmon).
The PDZ domains bind to the short carboxy-terminus of their target proteins within a groove created between the β2 strand and the α2 helix (Cabral et al., 1996 ▶; Doyle et al., 1996 ▶). The analogous groove of ZO-2 PDZ2 is conserved in the domain-swapped homodimer. The domain swapping is certainly not an artifact of crystallization as it also occurs in ZO-1 PDZ2 structures (Fanning et al., 2007 ▶; Chen et al., 2008 ▶). In fact, the structure of the ZO-1 PDZ2–connexin 43 complex revealed that the swapped β-strands constitute an integral part of the two grooves required for connexin 43 binding (Chen et al., 2008 ▶). Fig. 1 ▶(f) presents the superposition of ZO-2 PDZ2 with the ZO-1 PDZ2–ligand structure. This reveals that there are two grooves located near the dimer interface of ZO-2 PDZ2. Each groove is framed between the β2′ strand and the α2 helix. Overall, the residues in the grooves are conserved in the second PDZ domain of ZO-1 and ZO-2, with the exception of Arg63, which is Lys246 in ZO-1 PDZ2. Given the interaction between ZO-2 PDZ2 and the C-terminus of connexin 43 (Singh et al., 2005 ▶), we were surprised to discover that the grooves displayed good potency for binding to the C-terminus of connexin 43. In ZO-2 PDZ2, Tyr15′, Leu17′, Leu19′ and Ile66 form a deep hydrophobic cavity. This hydrophobic cavity might accord with the isoleucine at position 0 of the connexin 43 C-terminus. The binding specificity of PDZ domains is determined by the interaction of the first residue of the α2 helix with the side chain of the −2 residue of the ligand. Similarily, the leucine residue at position −2 of connexin 43 might contribute to the specificity by hydrophobic interaction with Ile59, the first residue of the α2 helix. Of course, there may be hydrogen-bonding interactions between the backbones of ZO-2 PDZ2 and the connexin 43 C-terminus. It has been reported that ZO-2 PDZ2 binds to the connexin 43 C-terminus with a much weaker affinity compared with ZO-1 PDZ2 (Chen et al., 2008 ▶). Structural superposition shows that Arg63 in ZO-2 PDZ2 (Lys246 in ZO-1 PDZ2) and the sequence differences in the β2 strand may be responsible for the different affinity for binding to the connexin 43 C-terminus.
In summary, the crystal structure of ZO-2 PDZ2 reveals that domain swapping is necessary for the formation of the heterodimer of ZO proteins. A comparison between ZO-2 PDZ2 and the ZO-1 PDZ2–ligand complex shows that the domain-swapping dimer generates binding grooves that are distinct from the canonical binding grooves. This domain-swapped structure and the potential ligand-binding grooves may provide a structural basis for the polymerization of membrane proteins and intracellular signalling complexes in which the ZO proteins are localized.
Supplementary Material
PDB reference: second PDZ domain from human zonula occludens 2, 3e17, r3e17sf
Acknowledgments
We thank Professor Yunyu Shi and Dr Jiawen Wu for providing the plasmid for ZO-2 PDZ2. Financial support for this project was provided by research grants from the Chinese National Natural Science Foundation (grant Nos. 30121001, 30025012 and 30571066), the Chinese Ministry of Science and Technology (grant Nos. 2006CB806500, 2006CB910200 and 2006AA02A318), the Chinese Academy of Sciences (grant No. KSCX2-YW-R-60) and the Chinese Ministry of Education (grant No. 20070358025) to LN and MT.
