Abstract
Base excision repair is critical for the maintenance of genome stability because it repairs at least 20,000 endogenously generated DNA lesions per cell per day. Several enzymes within the base excision repair pathway exhibit sequence context dependency during the excision and DNA synthesis steps of repair. New evidence is emerging that germ line and tumor-associated variants of enzymes in this repair pathway exhibit sequence context dependence that is different from their ancestral counterparts. We review what is known about the ancestral and variant base excision repair proteins within various sequence contexts. We suggest that altering the sequence context preferences of base excision repair proteins could give rise to rare cellular variants that might have a selective advantage in response to environmental exposure or to the tumor microenvironment.
Sequence context specific mutagenesis in the human genome
As the DNA sequence of the human genome began to emerge, investigators employed various types of analyses to categorize the types of mutations that were present in humans. In 1994, Hess and colleagues scored all mutations in the entire primate database of gene-pseudogene pairs [1]. They found that the highest rates of transition, transversion, and deletion were in a C-G pair with a 3′ GC neighbor. Using a novel methodology to estimate mutation rate and the Human Gene Mutation Database, which records mutations only if they result in a phenotype, Krawczak and coworkers showed that CpG sites were hypermutable compared to the overall base mutation rate [2]. They also demonstrated that CpG sites with 5′ pyrimidines and 3′ purines appeared to have the highest mutation rate in the human sequences that they compared. In a study of 2.6 million Single Nucleotide Polymorphisms (SNPs) in the human genome, Zhao and Boerwinkle also showed that the mutation rate is highest at CpG sites and that the bases immediately neighboring the 5′ and 3′ sites of the mutated site (SNP) were most likely to be C and G, respectively [3]. These observations were later confirmed in a study of 8.2 million SNPs. In this study, base substitutions were 6-fold overrepresented at CpG sites [4]. Thus, it is generally agreed that methylated CpG sites have the highest mutation rate in the human genome, and that the mutagenicity of these sites is influenced by the bases immediately preceding and following the mutated site.
Sequence context is an important aspect of base substitution mutagenesis. A likely and generally accepted scenario to explain the fact that CpG sites are hotspots of mutagenesis is that due to the inherent instability of DNA, 5′ methylcytosine deaminates to thymine, resulting in a T-G mispair. This mispair is usually recognized by Thymine DNA glycosylase (TDG), which initiates repair of the mispair by removing the thymine base. However, if the T-G mispair is refractory to repair, a mutation results. Sequence context is likely to influence the local structure of the DNA at the site of mutation. For example, in a study of the T-G mispair embedded with various sequence contexts, Mitra and colleagues found that a 5′ C and 3′ A induce the greatest conformational flexibility in the DNA molecule and suggest that this influences mutagenesis [5]. One possibility is that TDG may interact less stably with a T-G mispair that is embedded within a flexible local sequence, leading to mutagenesis instead of repair. The effects of sequence context on base substitution mutagenesis have been known since some of the earliest mutagenesis studies were performed. In this minireview, we attempt to summarize what is known about the sequence context specificities of the short patch base excision repair (BER) system, and relate them to base substitution mutagenesis. We then speculate on how aberrant BER alters the sequence context rules and how that might affect the cancer genome.
Endogenous Mutation Mechanisms
The rate of endogenous DNA damage has been estimated to be at least 20,000 lesions per cell per day [6],[7]. Such damage likely results from reactive oxygen species (ROS) that are generated as part of normal cellular metabolism. Base damage is also generated by exogenous agents, including ionizing radiation and methylating agents that are frequently used to treat a variety of human cancers. Specific DNA sequences such as microsatellites and CpG sites have been shown to exhibit increased susceptibility to mutation. The instability of microsatellites, which are stretches of simple repeated sequence, has been primarily attributed to DNA polymerase slippage during replication in the absence of damage and can also be attributed to non-canonical DNA structures [8], [9], [10]. The expansion or reduction of these repeated sequences has been extensively observed in tumor cells, highlighting the biological implications of their mutability. In addition to inherent instability during replication, frameshift frequencies within microsatellites are increased in the presence of oxidative damage relative to non-iterated sequences [11]. The CpG DNA sequence plays a key role in regulation of gene expression, as stretches of this short repeat are often found near promoter regions. Control of gene expression is facilitated by the methylation state of cytosine in these sequences; methylation at this position serves to suppress gene expression. Alteration of cytosine methylation and subsequent gene misregulation has been observed in tumors and likely contributes to tumorigenesis [12]. Methylation also affects stability of the cytosine nucleotide; 5-methylcytosine tends to undergo spontaneous deamination to thymidine, generating a T-G mispair. Maintaining the integrity of the methylation state of cytosine in a CpG context is critical to preserve appropriate gene expression.
