The haem oxygenase from H. pylori (HugZ) was overexpressed, purified, reconstituted with haem and crystallized. Selenomethionine-derivatized crystals diffracted X-rays to 1.8 Å resolution.
Keywords: haem oxygenases, Helicobacter pylori, HugZ, iron acquisition, pathogenesis
Abstract
The haem oxygenase HugZ is part of the iron-acquisition mechanism that is essential for the adaptive colonization of Helicobacter pylori, a major pathogen of gastroenteric diseases. The HugZ–haemin complex has been purified and crystallized and diffraction data sets have been collected to 1.8 Å resolution. The HugZ–haemin complex crystals belonged to space group P212121, with unit-cell parameters a = 88.40, b = 139.37, c = 152.97 Å.
1. Introduction
Iron is absolutely required for almost all life forms. It is incorporated, either alone or as part of iron–sulfur clusters or haem, into a wide range of proteins that are involved in many important processes. Most bacteria require a micromolar level of iron for survival, but the element is not usually biologically available to them (Wandersman & Delepelaire, 2004 ▶). Iron usually exists in one of two redox states: the Fe2+ (ferrous) or Fe3+ (ferric) forms. In aerobic inorganic environments, iron predominantly exists in the insoluble (approximate solubility of 10−18 M at pH 7) ferric form. In living organisms, free iron is maintained at very low concentrations by sequestration into carrier molecules such as lactoferrin, transferrin and ferritins or by binding to porphyrin in order to restrict microbe growth and to minimize the toxicity caused by the Fenton reaction, in which ferrous ions catalyzes the formation of hydroxyl radicals and superoxide anions, species that are highly deleterious to biomacromolecules. To overcome the iron shortage, bacteria have developed several acquisition mechanisms to import iron in various forms (Wandersman & Delepelaire, 2004 ▶).
It has been estimated that 95% of the iron in mammalian hosts is complexed in haemoproteins, primarily haemoglobin (Otto et al., 1992 ▶). Haemoproteins are thus the most important source of iron for pathogenic bacteria. For bacteria to acquire iron from haemoproteins, the haem must be removed from haemoproteins, transported into bacterial cells and degraded to release the iron (Wilks & Burkhard, 2007 ▶). While it has long been known that in eukaryotic cells haem oxygenases (HOs), in conjunction with aerobic electron donors, carry out the degradation of haem through oxidative cleavage, bacterial HOs have only recently been identified from several Gram-negative bacteria (Frankenberg-Dinkel, 2004 ▶). Most of the structurally characterized bacterial HOs are very similar to their eukaryotic counterparts in terms of molecular structure, even though their sequence homologies are low (Unno et al., 2007 ▶).
Recently, two HOs from pathogenic bacteria, Cj1613c from Campylobacter jejuni (Ridley et al., 2006 ▶) and HugZ from Helicobacter pylori (Guo et al., 2008 ▶), have been identified. These proteins were required for haem-iron acquisition of the respective bacteria. HugZ was further shown to be required for the adaptive colonization of H. pylori in hosts. With a sequence identity of 56% these two HOs are very similar to each other, but they do not have any sequence homology to canonical HOs. However, Cj1613c and HugZ do have weak sequence similarities to FMN-binding proteins with split-barrel folds. Studies on the solution behaviour of HugZ indicated that this protein exists as dimers, which is in agreement with the possibility that HugZ may adopt a split-barrel fold, since all proteins with this fold are dimers; this is distinct from canonical HOs, which are invariably monomers. HugZ has also been shown to have a unique δ-meso regiospecificity in the ring-opening reaction of haem (Guo et al., 2008 ▶), whereas canonical HOs are almost exclusively α-meso-specific. These experimental data indicate that HugZ is a unique HO and that its molecular structure and enzymatic mechanism are distinct from those of characterized HOs.
In order to better understand the structure–function relationship of this novel HO, we have embarked on crystallographic analysis of HugZ. We have purified HugZ to high homogeneity and have crystallized the protein in its complexed form with haemin. Here, we report the crystallization, diffraction data collection and preliminary crystallographic studies of the HugZ–haemin complex.
