Abstract
The positions of ≈4,800 individual miniature inverted-repeat transposable element (MITE)-like repeats from four families were mapped on the Caenorhabditis elegans chromosomes. These families represent 1–2% of the total sequence of the organism. The four MITE families (Cele1, Cele2, Cele14, and Cele42) displayed distinct chromosomal distribution profiles. For example, the Cele14 MITEs were observed clustering near the ends of the autosomes. In contrast, the Cele2 MITEs displayed an even distribution through the central autosome domains, with no evidence for clustering at the ends. Both the number of elements and the distribution patterns of each family were conserved on all five C. elegans autosomes. The distribution profiles indicate chromosomal polarity and suggest that the current genetic and physical maps of chromosomes II, III, and X are inverted with respect to the other chromosomes. The degree of conservation of both the number and distribution of these elements on the five autosomes suggests a role in defining specific chromosomal domains.
Caenorhabditis elegans is the first multicellular organism to have its genome essentially completely sequenced (1–4). Approximately 80% of the 100-megabase (Mb) genome is divided among five autosomes, with the remaining sequence on the X chromosome. Previous genetic and molecular analysis has indicated that the autosomes share a number of conserved features. All the autosomes contain central regions with high gene density and low levels of recombination (5). The bulk of the genes shared by Saccharomyces cerevisiae and C. elegans, the “central cluster,” are confined to their central autosomal regions (1). The autosomal arms flanking the central domains contain most of the inverted and tandem repetitive sequences (1–4) and have higher recombination rates (5).
The C. elegans genome contains a variety of mobile genetic elements. In addition to RNA- (6) and autonomous DNA-based mobile elements (7), miniature inverted-repeat transposable element (MITE)-like repeats are a common feature (8, 9). MITEs are small (<500-bp) elements found in high copy number in many eukaryotic genomes (9–11). Similarities in both terminal inverted-repeat (TIR) sequence and target site selection of MITEs to those of autonomous class 2 transposons have led to the suggestion that MITEs represent nonautonomous forms of DNA-based transposable elements (8, 11, 12). Although the precise evolutionary role(s) of MITEs remain(s) to be determined, it has been suggested that they provide regulatory sequences to a variety of genes (10, 13).
The computational survey presented in this paper indicates that conservation in autosome architecture extends to both the abundance and the distribution of specific sequence domains. The four repetitive elements characterized here have distinct distribution profiles that are conserved on all five autosomes. In addition, the X chromosome is distinct from the autosomes in that three of the four elements are underrepresented. Finally, the distribution profiles indicate conserved chromosomal polarity.
Although this type of analysis is currently limited to C. elegans, the potential for genome-level sequence characterization of chromosomal domains is clearly illustrated.
Materials and Methods
Computational Analysis.
Complete chromosomal sequences were downloaded from the available database (The Sanger Centre, http://www.sanger.ac.uk). Each chromosome was set locally as a separate blast (14) database. Repetitive element positions were identified by using a consensus sequence as a query in a local blast search of the individual chromosome databases. Returned element positions were verified by direct homology comparison with the consensus query sequence. For the Cele1, Cele2, and Cele14 elements, searches were based on previously reported consensus sequences (8, 9). Cele42 repeat-consensus sequences were identified as described below. After accumulation of element positions on the individual chromosomes, chromosome lengths were normalized and maps of chromosomes II, III, and X were inverted with respect to I, IV, and V.
The Cele42 MITE-like repeats were identified in C. elegans genomic sequences by using a previously described algorithm (15). The actual boundaries of the repeats were determined by direct comparison of related repeats from multiple loci using macvector (Oxford Molecular Group, Oxford, U.K.).
Statistical Analysis.
To determine whether the distribution profiles of individual elements differed on the separate chromosomes, an row × column test of independence was used. The distribution of Cele14 elements on all chromosomes was statistically analyzed, with analysis of the remaining elements limited to the autosomes (because of the small number of elements on the X chromosome).
