Perdeuterated type III antifreeze protein has been expressed, purified and crystallized. Preliminary neutron data collection showed diffraction to 1.85 Å resolution from a 0.13 mm3 crystal.
Keywords: type III antifreeze proteins, neutron diffraction, perdeuteration
Abstract
The highly homologous type III antifreeze protein (AFP) subfamily share the capability to inhibit ice growth at subzero temperatures. Extensive studies by X-ray crystallography have been conducted, mostly on AFPs from polar fishes. Although interactions between a defined flat ice-binding surface and a particular lattice plane of an ice crystal have now been identified, the fine structural features underlying the antifreeze mechanism still remain unclear owing to the intrinsic difficulty in identifying H atoms using X-ray diffraction data alone. Here, successful perdeuteration (i.e. complete deuteration) for neutron crystallographic studies of the North Atlantic ocean pout (Macrozoarces americanus) AFP in Escherichia coli high-density cell cultures is reported. The perdeuterated protein (AFP D) was expressed in inclusion bodies, refolded in deuterated buffer and purified by cation-exchange chromatography. Well shaped perdeuterated AFP D crystals have been grown in D2O by the sitting-drop method. Preliminary neutron Laue diffraction at 293 K using LADI-III at ILL showed that with a few exposures of 24 h a very low background and clear small spots up to a resolution of 1.85 Å were obtained using a ‘radically small’ perdeuterated AFP D crystal of dimensions 0.70 × 0.55 × 0.35 mm, corresponding to a volume of 0.13 mm3.
1. Introduction
Antifreeze proteins (AFPs), which were first discovered in the blood of polar fishes (DeVries, 1983 ▶), allow many other living organisms, including arthropods (Duman, 1982 ▶), bacteria, fungi (Olsen et al., 1998 ▶) and plants (Urrutia et al., 1992 ▶; Griffith et al., 1992 ▶), to survive subzero temperatures. In polar fishes, five types of these proteins have been biochemically characterized. These include antifreeze glycoproteins (AFGPs) and AFP types I, II, III and IV (for reviews, see Davies & Sykes, 1997 ▶; Venketesh & Dayananda, 2008 ▶).
There is a strong commercial interest in AFPs owing to their potential applications, which range from simply increasing the freeze tolerance of crop plants and extending the harvest season in cooler climates to improving cryosurgery (Feeney et al., 1996 ▶; Fletcher et al., 1999 ▶; Zhang et al., 2007 ▶). With a neutron crystallographic study, we therefore hope to contribute to a better understanding of the antifreezing mechanism and support the engineering of medical and other applications of AFPs.
The three-dimensional structure of type III AFP, a small globular protein, was first determined by NMR (Sonnichsen et al., 1993 ▶), followed by X-ray crystallography (Jia et al., 1996 ▶; 1.25 Å resolution at 277 K). Steric mutations (DeLuca et al., 1998 ▶) and further crystal structures (Yang et al., 1998 ▶; Graether et al., 1999 ▶; Antson et al., 2001 ▶) have completed the initial model of a flat ice-binding surface (IBS) that makes hydrogen bonds with ice, with additional interactions based on hydrophobic side chains that might play an important role in ice binding. The human genome sequencing project has identified a C-terminal domain (Baardsnes et al., 2001 ▶) in sialic acid synthase (SAS) which is homologous to the type III AFPs (International Human Genome Sequencing Consortium, 2001 ▶).
Recently, the ultrahigh-resolution X-ray crystal structure (Ko et al., 2003 ▶; 0.62 Å resolution at 110 K) of AFP from the antarctic eel pout Lycodichthys dearborni has provided interesting fine-scale features regarding the microheterogeneity of the protein and the solvent structure: the flat and mostly neutral IBS interacts with ice through one disordered peptide bond and four side chains modelled with dual conformation together with several water molecules that are split into pairs of very close locations.