2. Materials and methods
2.1. Expression and purification of His-tagged HugZ
The pET-22b plasmid containing the gene for HugZ was constructed as described previously (Guo et al., 2008 ▶). The HugZ C-terminus was fused to a His6 tag preceded by Leu and Glu in order to facilitate purification. The plasmid was transformed into Escherichia coli BL21(DE3) strain competent cells, which were then grown on an LB plate supplemented with 100 µg ml−1 ampicillin at 310 K overnight. A 50 ml inoculum in LB media was prepared from a single colony of transformed cells. 800 ml LB media was inoculated with 8 ml prepared culture and incubated until the OD600 of the culture reached 0.8. Isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 10 µM to induce the expression of recombinant HugZ. The bacteria were grown at 295 K for an additional 12 h, after which the cells were harvested by centrifugation (Hitachi, Himac CRT, R5S2 rotor) at 4000 rev min−1 for 30 min and stored at 253 K until use. The LB medium used contained 100 µg ml−1 ampicillin.
After being thawed on ice, the cell pellets were suspended in 30 ml lysis buffer (50 mM phosphate buffer pH 8.0, 300 mM NaCl and 10 mM imidazole) supplemented with 1 mM phenylmethylsulfonyl fluoride. Lysozyme (BBI) was added to the suspension to a final concentration of 0.5 mg ml−1 and the mixture was then stirred on ice for half an hour. The cells were then lysed by sonication and centrifuged at 16 000 rev min−1 for 30 min (Sigma 3K30, 12150 rotor). The resulting supernatant was applied onto a nickel–nitriloacetic acid resin column (Novagen) previously equilibrated with lysis buffer. The column was washed with ten column volumes of lysis buffer followed by five column volumes of wash buffer (50 mM phosphate buffer pH 8.0, 300 mM NaCl, 20 mM imidazole). The His-tagged HugZ was eluted with elution buffer (50 mM phosphate buffer pH 8.0, 300 mM NaCl, 250 mM imidazole). The purity of the eluate was evaluated by SDS–PAGE to be >98% HugZ. The eluate was concentrated by ultrafiltration (Amicon Ultra-15) and loaded onto four HiTrap Desalting columns (GE Healthcare) connected in tandem and pre-equilibrated with 50 mM Tris pH 8.0 buffer containing 150 mM NaCl. The protein fractions were pooled and stored at 277 K and the concentration was determined via absorbance at 280 nm, assuming a molar extinction coefficient of 13 535 M −1 cm−1.
The selenomethionine-substituted derivative of HugZ was obtained by expression of HugZ in the methionine-metabolism defective E. coli strain B834(DE3). Transformed cells were first grown in LB medium containing 100 µg ml−1 ampicillin until the OD600 of the culture reached 0.8. The cells were harvested by low-speed centrifugation, resuspended in minimal medium containing 100 µg ml−1 ampicillin and incubated for 2 h. Selenomethionine (SeMet) was added to a final concentration of 50 µg ml−1. The induction and purification of SeMet-derivatized HugZ were performed as described above for the native protein.
2.2. Reconstitution of HugZ with haemin
The HugZ–haemin complex was prepared as described previously (Wilks & Schmitt, 1998 ▶). A small volume of haemin (5 mM in DMSO) was added to purified HugZ solution to a final haemin:HugZ molar ratio of 2:1. The mixture was incubated at 277 K overnight with shaking. After centrifugation, the soluble fraction was concentrated to 1 ml and applied onto a HiLoad 16/60 Superdex 200 prep-grade gel-filtration column (GE Healthcare) pre-equilibrated with 20 mM Tris pH 8.0 buffer containing 5 buffer containing 5 mM sodium azide. The peak fractions were collected and assayed by SDS–PAGE. The majority of the HugZ–haemin eluted as dimers (59 kDa; monomer 29.5 kDa) based on retention time. The fractions containing pure dimeric HugZ–haemin complex were pooled and concentrated by ultrafiltration for crystallization. The concentration of the HugZ–haemin complex was determined from the absorbance at the Soret spectrum peak (412 nm), assuming an extinction efficient of 139.6 mM −1 cm−1.