For analysis of the Cele14, Cele42, and Cele1 elements, the chromosomes were divided into thirds to allow for comparison of the two chromosomal arms and the central region. A log-likelihood ratio test (G test) for heterogeneity was used. For the Cele14 elements, the chromosomal distributions over all five autosomes and the X chromosome were found to be homogeneous (G = 12.5; df = 10; P = 0.253). The Cele42 elements were found to be homogeneously distributed on the five autosomes (G = 6.37; df = 8; P = 0.606). The homogeneity in distribution of these two elements allows for the pooling of data from the different chromosomes. For the Cele14 and Cele42 elements, randomness was evaluated by using a simple goodness-of-fit test of the pooled data versus a hypothetical random distribution across the chromosome (16). An acceptable G value (>0.05) with the log-likelihood ratio would support a random distribution.
In contrast to the Cele14 and Cele42 elements, the distributions of the Cele1 elements over the autosome thirds was found to be heterogeneous (G = 20.1; df = 8; P = 0.010). However, the distributions of Cele1 elements on chromosomes I, II, III, and V were found to be homogeneous (G = 9.14; df = 6; P = 0.166), allowing the pooling of the data from these chromosomes to characterize randomness, as described above.
Because the distribution profile of the Cele2 elements differed so markedly from the others (Fig. 1B), heterogeneity testing was carried out by dividing the chromosome into two segments (0–0.8 and 0.8–1; Fig. 1). Log-likelihood ratio testing revealed a homogeneous distribution pattern (G = 4.51; df = 4; P = 0.341) across the autosomes, allowing the pooling of autosomal data to characterize randomness.
Figure 1.
Distribution of repetitive elements on the C. elegans chromosomes. The figure shows the distributions of Cele14 (A), Cele2 (B), Cele1 (C), and Cele42 (D) MITE-like repeats. The number of elements on each chromosome is shown on the right. Repetitive-element positions were identified by using a consensus sequence, chromosome lengths were normalized, and maps of chromosomes II, III, and X were inverted with respect to chromosomes I, IV, and V.
Results
Distribution Profiles of the Cele14 Elements.
Four repetitive-DNA elements, which represent the most abundant computationally identified (15) repeats on the C. elegans autosomes, were selected for distribution analysis. We have previously identified the Cele14 elements (8) as ≈180-bp repeats defined by 58-bp imperfect TIRs. The inverted sequences defining the Cele14 elements are related to the repeats defining mariner transposons (8) [the TIRs are entered into a C. elegans database (ACEDB) as CeRep24].
The chromosomal positions of ≈2,000 Cele14 elements are shown in Fig. 1A. The C. elegans chromosomes have lengths of ≈13 Mb (I and III), 15 Mb (II), 17 Mb (IV), 22 Mb (V), and 17 Mb (X); therefore, the lengths were normalized before plotting repeat positions to allow for direct comparison of the profiles (Fig. 1). In addition, the profiles of chromosomes II, III, and X are reversed relative to I, IV, and V. As shown in Fig. 1, the total number of repeats and their distribution profiles on the different chromosomes are surprisingly similar.
Several investigators have noted the association of specific repetitive elements with autosomal arms (2–5). As shown in Fig. 1A, the Cele14 elements have a distribution profile similar to CeRep3 (2, 3), with high-density domains on both chromosomal arms. However, on each chromosome the density of the Cele14 elements on one arm is ≈2-fold higher than on the other. The Cele14 profile observed on the X chromosome is similar to the autosomes (Fig. 1A). The homogeneous distributions of these elements on all chromosomes are statistically supported by log-likelihood ratio analysis (see Materials and Methods). This homogeneity (G = 12.5; df = 10; P = 0.253) depends on reversing the orientations of chromosomes II, III, and X. Direct analysis of the profiles (i.e., chromosomes II, III, and X not reversed) indicates heterogeneity among the chromosomes (G = 39.6; df = 10; P < 0.001). To test for randomness, the distribution data from the six chromosomes (Fig. 1A) were pooled and compared with a random profile. The hypothesis of random distribution is not supported by goodness-of-fit testing (G = 172; df = 2; P < 0.001).