Despite the many extensive X-ray structural studies that have been carried out to date, additional work is required both at the residue-protonation and hydration levels to completely elucidate the antifreeze mechanism. To conclusively answer these questions, we performed neutron diffraction at 293 K on crystals of perdeuterated antifreeze protein (AFP D). As these crystals could not be grown to the usual volume for neutron diffraction (>1 mm3), perdeuteration (Shu et al., 2000 ▶) is necessary as it improves the diffraction power and decreases the incoherent scattering background. This was highlighted by our previous experience with human aldose reductase (h-AR) on LADI-I. Using a D2O-soaked crystal of the hydrogenated protein (0.15 mm3 volume) we achieved a resolution of only 4.5 Å, whereas a crystal of equivalent volume of the perdeuterated protein diffracted to a useful resolution of 2.2 Å, enabling the location of most H atoms (as deuterium; Hazemann et al., 2005 ▶; Blakeley, Ruiz et al., 2008 ▶). Here, we report the perdeuteration protocol and the crystallization in D2O of AFP D from the North Atlantic ocean pout Macrozoarces americanus. Preliminary neutron Laue diffraction at 293 K on LADI-III clearly showed the capability of these AFP D (MW = 7 kDa) crystals to achieve a high resolution of around 1.85 Å with a ‘radically small’ crystal volume of 0.13 mm3.
2. Experimental methods
2.1. Expression and purification of perdeuterated AFP
The synthetic gene of the type III antifreeze protein AFP (isoform HPLC12) from the North Atlantic ocean pout M. americanus was subcloned into a pET-28a (Novagen) vector that confers kanamycin resistance. The sequence was then adjusted to the protein sequence of PDB entry 1hg7. Recombinant AFP D was obtained by expression in Escherichia coli BL21 (DE3) at the ILL–EMBL Deuteration Facility in Grenoble, France. Cells were grown in deuterated minimal medium (Meilleur et al., 2004 ▶; Artero et al., 2005 ▶; Hazemann et al., 2005 ▶; Di Costanzo et al., 2007 ▶; Wood et al., 2008 ▶) containing 25 mg l−1 kanamycin. The pD value of the deuterated buffers was measured using a H2O-calibrated pH meter. The conversion of pH to pD was accomplished by adding a constant of 0.4 (Kręzel & Bal, 2004 ▶). For the preparation of fully deuterated medium, mineral salts were dried out in a rotary evaporator (Heidolph) at 333 K and labile protons were exchanged for deuterons by dissolving in a minimal volume of D2O and re-dried. Perdeuterated d8-glycerol (Eurisotop, France) was used as a carbon source. The adaptation of BL21 (DE3) cells to deuterated minimal medium was achieved by an adaptation process on minimal medium agar plates (Artero et al., 2005 ▶). 1 l deuterated medium was inoculated with a 100 ml preculture of adapted cells in a 3 l fermenter (Labfors, Infors). During the batch and fed-batch phases, the pH was adjusted to 6.9 by addition of NaOD (Eurisotop, France) and the temperature was adjusted to 303 K. The gas-flow rate of sterile filtered air was 0.5 l min−1. Stirring was adjusted to ensure a dissolved oxygen tension (DOT) of 30%. The fed-batch phase was initiated when the optical density at 600 nm reached a value of 4. d8-Glycerol was added to the culture to keep the growth rate stable during fermentation. When the OD600 reached a value of 12, antifreeze overexpression was induced by the addition of 1 mM IPTG and incubation was continued for 20 h. Cells were then harvested and stored at 193 K. 10 g frozen cell paste was resuspended in 10 ml 8 M guanidine hydrochloride in D2O, stirred on ice for 1 h and sonicated to increase the yield of soluble proteins. After centrifugation at 19 000 rev min−1 for 20 min at 277 K, the supernatant containing the soluble proteins was added dropwise to 50 ml ice-cold refolding buffer (50 mM K2PO4, 100 mM NaCl in D2O pD 11.1). The AFP solution was then dialysed against 300 ml 50 mM sodium acetate pH 3.7. Insoluble material was removed by centrifugation and the supernatant was filtered through a 0.2 µm membrane. The pD of the protein solution was lowered to 3.1 using citric acid and the antifreeze protein was purified on a 5 ml SP Sepharose column (GE Healthcare). This step removes contaminating proteins; AFP D is recovered in the flowthrough. Protein purity was assessed by SDS–PAGE. AFP D was concentrated on 5K Amicon Ultra centrifugal filter units (Millipore) to a final concentration of 10 mg ml−1 in 50 mM sodium acetate pD 5.2.
2.2. Determination of deuteration level
The molecular weight of AFP D and its deuteration level were determined by MALDI mass spectrometry. Laser desorption/ionization mass-spectrometric analysis was performed with a Perseptive Biosystems (Framingham, Massachusetts, USA) Voyager Elite XL time-of-flight mass spectrometer operating with a pulsed nitrogen laser at 337 nm. External calibration was performed using the calibration mixture 3 standards from the Applied Biosystems Corporation with m/z values of 5734.59, 11 674.48 and 16 952.56 Da. Samples of concentrated native AFP diluted in 0.1% trifluoroacetic acid (Sigma; final concentration of about 10 mM) were mixed with an equal volume of a saturated solution of sinapinic acid (Fluka) prepared in a 50%(v/v) solution of acetonitrile/0.3% aqueous trifluoroacetic acid directly on the stainless-steel sample plate and air-dried prior to analysis.