2.3. Crystallization of the HugZ–haemin complex
The concentration of the HugZ–haemin complex used in crystallization was approximately 27.5 mg ml−1. Crystallization experiments were set up via the hanging-drop vapour-diffusion method by mixing equal volumes of protein and reservoir solutions and suspending the drop over 0.5 ml reservoir solution at 293 K. The crystallization condition was optimized by varying the type and the concentration of precipitant (PEG), salts, buffers and organic compounds and the pH. The optimization of the crystallization conditions for SeMet-derivatized protein was conducted on the basis of that for the native protein with minor modifications.
2.4. Data collection and processing
All diffraction data sets were collected on beamline 17A of the Photon Factory (KEK, Japan) using an ADSC Quantum 270 CCD detector. For cryoprotection, crystals were soaked in reservoir solution containing ethylene glycol [the concentration was increased to 20%(v/v) stepwise] for 1 min before being mounted on nylon cryoloops and flash-frozen in a liquid-nitrogen stream at 95 K. A high-resolution data set was collected from a selenomethionine-derivatized crystal at 1.00000 Å wavelength with a crystal-to-detector distance of 244 mm and 1° oscillation per frame. The exposure time for each frame was 4 s and the total oscillation range was 180°. Multiple-wavelength anomalous diffraction (MAD; Taylor, 2003 ▶) data sets were collected from the same crystal at three wavelengths: 0.97912 Å for the peak, 0.97931 Å for the inflection point and 0.96408 Å for the high-energy remote, and with the data-collection conditions 300 mm crystal-to-detector distance, 1° oscillation per frame, 3 s exposure time per frame and a 240° oscillation range for the peak data set and a 120° oscillation range for the inflection-point and remote data sets. MOSFLM (v.7.0.4; Leslie, 2006 ▶) and SCALA (v.6.0) from the CCP4 program suite (v.6.0.2; Collaborative Computational Project, Number 4, 1994 ▶) were used for the indexing, integration and scaling of the diffraction data sets.
3. Results
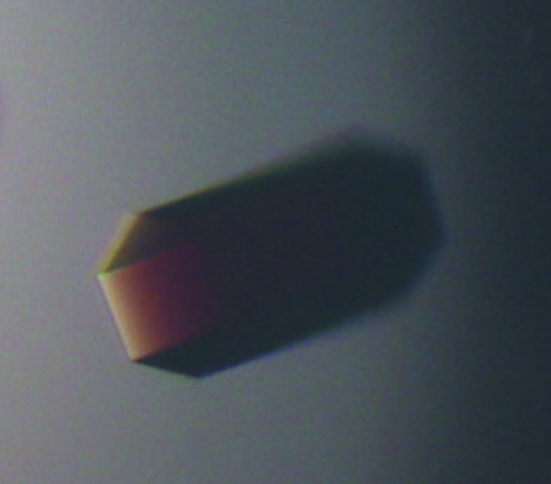
During the initial crystallization-condition screening experiments, thin hexagonal plate-shaped crystals were observed after 3 d. As a result of optimization, crystals with hexagonal prismatic shape that were suitable for high-resolution diffraction data collection were obtained using the hanging-drop vapour-diffusion method by mixing 1 µl 25 mg ml−1 HugZ–haemin solution (in 20 mM Tris pH 8.0, 5 mM sodium azide) and 1 µl reservoir solution consisting of 16%(w/v) PEG 3350, 3% Tacsimate, 0.1 M imidazole pH 7.5, 12% methanol. High-quality selenomethionine-derivative crystals (Fig. 1 ▶) were also obtained by the same method with reservoir solution consisting of 14% PEG 3350, 2% Tacsimate, 0.1 M imidazole pH 7.5, 0.1 M sodium cacodylate pH 6.5 and 12% methanol.
Figure 1.
Crystal of SeMet-derivatized HugZ–haemin complex (0.5 × 0.1 × 0.1 mm).
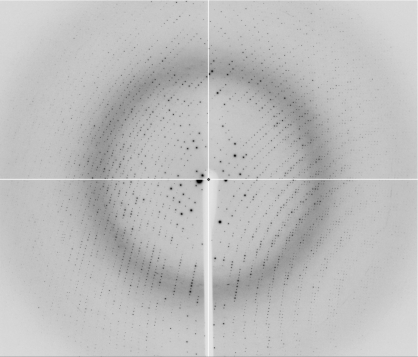
The data-collection statistics for the SeMet-derivatized crystal are given in Table 1 ▶. The crystal diffracted to a resolution of 1.8 Å (Fig. 2 ▶) and belonged to space group P212121, with unit-cell parameters a = 88.40, b = 139.37, c = 152.97 Å. The asymmetric unit is estimated to contain six HugZ–haemin complexes, with a corresponding Matthews coefficient of 2.66 Å3 Da−1 and a solvent content of 53.75%.