Distribution Profiles of the Cele2, Cele1, and Cele42 Elements.
Although the Cele14 distribution reflects the previously observed high density of repeats at chromosome ends (2–5), other MITE-like repeats show strikingly different patterns. The Cele2 elements are ≈300 bp in length and are defined by 90-bp TIRs (9) (these elements are partially defined by CeRep18 in ACEDB). The Cele2 repeats are evenly distributed across 80% of each autosome, with significantly decreased density on one arm of chromosomes I, II, III, and V (Fig. 1B). This decreased density is observed on both arms of chromosome IV. Very few Cele2 repeats are located on the X chromosome (Fig. 1B). The Cele2 distribution profiles (as defined in Materials and Methods) on the different chromosomes were found to be homogeneous and nonrandom (goodness-of-fit testing relative to random distribution; G = 49.4; df = 1; P < 0.001).
The Cele1 elements are 300-bp repeats defined by 120-bp TIRs (9) (the TIR of this element is listed as CeRep14 in ACEDB). The distribution profiles of these elements are different from those of either the Cele14 or the Cele2 repeats. The Cele1 repeats have a high density on one autosomal arm (≈80% of the total), with a more even distribution across the remaining 60% of the chromosomes (Fig. 1C). Goodness-of-fit testing of the pooled profiles from chromosomes I, II, III, and V (see Materials and Methods) relative to a random profile suggests a nonrandom distribution of these elements (G = 65.8; df = 2; P < 0.001).
The structure of the final MITE-like element used in this study, Cele42, is shown in Fig. 2. These are ≈240-bp elements defined by 23-bp imperfect TIRs. On all five autosomes, ≈90% of the Cele42 elements is localized to a single arm (Fig. 2D). The core 60% of each autosome is virtually devoid of these elements, with the remaining 10% localized to the other arm. The homogeneous distribution of these elements on all autosomes (G = 6.37; df = 8; P = 0.606) allows the pooling of data and comparison to a random distribution profile. A hypothetical random distribution is not supported by goodness-of-fit testing (G = 87.6; df = 2; P < 0.001). As in the case of the Cele14 repeats, the failure to reverse chromosomes II and III results in a heterogeneous pattern among the autosomes (G = 66.5; df = 8; P < 0.001).
Figure 2.
Structure of the Cele42 MITE-like elements. The Cele42 elements were identified in C. elegans genomic sequences by using a previously described search algorithm (15). Arrows indicate inverted-repeated domains. Heterogeneity is indicated with the International Union of Biochemistry ambiguity code (26). Positions in GenBank accession numbers for the nucleotides shown are as follows: AF043698, 8180–8418; AL032647, 5822–6045; and AF043701, 4596–4825.
Similar to the Cele2 repeats, the Cele42 and Cele1 repeats occur in limited numbers on the X chromosome (Fig. 2 C and D). The low density of Cele1, Cele2, and Cele42 repeats on the X chromosome may be related to the presence of a number of repeats specific to this chromosome (1, 17).
The data in Fig. 1 indicate that both the distribution and the number of elements from each family on individual autosomes are conserved, with variation largely independent of chromosome length. In addition, the distinct distribution profiles for these different families of repeats indicate that the current genetic and physical maps of chromosomes II, III, and X have an inverted polarity relative to the other chromosomes.
Conservation of Distribution Profiles on the Autosomes.