Sequence context also affects the stability of DNA duplexes containing abasic sites. Using differential scanning calorimetry and temperature-dependent UV absorbance spectrophotometry, Gelfand and colleagues show that the presence of an abasic site has significant enthalpic consequences on DNA stability, and that sequence context affects the magnitude of destabilization [13]. Molecular modeling also indicates that flanking sequence around an abasic site determines the ability of the unpaired base to become extrahelical [14]. Intrinsic thermodynamic differences between various DNA sequences containing abasic sites present the BER system with a variety of substrates that may have different repair efficiencies, despite containing a common lesion.
Eukaryotic Base Excision Repair
The base excision repair (BER) pathway is the primary cellular defense against the majority of endogenous DNA damage. Resolving such a high level of damage is critical to maintaining genomic integrity and ultimately, cellular viability. Cells that are BER deficient or have aberrant BER function risk high levels of mutation that may lead to cellular transformation and cancer [15]. The BER pathway facilitates the resolution of such DNA damage, including abasic sites, oxidative lesions and alkylated and alternative bases [16]. In short patch repair, thought to be the major BER repair pathway, a damage-specific DNA glycosylase works to remove the damaged base, leaving an abasic site. A monofunctional DNA glycosylase generates an abasic site by hydrolyzing the N-glycosidic bond between the base and the deoxyribose group [17]. AP endonuclease 1 (APE1) then nicks the DNA backbone, leaving a free 5′-terminal deoxyribose phosphate (dRP) and a 3′-hydroxyl [18]. In addition to base removal, bifunctional glycosylases also have innate AP-lyase activity and form an abasic site with a 3′ dRP group and a 5′ phosphate [17]. APE1 removes the 3′ dRP, generating a 3′-hydroxyl and this is the substrate for DNA polymerase β (Pol β). In mammalian cells, Pol β binds the nicked backbone and its own dRP lyase domain removes the 5′ dRP [19]. Pol β then fills in the abasic site with the complementary Watson-Crick base and the resultant substrate is sealed by the DNA ligase III/XRCC1 complex, completing the repair process [20]. Alternatively, the BER pathway may also be initiated by the NEIL1/2 DNA glycosylases, which remove the damaged base and leave behind free phosphates at both the 3′ and 5′ ends [21]. PNK acts on the resulting DNA substrate, removing the 3′-phosphate. DNA Pol β then binds to the site of damage and fills the gap with the complementary nucleotide. XRCC1/DNA ligase III again completes repair by sealing the nicked DNA backbone.
BER Proteins and Sequence Context
The efficiency of the BER pathway is affected by local DNA sequence context. This pathway is responsible for repairing a variety of lesions, and employs a range of glycosylases to initiate the process. The glycosylase used is dependent on the type of damage present in the DNA, and the activity of many DNA glycosylases is affected by the local DNA sequence. The catalytic efficiency of DNA pol β is also dependent on local DNA context. Differences in repair rates based on sequence context may lead to specific sequences being mutated at disproportionately higher rates. If such sequences are found within growth control genes or regulatory sequences, sequence context based mutagenesis could have implications for tumor development or progression. BER proteins demonstrated to function with specificity for sequence context are discussed below.