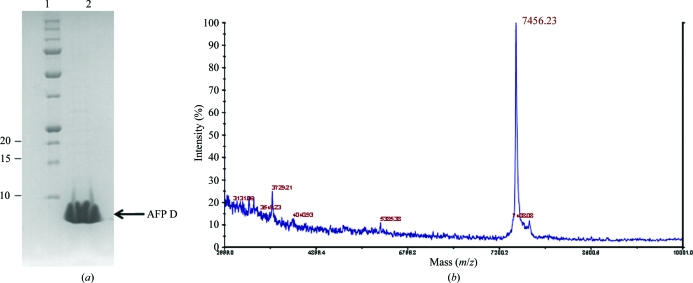
The values expressed are average mass and correspond to the [M + H]+ ion. For technical reasons, the deuteration level of AFP was determined in hydrogenated solutions. The measured molecular weight obtained (7456 Da; Fig. 1 ▶ b) when deuterated AFP was refolded in H2O buffer is very close to the theoretical value (7452 Da). This value takes into account the 115 exchangeable H atoms, indicating that more than 99% of the 418 non-exchangeable (carbon-bound) H atoms were deuterated. When deuterated AFP was refolded in deuterated buffer instead of hydrogenated buffer, a small increase in molecular weight (25 Da) was observed. This indicates that some of the 115 potentially exchangeable deuterons (oxygen- or nitrogen-bound) were trapped in the AFP structure and did not exchange during the mass-spectrometric analysis. To avoid trapping H atoms during the preparation of deuterated AFP, all denaturation and renaturation steps for preparative purposes were consequently carried out in deuterated solutions. We conclude that AFP expressed under the conditions described above is completely deuterium-labelled (perdeuterated).
Figure 1.
(a) 12% SDS Tris–tricine polyacrylamide gel electrophoresis. The standard protein bands of Precision Plus prestained molecular-weight standards (Bio-Rad) are indicated in lane 1. 28 µg purified AFP D is shown in lane 2. (b) MALDI mass spectrum of perdeuterated AFP prepared in H2O.
2.3. Crystallization
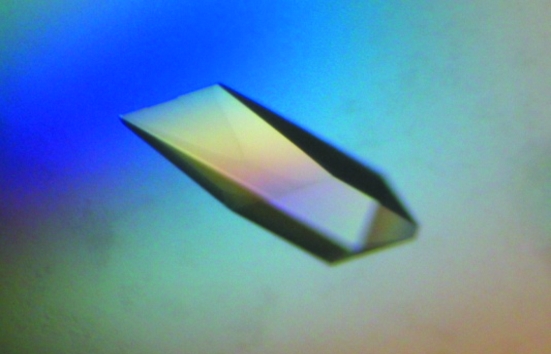
AFP D was concentrated to ∼9.6 mg ml−1 in 50 mM sodium acetate pD 5.2. All crystallization experiments were carried out by the sitting-drop vapour-diffusion method in 24-well sitting-drop Cryschem plates (Hampton Research). The 50 µl sitting drop was prepared at 285 K using 16 µl protein sample and 34 µl reservoir solution and was equilibrated against 800 µl reservoir solution (2.2 M ammonium sulfate, 9% d8-glycerol, 50 mM sodium acetate pD 5.2, temperature 285 K). Under these conditions, rectangular prismatic crystals appeared after three weeks and grew to maximum dimensions of 0.35 × 0.55 × 0.7 mm, corresponding to a crystal volume of 0.13 mm3 (see Fig. 2 ▶).
Figure 2.
Picture of the largest perdeuterated AFP crystal taken under polarized light. The crystal has dimensions 0.70 × 0.55 × 0.35 mm and a volume of 0.13 mm3. The protein crystallizes in space group P212121, with unit-cell parameters a = 32.5, b = 39.4, c = 45.3 Å at 293 K.