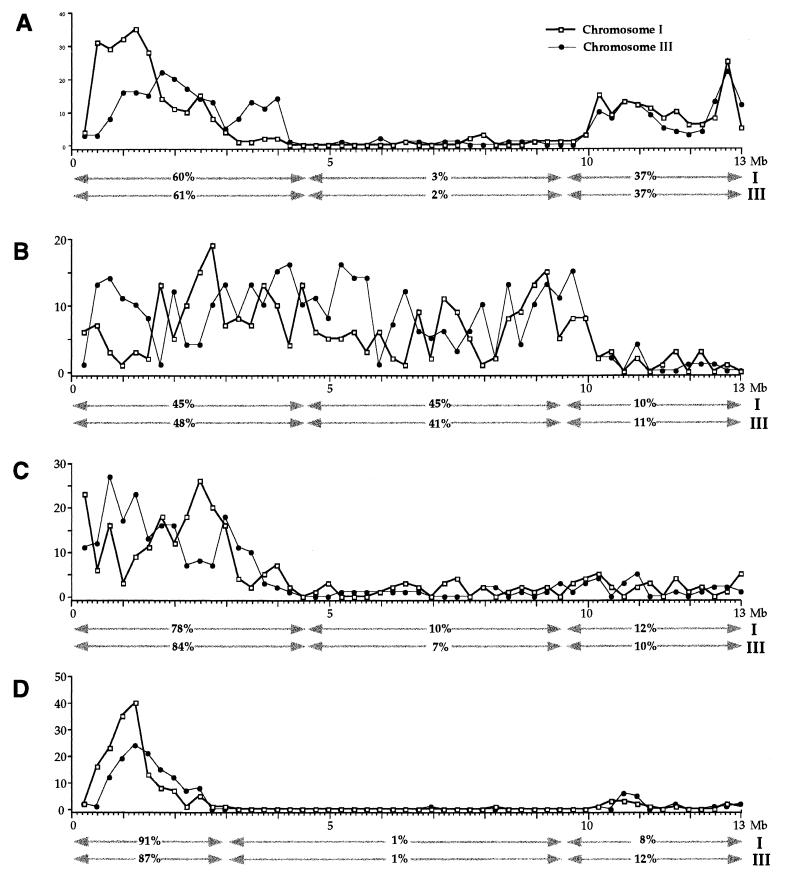
As chromosomes I and III are of similar length, a direct comparison of repeat-distribution profiles can be made (Fig. 3). The distribution of Cele14 elements on the two autosomes is essentially identical on the right arm and very similar over the remainder of the chromosomes (Fig. 3A). The Cele14 profile on the left arm of chromosome III seems to be offset by ≈0.5 Mb relative to chromosome I.
Figure 3.
Distribution of repetitive elements on C. elegans chromosomes I and III. The figure shows the distributions of Cele14 (A), Cele2 (B), Cele1 (C), and Cele42 (D) MITE-like repeats. The number of the individual repeat family members per 200 kb is plotted relative to the 13 Mb of sequence downloaded from each chromosome (see Figs. 1 and 2). The distribution of elements on chromosome III is inverted relative to standard genetic and physical maps.
Similar to the distribution of the Cele14 elements, the distribution of Cele2, Cele42, and Cele1 repeats is essentially identical on chromosomes I and III (Fig. 3 B–D). Examination of the Cele42 element distribution reveals highly conserved left and right boundary positions for the low-density domain (Fig. 3D).
Discussion
Although a variety of repetitive-DNA sequences have been identified in C. elegans, MITEs, small (<500-bp) elements defined by TIRs, are among the most common (1, 4, 8, 9, 17). Given both the sequence similarity of the TIRs of many of these elements to known class 2 transposons and the target site selection (8, 11, 12, 17), in all probability these repeats represent nonautonomous forms of autonomous transposons.
The essential completion of the genomic sequence of C. elegans allows accurate determination of the distribution of these types of elements throughout the genome of a multicellular organism. Previous characterization of the distribution of inverted-repeat sequences on the C. elegans chromosomes indicated a nonrandom pattern (1, 4). Inverted-repeated domains were observed to be largely localized to chromosomal arms in regions where genes similar to those in yeast (“core genes”) are limited and where recombination rates are high (5). However, the “nonrandomness” of the distribution pattern becomes much more striking if the positions of specific families of repeats are plotted. The primary observations reported in this paper are (i) the different elements show distinct distribution profiles, (ii) these profiles are highly conserved on all autosomes, (iii) the X chromosome is distinct from the autosomes in that three of the four elements are underrepresented, and (iv) the profiles indicate conserved chromosomal polarity. No obvious association of the distribution profiles with specific base frequencies across the chromosomes was observed.