MPG
Human N-methylpurine-DNA glycosylase (MPG) is responsible for removal of methylated purine DNA adducts, including 3-methyladenine (3-meA) and 7 methylguanine (7-meG). Ye and colleagues mapped the repair of 3-meA and 7-meG adducts both in vivo and in vitro in the promoter and first exon of the PGK1 gene [22]. Repair of both of these adducts in normal fibroblast cells was found to be heterogeneous; 7-meG and 3meA persist in DNA for varying periods of time depending on their DNA sequence context. This variation may have implications for genome stability, as lesions that persist longer in DNA have a greater chance of leading to transcriptional pausing or resulting in mutagenesis. In vitro excision of 149 different 7-meG lesions within the same sequence using recombinant MPG indicates that the first order rate of removal for 7-meG varies up to 185-fold depending on the lesion’s position within the DNA sequence. However, removal of 3-meA by MPG in vitro did not depend on sequence context. Therefore, any sequence context effect observed in vivo for removal of 3-meA is not attributed to MPG and is likely the result of subsequent repair processes.
MPG also appears to excise hypoxanthine (Hx) from DNA in a sequence context dependent manner, with up to a 17-fold difference in the steady-state rate constant ratio kcat/KM determined in vitro [23]. While Hx excision was shown to be dependent on both DNA binding and catalysis, DNA binding was found to be the primary contributor to sequence specific excision differences. The 17-fold differences in steady-state rate constants are attributed to greater variability in DNA binding affinity.
UDG
Uracil DNA glycosylase (UDG) recognizes and removes uracil from DNA; this activity has been shown to be dependent upon sequence context [24], [25]. Initial studies using 40 different uracil sites found more than a 10-fold difference in rates of removal within double-stranded DNA [24]. The crystal structure of UDG complexed with double-stranded DNA indicates that the DNA is dramatically bent in the local region where the uracil residue is located [25]. Molecular dynamic simulations performed on two distinct DNA sequences known to have significant differences in UDG activity indicate that the more flexible the local sequence, the more efficiently the uracil is removed [25]. Kinetic constants of DNA binding and catalysis were compared between the two sequences; the activity disparity is attributable to DNA binding.
MBD4
Methyl-CpG-binding protein 4 (MBD4), a thymine/uracil glycosylase, contains a methyl-CpG binding domain and lacks endonuclease activity [26]. MBD4 exhibits a preference for mismatches within a CpG context. Mice deficient in MBD4 exhibit a 3-fold increase in C→T transitions specifically at CpG sites, and also show reduced survival in a background of spontaneous intestinal neoplasia [27]. This observation highlights the biological significance of a glycosylase that is more efficient in repairing damage in a CpG sequence context.
TDG
Human thymine glycosylase (TDG) removes thymidine from G:T mispairs that result from the deamination of 5′ methylcytosine (m5C) or cytosine. In vitro incision assays have demonstrated that the activity of TDG has been shown to be most efficient when the mispair is in a CpG context [28]. Excision in a CpG context was 3–12 fold greater compared to ApG, CpC and TpC. The reported crystal structure of the catalytic domain of TDG complexed with abasic DNA provided insight regarding observed CpG specificity [29]. In a CpG context, the enzyme’s insertion loop interacts with the guanine involved in the mispair and the guanine 3′ to the mispaired thymidine residue; this interaction stabilizes the loop and promotes nucleotide flipping and/or chemistry.
NEIL1 and 2
The Nei-like NEIL1 and NEIL2 glycosylases remove damaged bases from DNA and provide the substrate for an APE1-independent BER pathway. NEIL1 works on thymine glycol, formamidopyrimidine-A and –G; both NEIL1 and NEIL2 excise 5-hydroxyuracil (5-OHU). Both NEIL1 and NEIL2 exhibit a preference for repairing 5-OHU within the single stranded DNA in a bubble structure compared to double-stranded DNA [30].