The crystallization conditions of AFP D in D2O were adapted from those of AFP H in aqueous solution by a grid search for the optimal conditions for obtaining large crystals. The differences between the AFP D and AFP H crystallization conditions were as follows: (i) temperature of 285 K versus 295 K, (ii) pD 5.2 (pH 4.8) versus pH 4.5, (iii) 50 mM versus 20 mM sodium acetate concentration in the reservoir and (iv) 1.5 M versus 1.1 M initial concentration of ammonium sulfate in the drop.
2.4. Preliminary neutron Laue diffraction
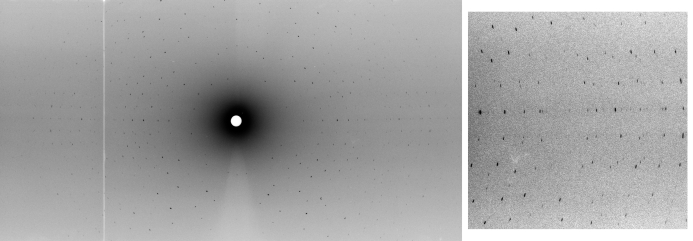
Preliminary tests of neutron Laue diffraction on the new Laue diffractometer LADI-III showed that the crystal of volume 0.13 mm3 (Fig. 2 ▶) diffracted to 1.85 Å resolution with 24 h exposures (Fig. 3 ▶). Preliminary data treatment showed an overall R merge factor of 14%. Details of the complete neutron data collection will be published subsequently.
Figure 3.
Left: a neutron Laue full diffraction pattern from the 0.13 mm3 perdeuterated AFP crystal using LADI-III and a 24 h exposure time. Right: close-up of a high-resolution region.
3. Conclusions
This lowering of the perdeuterated crystalline sample volume required to obtain a usable neutron diffraction data set is a significant advance which confirms our previous experience with fully deuterated human aldose reductase (h-AR D; Blakeley, Ruiz et al., 2008 ▶). Of these two cases of perdeuterated proteins with small crystal volumes measured to date on the LADI-I (h-AR D) and LADI-III (AFP D) instruments, the antifreeze crystals are 15% smaller (0.13 mm3 versus 0.15 mm3) and diffract to higher resolution (1.85 Å versus 2.2 Å) than the h-aldose reductase crystals. These improvements are possibly a consequence of the AFP D crystals having a 4.5 times better ratio of crystal volume to asymmetric unit volume (Blum et al., 2007 ▶) and the use of LADI-III versus LADI-I. Thus, the use of perdeuterated samples and the improvement of the instrumentation have made neutron protein crystallography more accessible to the structural biology community by extending the size and complexity of the biochemical systems that can be studied (for recent reviews, see Niimura & Bau, 2008 ▶; Blakeley, Langan et al., 2008 ▶).
Acknowledgments
We thank Bernard Dublet and Eric Forest (Institut de Biologie Structurale, IBS, Grenoble, France) for the mass-spectrometric analysis, Susana Teixeira and Jean-Baptiste Artero from the ILL for technical advice and François Dauvergne for technical support on LADI-III. This work was supported by the Human Frontiers Science Program grant RGP0021/2006-C and benefited from the activities of the DLAB consortium funded by the European Union under contract HPRI-2001-50065 and from United Kingdom Engineering and Physical Sciences Research Council (EPSRC) funded activity within the ILL–EMBL Deuteration Laboratory under grant GR/R99393/01. EIH is a member of the ‘Carrera del Investigador’ Conicet-Argentina. This work was supported by the Centre National de la Recherche Scientifique (CNRS), by the Institut National de la Santé et de la Recherche Médicale and the Hôpital Universitaire de Strasbourg (HUS).