References
- Anderson, J. M., Fanning, A. S., Lapierre, L. & Van Itallie, C. M. (1995). Biochem. Soc. Trans.23, 470–475. [DOI] [PubMed]
- Angst, B. D., Marcozzi, C. & Magee, A. I. (2001). J. Cell Sci.114, 625–626. [DOI] [PubMed]
- Bazzoni, G., Martinez-Estrada, O. M., Orsenigo, F., Cordenonsi, M., Citi, S. & Dejana, E. (2000). J. Biol. Chem.275, 20520–20526. [DOI] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed]
- Cabral, J. H. M., Petosa, C., Sutcliffe, M. J., Raza, S., Byron, O., Poy, F., Marfatia, S. M., Chishti, A. H. & Liddington, R. C. (1996). Nature (London), 382, 649–652. [DOI] [PubMed]
- Chen, J., Pan, L., Wei, Z., Zhao, Y. & Zhang, M. (2008). EMBO J.27, 2113–2123. [DOI] [PMC free article] [PubMed]
- Doyle, D. A., Lee, A., Lewis, J., Kim, E., Sheng, M. & MacKinnon, R. (1996). Cell, 85, 1067–1076. [DOI] [PubMed]
- Elkins, J. M., Papagrigoriou, E., Berridge, G., Yang, X., Phillips, C., Gileadi, C., Savitsky, P. & Doyle, D. A. (2007). Protein Sci.16, 683–694. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fanning, A. S., Jameson, B. J., Jesaitis, L. A. & Anderson, J. M. (1998). J. Biol. Chem.273, 29745–29753. [DOI] [PubMed]
- Fanning, A. S., Lye, M. F., Anderson, J. M. & Lavie, A. (2007). J. Biol. Chem.282, 37710–37716. [DOI] [PMC free article] [PubMed]
- Giepmans, B. N. & Moolenaar, W. H. (1998). Curr. Biol.8, 931–934. [DOI] [PubMed]
- Glaunsinger, B. A., Weiss, R. S., Lee, S. S. & Javier, R. (2001). EMBO J.20, 5578–5586. [DOI] [PMC free article] [PubMed]
- Haskins, J., Gu, L., Wittchen, E. S., Hibbard, J. & Stevenson, B. R. (1998). J. Cell Biol.141, 199–208. [DOI] [PMC free article] [PubMed]
- Hillier, B. J., Christopherson, K. S., Prehoda, K. E., Bredt, D. S. & Lim, W. A. (1999). Science, 284, 812–815. [PubMed]
- Holm, L. & Sander, C. (1993). J. Mol. Biol.233, 123–138. [DOI] [PubMed]
- Hung, A. Y. & Sheng, M. (2002). J. Biol. Chem.277, 5699–5702. [DOI] [PubMed]
- Hunter, A. W., Barker, R. J., Zhu, C. & Gourdie, R. G. (2005). Mol. Biol. Cell, 16, 5686–5698. [DOI] [PMC free article] [PubMed]
- Im, Y. J., Lee, J. H., Park, S. H., Park, S. J., Rho, S. H., Kang, G. B., Kim, E. & Eom, S. H. (2003). J. Biol. Chem.278, 48099–48104. [DOI] [PubMed]
- Im, Y. J., Park, S. H., Rho, S. H., Lee, J. H., Kang, G. B., Sheng, M., Kim, E. & Eom, S. H. (2003). J. Biol. Chem.278, 8501–8507. [DOI] [PubMed]
- Itoh, M., Morita, K. & Tsukita, S. (1999). J. Biol. Chem.274, 5981–5986. [DOI] [PubMed]
- Itoh, M., Nagafuchi, A., Moroi, S. & Tsukita, S. (1997). J. Cell Biol.138, 181–192. [DOI] [PMC free article] [PubMed]
- Jones, S. & Thornton, J. M. (1996). Proc. Natl Acad. Sci. USA, 93, 13–20.
- Kausalya, P. J., Phua, D. C. & Hunziker, W. (2004). Mol. Biol. Cell, 15, 5503–5515. [DOI] [PMC free article] [PubMed]
- Li, X., Olson, C., Lu, S., Kamasawa, N., Yasumura, T., Rash, J. E. & Nagy, J. I. (2004). Eur. J. Neurosci.19, 2132–2146. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Penes, M. C., Li, X. & Nagy, J. I. (2005). Eur. J. Neurosci.22, 404–418. [DOI] [PubMed]
- Sheng, M. & Sala, C. (2001). Annu. Rev. Neurosci.24, 1–29. [DOI] [PubMed]
- Singh, D. & Lampe, P. D. (2003). Cell Commun. Adhes.10, 215–220. [DOI] [PubMed]
- Singh, D., Solan, J. L., Taffet, S. M., Javier, R. & Lampe, P. D. (2005). J. Biol. Chem.280, 30416–30421. [DOI] [PMC free article] [PubMed]
- Songyang, Z., Fanning, A. S., Fu, C., Xu, J., Marfatia, S. M., Chishti, A. H., Crompton, A., Chan, A. C., Anderson, J. M. & Cantley, L. C. (1997). Science, 275, 73–77. [DOI] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Traweger, A., Fuchs, R., Krizbai, I. A., Weiger, T. M., Bauer, H. C. & Bauer, H. (2003). J. Biol. Chem.278, 2692–2700. [DOI] [PubMed]
- Utepbergenov, D. I., Fanning, A. S. & Anderson, J. M. (2006). J. Biol. Chem.281, 24671–24677. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Wittchen, E. S., Haskins, J. & Stevenson, B. R. (1999). J. Biol. Chem.274, 35179–35185. [DOI] [PubMed]
- Wu, J., Yang, Y., Zhang, J., Ji, P., Du, W., Jiang, P., Xie, D., Huang, H., Wu, M., Zhang, G. & Shi, Y. (2007). J. Biol. Chem.282, 35988–35999. [DOI] [PubMed]
- Zahraoui, A., Louvard, D. & Galli, T. (2000). J. Cell Biol.151, F31–F36. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: second PDZ domain from human zonula occludens 2, 3e17, r3e17sf
PDB reference: second PDZ domain from human zonula occludens 2, 3e17, r3e17sf