Pol β
Pol β is the primary polymerase responsible for gap filling and removal of the dRP group during BER. The catalytic efficiency of this polymerization by the wild-type protein is linked to the DNA sequence surrounding the abasic site. Base stacking forces within DNA and between DNA and protein facilitate stability of the duplex within the active site of Pol β. Crystal structures of DNA complexed with Pol β show that protein-DNA interactions are formed up to 4–5 base pairs upstream of the active site [31]. These interactions help form the nucleotide-binding pocket and therefore contribute to polymerase fidelity. Beard and colleagues evaluated the effects of altering the duplex DNA sequence upstream of the active site by incorporation of abasic sites, mismatches or extra nucleotides [32]. An upstream abasic site disturbs base stacking interactions and produces a significant loss of catalytic efficiency of Pol β. This observed effect is greater when the abasic site is in the primer rather than the template. Introduction of a frameshift intermediate in the form of an extrahelical unpaired nucleotide affects efficiency in a similar fashion; this effect is maintained with an extra base placed up to five nucleotides upstream of the abasic site to be repaired. The base composition of the primer terminus also affects the catalytic efficiency of Pol β primers with a terminal pyrimidine are more efficiently extended than those with a purine [32]. These results indicate that the sequence context immediately upstream of an abasic site affects the rate of repair.
Misincorporation of dATP directly opposite an 8-oxo-dG lesion by Pol β is also dependent on sequence context [33]. Using a 5 base pair gapped substrate with a central 8-oxo-dG lesion, Efrati and colleagues found that the downstream sequence affects incorporation opposite 8-oxo-dG by means of dNTP-stabilized misalignment. Alteration of downstream sequence affects the identity of the dNTP selected by Pol β during such misalignment. Surprisingly, the presence of 8-oxo-dG in this sequence context was also found to reduce Pol β fidelity on the surrounding sequence. Thus, the fidelity of Pol β appears to be affected by the sequence surrounding a lesion, as well as the lesion itself when extending multiple bases.
The Cancer Genome and Sequence Context Dependent Mutagenesis
BER variants and sequence context dependence
Several germline SNPs in BER genes were documented in individuals who did not have cancer or other diseases at the time of sample collection as part of the Environmental Genome Project (http://www.genome.utah.edu/genesnps/). BER variants have been identified in human tumors; for example see [34], [35], [36]. Importantly, the majority of the germline and tumor associated variants have not been characterized in detail. We will focus on a few examples for which a shift in sequence context dependence has been demonstrated.
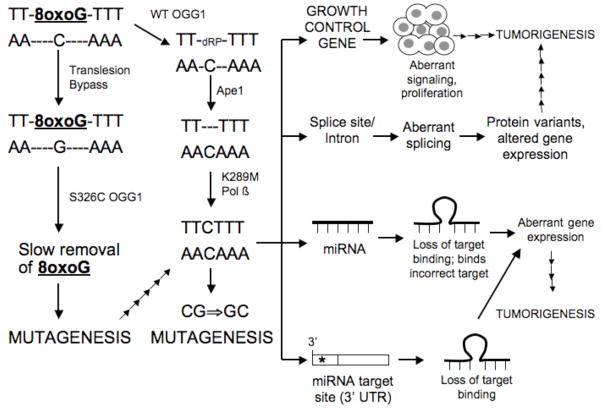
The OGG1 S326C variant is present in the germline of at least 18.5% of individuals who have been tested. The presence of this variant has been shown to be associated with increased incidence of oral, lung, and prostate cancer [37], [38], [39], [40], [41], [42], [43]. OGG1 is a bifunctional DNA glycosylase and is critical for the removal of the 8oxoG base from DNA. S326C has been shown to dimerize, unlike the WT protein, and generally to have a lower catalytic efficiency than WT OGG1, as shown in Figure 1 [44]. However, subtle changes in sequence context dependent incision rates were also identified in that the sequence context dependent recognition of S326C is completely different to that of WT OGG1. The rates of excision and AP site cleavage of 8oxoG opposite T and G are 3.7 and 5.6 lower than that of WT OGG1. Thus, if a T or G is misincorporated opposite 8oxoG, for example, by a translesion DNA polymerase, cells with the S326C variant will be less likely to remove the misincorporated base versus cells with WT OGG1. After DNA replication occurs, a mutation will be generated that could rarely be observed in cells with the WT OGG1. To the best of our knowledge, little is known about the how the surrounding sequence context influences base excision and AP site incision by the S326C variant, so even more variation in repair rates at specific sites could be present in cells expressing this variant.