References
- Antson, A. A., Smith, D. J., Roper, D. I., Lewis, S., Caves, L. S., Verma, C. S., Buckley, S. L., Lillford, P. J. & Hubbard, R. E. (2001). J. Mol. Biol.305, 875–879. [DOI] [PubMed]
- Artero, J.-B., Härtlein, M., McSweeney, S. & Timmins, P. (2005). Acta Cryst. D61, 1541–1549. [DOI] [PubMed]
- Baardsnes, J., Jelokhani-Niaraki, M., Kondejewski, L. H., Kuiper, M. J., Kay, C. M., Hodges, R. S. & Davies, P. L. (2001). Protein Sci.10, 2566–2576. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Langan, P., Niimura, N. & Podjarny, A. (2008). Curr. Opin. Struct. Biol.18, 1–8. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Ruiz, F., Cachau, R., Hazemann, I., Meilleur, F., Mitschler, A., Ginell, S., Afonine, P., Ventura, O. N., Cousido-Siah, A., Haertlein, M., Joachimiak, A., Myles, D. & Podjarny, A. (2008). Proc. Natl. Acad. Sci USA, 105, 1844–1848. [DOI] [PMC free article] [PubMed]
- Blum, M.-M., Koglin, A., Rüterjans, H., Schoenborn, B., Langan, P. & Chen, J. C.-H. (2007). Acta Cryst. F63, 42–45. [DOI] [PMC free article] [PubMed]
- Davies, P. L. & Sykes, B. D. (1997). Curr. Opin. Struct. Biol.7, 828–834. [DOI] [PubMed]
- DeLuca, C. I., Davies, P. L., Ye, Q. & Jia, Z. (1998). J. Mol. Biol.275, 515–525. [DOI] [PubMed]
- DeVries, A. C. (1983). Annu. Rev. Physiol.45, 245–260. [DOI] [PubMed]
- Di Costanzo, L., Moulin, M., Haertlein, M., Meilleur, F. & Christianson, D. W. (2007). Arch. Biochem. Biophys.465, 82–89. [DOI] [PMC free article] [PubMed]
- Duman, J. G. (1982). Cryobiology, 19, 613–627. [DOI] [PubMed]
- Feeney, R. E., Osuga, D. T. & Yeh, Y. (1996). Agric. Food Chem.3, 155–174.
- Fletcher, G. L., Goddard, S. V. & Wu, Y. L. (1999). Chemtech, 29, 17–28.
- Graether, S. P., DeLuca, C. I., Baardsnes, J., Hill, G. A., Davies, P. L. & Jia, Z. (1999). J. Biol. Chem.274, 11842–11845. [DOI] [PubMed]
- Griffith, M., Ala, P., Yang, D. S. C., Hon, W. C. & Moffatt, B. A. (1992). Plant Physiol.100, 593–596. [DOI] [PMC free article] [PubMed]
- Hazemann, I., Dauvergne, M. T., Blakeley, M. P., Meilleur, F., Haertlein, M., Van Dorsselaer, A., Mitschler, A., Myles, D. A. A. & Podjarny, A. (2005). Acta Cryst. D61, 1413–1417. [DOI] [PubMed]
- International Human Genome Sequencing Consortium (2001). Nature (London), 409, 860–921.
- Jia, Z., DeLuca, C. I., Cha, H. & Davies, P. L. (1996). Nature (London), 384, 285–288. [DOI] [PubMed]
- Ko, T.-P., Robinson, H., Gao, Y. G., Cheng, C. H., DeVries, A. L. & Wang, A. H. (2003). Biophys. J.84, 1228–1237. [DOI] [PMC free article] [PubMed]
- Kręzel, A. & Bal, W. (2004). J. Inorg. Biochem.98, 161–166. [DOI] [PubMed]
- Meilleur, F., Contzen, J., Myles, D. A. & Jung, C. (2004). Biochemistry, 43, 8744–8753. [DOI] [PubMed]
- Niimura, N. & Bau, R. (2008). Acta Cryst. A64, 12–22. [DOI] [PubMed]
- Olsen, T., Sass, S., Li, N. & Duman, J. (1998). J. Exp. Biol.201, 1585–1594. [DOI] [PubMed]
- Shu, F., Ramakrishnan, V. & Schoenborn, B. P. (2000). Proc. Natl Acad. Sci. USA, 97, 3872–3877. [DOI] [PMC free article] [PubMed]
- Sonnichsen, F. D., Sykes, H. D., Chao, H. & Davies, P. L. (1993). Science, 259, 1154–1157. [DOI] [PubMed]
- Urrutia, M. E., Duman, J. G. & Knight, C. A. (1992). Biochim. Biophys. Acta, 1121, 199–206. [DOI] [PubMed]
- Venketesh, S. & Dayananda, C. (2008). Crit. Rev. Biotechnol.19, 57–82. [DOI] [PubMed]
- Wood, K., Froelich, A., Paciaroni, A., Moulin, M., Haertlein, M., Zaccai, G., Tobias, G. J. & Weik, M. (2008). J. Am. Chem. Soc.130, 4586–4587. [DOI] [PubMed]
- Yang, D. S., Hon, W. C., Bubanko, S., Xue, Y., Seetharaman, J., Hew, C. L. & Sicheri, F. (1998). Biophys. J.74, 2142–2151. [DOI] [PMC free article] [PubMed]
- Zhang, C., Zhang, H. & Wang, L. (2007). Food Res. Int.40, 763–769.