Table 1. Data statistics for the SeMet-derivatized HugZ–haemin crystal.
Values in parentheses are for the highest resolution shell.
| MAD | ||||
|---|---|---|---|---|
| High resolution | Inflection | Peak | Remote | |
| Resolution (Å) | 47.89–1.80 (1.90–1.80) | 57.83–2.32 (2.45–2.32) | 57.83–2.32 (2.45–2.32) | 57.83–2.32 (2.45–2.32) |
| Wavelength (Å) | 1.00000 | 0.97931 | 0.97912 | 0.96408 |
| No. of unique reflections | 165702 | 82283 | 82219 | 82233 |
| Completeness (%) | 94.9 (72.8) | 100 (99.9) | 100 (100) | 99.9 (99.9) |
| Redundancy | 6.7 (4.5) | 4.9 (4.9) | 9.9 (9.7) | 4.9 (4.9) |
| Average I/σ(I) | 8.6 (4.3) | 9.3 (5.7) | 7.8 (5.2) | 9.0 (5.3) |
| Rmerge† (%) | 4.9 (17.2) | 5.0 (11.5) | 6.0 (12.7) | 5.2 (12.7) |
| Rp.i.m.‡ (%) | 2.1 (10) | 3.0 (6.5) | 2.4 (4.8) | 3.0 (7.0) |
R
merge = 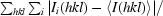
 , where Ii(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is its average.
, where Ii(hkl) is the ith intensity measurement of reflection hkl and 〈I(hkl)〉 is its average.
R
p.i.m. = 
 , where Ii(hkl) is the ith intensity measurement of reflection hkl, N is the multiplicity of reflection hkl and 〈I(hkl)〉 is its average
, where Ii(hkl) is the ith intensity measurement of reflection hkl, N is the multiplicity of reflection hkl and 〈I(hkl)〉 is its average
Figure 2.
Diffraction pattern of the SeMet-derivatized HugZ–haemin crystal.
The high quality of the data that we have collected has laid a solid foundation for our future structure determination of the HugZ–haemin complex. Attempts to solve the structure of the HugZ–haemin complex by the MAD method using the 2.3 Å resolution data sets are in progress.
Acknowledgments
The diffraction data sets were collected on beamline 17A of KEK, Photon Factory, Tsukuba, Japan. We are grateful for the support from KEK (08G007). This work was supported by funding from the National Science Foundation of China (30400019), the National 973 Program (2006CB806502) and the National 863 Program (2006AA02A322).
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Frankenberg-Dinkel, N. (2004). Antioxid. Redox Signal.6, 825–834. [DOI] [PubMed]
- Guo, Y., Guo, G., Mao, X., Zhang, W., Xiao, J., Tong, W., Liu, T., Xiao, B., Liu, X., Feng, Y. & Zou, Q. (2008). BMC Microbiol.8, 226. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- Otto, B. R., Verweijvanvught, A. M. J. J. & Maclaren, D. M. (1992). Crit. Rev. Microbiol.18, 217–233. [DOI] [PubMed]
- Ridley, K. A., Rock, J. D., Li, Y. & Ketley, J. M. (2006). J. Bacteriol.188, 7862–7875. [DOI] [PMC free article] [PubMed]
- Taylor, G. (2003). Acta Cryst. D59, 1881–1890. [DOI] [PubMed]
- Unno, M., Matsui, T. & Ikeda-Saito, M. (2007). Nat. Prod. Rep.24, 553–570. [DOI] [PubMed]
- Wandersman, C. & Delepelaire, P. (2004). Annu. Rev. Microbiol.58, 611–647. [DOI] [PubMed]
- Wilks, A. & Burkhard, K. A. (2007). Nat. Prod. Rep.24, 511–522. [DOI] [PubMed]
- Wilks, A. & Schmitt, M. P. (1998). J. Biol. Chem.273, 837–841. [DOI] [PubMed]