Figure 1. S326C OGG1 and K289M Pol β Variants Induce Sequence Context Dependent Mutations.
8oxoG arises within the DNA. If not excised before DNA replication, a DNA polymerase may bypass it and insert the incorrect base. If 8oxoG is paired with G, the S326C OGG1 will remove it much slower than WT OGG1. This could lead to additional bypass and mutagenesis, which ultimately results in tumorigenesis. 8oxoG could also be excised, especially in the presence of WT OGG1. This would generate a 3′ dRP, which would be removed by Ape1. If K289M filled in the gap, it could insert the wrong base within the sequence context shown, leading to a mutation. If the mutation arises in a growth control gene such as Ras, aberrant signaling and proliferation would lead to tumorigenesis. The mutation would be just as likely to arise in noncoding DNA, such as a splice site, miRNA-encoding sequence, or a miRNA target in a 3′ UTR. This could effect gene expression, and ultimately lead to tumorigenesis or more aggressive disease. Importantly, because the mutations that arise are likely to be within sequence contexts that are not usually amenable to mutation in cells with WT BER enzymes, rare cellular variants might result and be selected by the environment.
Approximately 30% of human tumors express Pol β variants [34]. We have shown that the I260M and K289M prostate and colon cancer-associated variants, respectively, induce cellular transformation in immortalized mouse cells [45]. Importantly, these variants induce specific types of base substitutions at increased frequencies within certain sequence contexts [46], [47]. For example, when synthesizing DNA in vitro, I260M predominately expands a TC dinucleotide repeat sequence within the HSV-tk target. WT Pol β induces mainly deletions within this same sequence [47]. The K289M variant induces C to G base substitutions within the AACAAA sequence, which resembles a mutation hotspot within the APC gene, a gene that is frequently mutated in colon carcinoma, as shown in Figure 1 [46]. This was validated in an in vitro assay, suggesting that the sequence context dependent misincorporation exhibited by K289M is an inherent property of the enzyme. It is likely that if these variants are present in cells, they will have a higher propensity than WT Pol β to make errors during the gap filling step of BER, eventually resulting in mutations that are rarely observed in cells with WT Pol β.
Cancer-associated consequences of mutagenesis
Mutations in BER genes, like the ones described above, can lead to deficient or aberrant repair of the coding regions of the genome. In the case of cancer, this could be manifested as a mutation in a key growth control gene, such as Ras, as shown in Figure 1. For example, mice that are deficient in the OGG1 enzyme appear to accumulate mutations in the Ras oncogene [48]. Similar results might be obtained for the S326C or other OGG1 variants if such a study were performed. This would ultimately result in altered cellular signaling that could lead to a lack of response of cells to growth controls, leading to uncontrolled proliferation and, eventually, cancer. This scenario could be imagined for several different oncogenes, other types of growth control genes, and genes that function in the control of metastasis.
Proteins rarely act alone and usually function as parts of complexes. Alteration of residues that are on the surface of proteins could lead to defects in protein-protein interactions and complex formation. This could result in less interaction of a specific protein with its partners, or sequestration of other members of the complex. In both cases, the processes that require the altered proteins will likely be compromised.
Mutations induced by BER variants could also occur in the noncoding regions of the genome. These types of mutations could affect splicing if they occurred at a splice site or within introns themselves, which are also important during the splicing reaction, as shown in Figure 1. Mutations in noncoding DNA could also alter microRNAs (miRNAs), which have been shown to have an important role in the regulation of gene expression [for example see Didiano [49]]. This could result in inefficient binding of a miRNA to its target sequence, and result in a deficiency in posttranscriptional regulation, which could lead to over-or underexpression of a protein in the cell, as shown in Figure 1. Mutations in the 3′ UTRs of genes, the sites of miRNA target sequences, could have similar effects. In fact, the miRNA target appears to be defined not only by the sequence to which the miRNA binds, but also by the surrounding sequence context, so mutations anywhere within the 3′ UTRs of genes could effect gene expression.