The members of the repeat families examined in this paper are laid out in precise, and family-specific, patterns on the C. elegans autosomes (Figs. 1 and 3). The distribution profiles indicate that all of the autosomes are similarly organized with respect to the densities of specific MITE-like repeats. This finding suggests a conserved, autosomal domain arrangement along the lengths of the chromosomes, either established or reflected by the distribution of these elements. For example, the distribution profiles of the Cele1, Cele2, and Cele42 repeats define at least four distinct domains on the autosomes (Figs. 1 and 3). The first 20% of each autosome contains all three elements. The region from 20% to 45% is enriched in Cele1 and Cele2 repeats, the region from 45% to 80% contains largely Cele2 repeats, and the region from 80% to 100% is enriched in Cele1 repeats. This conserved, autosomal, repetitive-DNA density profile, potentially distinguishing distinct chromosomal domains, could act as a template for rapid scanning of complete chromosomes. A first step in distinguishing such a system would be the identification and characterization of nuclear proteins with the potential to bind specifically to the MITE sequences.
One of the more striking features of the distribution profiles shown in Fig. 1 is the virtual absence of Cele1, Cele2, and Cele42 elements on the X chromosome. The X chromosome differs from the autosomes in the requirement for dosage compensation, i.e., the interphase expression levels of X-linked genes in XX hermaphrodites must be halved relative to X0 males (18–20). If the conserved, autosomal density profiles of the Cele1, Cele2, and Cele42 families (which collectively make up over 1% of the autosome sequences) are associated with gene expression levels in specific domains, their absence from the X chromosome may be a reflection of the specific regulatory requirements imposed on X-linked transcription units.
Finally, the conserved, nonuniform distribution of these elements on the two autosomal arms indicates chromosomal polarity. Of the repeat families examined, only the Cele14 elements display a pattern similar to the overall distribution of inverted-repeated domains, i.e., accumulation on autosomal arms (1, 4). However, even in the case of these repeats, the distribution is weighted toward one of the arms (Fig. 1A). This asymmetric distribution is even more striking in the case of the Cele42 repeats (Fig. 1D). The chromosomal arm with a higher density of Cele14 and Cele42 repeats also has a high density of Cele1 elements (Fig. 1C). This polarity may be a reflection of the complex centromeric activity in this organism. Although holocentric during mitotic divisions, C. elegans seems to be monocentric during meiosis (21), with centromeric functions limited to chromosomal termini (22, 23). The completion of genome sequences for organisms such as Arabidopsis thaliana will be required to determine whether the chromosomal conservation of MITE number and the distribution observed in C. elegans is also found in organisms with more traditional centromere functions.
The distinct distribution profiles of the MITE-like repeats are difficult to explain. If these elements represent nonautonomous forms of autonomous class 2 transposons (8, 11, 12), a pattern of local transposition (24) over an extended time period would be expected to result in random distribution over the chromosomes. Regardless of the transposition mechanism, similar types of elements would be expected to exhibit similar profiles. However, the Cele1 and Cele2 elements, which are of similar size and structure, have radically different chromosomal distributions (Figs. 1 and 3). The profiles observed here may reflect a differential preference among the individual elements (25) for the highly repetitive heterochromatin-like sequence domains found on the autosomal arms (1). However, even though the X chromosome has a much more even gene and general repetitive-DNA distribution than is observed on the autosomes (1, 4, 5), the distribution profile of Cele14 elements on the X chromosome is highly similar to that on the autosomes (Fig. 1A). This finding suggests that the observed distribution profiles do not result from simple targeting of these elements to regions containing abundant repetitive DNA.
The characterization of chromosomal repetitive-DNA profiles of additional organisms will be important in determining whether the architectural features described in this paper represent a general feature of these types of genomes, as well as in elucidating potential roles in establishing or reflecting specific chromosomal domains.
Acknowledgments
We thank J. M. Burke for statistical analysis design.
Abbreviations
- MITE
miniature inverted-repeated transposable element
- Mb
Megabase
- TIR
terminal inverted repeat
References
- 1.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 2.Felsenstein K M, Emmons S W. Mol Cell Biol. 1988;8:875–883. doi: 10.1128/mcb.8.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cangiano G, La Volpe A. Nucleic Acids Res. 1993;21:1133–1139. doi: 10.1093/nar/21.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson R K. Trends Genet. 1999;15:51–58. doi: 10.1016/s0168-9525(98)01666-7. [DOI] [PubMed] [Google Scholar]
- 5.Barnes T M, Kohara Y, Coulson A, Hekimi S. Genetics. 1995;141:159–179. doi: 10.1093/genetics/141.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Youngman S, van Luenen H G, Plasterk R H. FEBS Lett. 1996;380:1–7. doi: 10.1016/0014-5793(95)01525-6. [DOI] [PubMed] [Google Scholar]
- 7.Berg D E, Howe M M, editors. Mobile DNA. Washington, DC : Am. Soc. Microbiol.; 1989. [Google Scholar]
- 8.Oosumi T, Garlick B, Belknap W R. J Mol Evol. 1996;43:11–18. doi: 10.1007/BF02352294. [DOI] [PubMed] [Google Scholar]
- 9.Oosumi T, Garlick B, Belknap W R. Proc Natl Acad Sci USA. 1995;92:8886–8890. doi: 10.1073/pnas.92.19.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wessler S R, Bureau T E, White S E. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 11.Oosumi T, Belknap W R, Garlick B. Nature (London) 1995;378:672. doi: 10.1038/378672a0. [DOI] [PubMed] [Google Scholar]
- 12.Wessler S R. Physiol Plant. 1998;103:581–586. [Google Scholar]
- 13.Oosumi T, Belknap W R. J Mol Evol. 1997;45:137–144. doi: 10.1007/pl00006213. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Surzycki S A, Belknap W R. J Mol Evol. 1999;48:684–691. doi: 10.1007/pl00006512. [DOI] [PubMed] [Google Scholar]
- 16.Wilks S S. Ann Math Statist. 1935;6:190–196. [Google Scholar]
- 17.Rezsohazy R, van Luenen H G, Durbin R M, Plasterk R H. Nucleic Acids Res. 1997;25:4048–4054. doi: 10.1093/nar/25.20.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucchesi J C. Curr Opin Genet Dev. 1998;8:179–184. doi: 10.1016/s0959-437x(98)80139-1. [DOI] [PubMed] [Google Scholar]
- 19.Dawes H E, Berlin D S, Lapidus D M, Nusbaum C, Davis T L, Meyer B J. Science. 1999;284:1800–1804. doi: 10.1126/science.284.5421.1800. [DOI] [PubMed] [Google Scholar]
- 20.Lieb J D, Albrecht M R, Chuang P T, Meyer B J. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- 21.Wicky C, Rose A M. BioEssays. 1996;18:447–452. doi: 10.1002/bies.950180606. [DOI] [PubMed] [Google Scholar]
- 22.McKim K S, Howell A M, Rose A M. Genetics. 1988;120:987–1001. doi: 10.1093/genetics/120.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKim K S, Peters K, Rose A M. Genetics. 1993;134:749–768. doi: 10.1093/genetics/134.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machida C H, Onouchi H, Koizumi J, Hamada S, Semiarti E, Torikai S, Machida Y. Proc Natl Acad Sci USA. 1997;94:8675–8680. doi: 10.1073/pnas.94.16.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terrinoni A, Franco C D, Dimitri P, Junakovic N. J Mol Evol. 1997;45:145–153. doi: 10.1007/pl00006214. [DOI] [PubMed] [Google Scholar]
- 26.Nomenclature Committee of the International Union of Biochemistry. Eur J Biochem. 1985;150:1–5. doi: 10.1111/j.1432-1033.1985.tb08977.x. [DOI] [PubMed] [Google Scholar]