Cancer is associated with a mutator phenotype
In 1974, Loeb proposed that cancer results from a mutator phenotype [50]. The basic idea is that the numbers of mutations found in tumors is much larger than what is found in somatic cells, as has recently been shown to be the case [51]. The low numbers of mutations in somatic cells is indicative of a low mutation rate and, conversely, the high numbers of mutations in tumor cells is suggestive of the presence of a high mutation rate. The mutator phenotype was originally suggested to be driven by mutations that arose in proteins that function to maintain genome stability; these early mutations likely arise as part of a stochastic process during DNA replication or repair. One example of this is that mutations in certain mismatch repair genes are associated with Heterologous Nonpolyposis Colon Cancer (HNPCC) [52]. Mismatch repair functions to maintain genome stability by repairing mistakes made during DNA replication and recombination. Thus, in the absence of mismatch repair, a strong mutator phenotype is present and has the possibility to lead to an even higher mutation rate should other genes that function in genome stability maintenance sustain mutations.
The environment selects for novel variants
Mutagenesis drives evolution because it produces many types of variants, one or more of which emerge via natural selection. As noted above, BER functions in the removal and correct repair of at least 20,000 endogenous lesions per cell per day. These lesions are likely to occur in regions of the genome that code for proteins and in noncoding regions. Thus, BER proteins are likely to act upon the DNA of both coding and noncoding regions of the genome. As discussed above, many of the BER proteins are known to exhibit sequence context dependent catalysis. We suggest that alterations of BER proteins, resulting from germline or somatic SNPs, could change their sequence context dependence; some examples are already known. This could have important consequences for mutation of both coding and noncoding regions of the human genome. Novel cellular variants that are not usually present could arise and be at a selective advantage to respond to selective pressure from the environment. Alteration of sequence context dependent repair could be an underlying mechanism for predisposition to cancer in people carrying germline variants in BER genes, depending upon the environment to which an individual is exposed. BER protein variants that arise in somatic cells could also be capable of altering sequence context dependent repair, and this could lead to tumorigenesis or more aggressive disease if these variants are generated and selected by the tumor microenvironment.
In summary, BER is critical for genome stability maintenance. This process is responsible for the repair of a variety endogenous damage. It appears that many of the proteins that function in BER exhibit sequence context dependent repair during binding and catalysis. Several variants of these genes are present in people that could predispose them to cancer. BER variants also arise in tumors. These BER variants could exhibit sequence context dependent repair. Therefore, the mutator phenotype that is an underlying mechanism of cancer, is likely to be driven not only by mutations in genes that result in a lack of repair or growth control, but also by mutations that result in changes to sequence context dependency. These types of mutations, which are likely to occur in both coding and noncoding regions of the genome, could result in rare variants that can be selected by the cellular environment and lead to cancer. These rare variants might also lead to resistance to various cancer therapies.
References
- 1.Hess ST, Blake JD, Blake RD. Wide variations in neighbor-dependent substitution rates. J Mol Biol. 1994;236(4):1022–1033. doi: 10.1016/0022-2836(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 2.Krawczak M, Ball EV, Cooper DN. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am J Hum Genet. 1998;63(2):474–488. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Z, Boerwinkle E. Neighboring-nucleotide effects on single nucleotide polymorphisms: a study of 2.6 million polymorphisms across the human genome. Genome Res. 2002;12(11):1679–1686. doi: 10.1101/gr.287302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Z, Zhang F. Sequence context analysis of 8.2 million single nucleotide polymorphisms in the human genome. Gene. 2006;366(2):316–324. doi: 10.1016/j.gene.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 5.Mitra R, Pettitt BM, Blake RD. Conformational states governing the rates of spontaneous transition mutations. Biopolymers. 1995;36(2):169–179. doi: 10.1002/bip.360360206. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 7.Sung JS, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006;273(8):1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 8.Streisinger G, Okada Y, Emrich J, et al. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat Res. 2006;598(1–2):103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Vasquez KM. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc Natl Acad Sci U S A. 2004;101(37):13448–13453. doi: 10.1073/pnas.0405116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson AL, Chen R, Loeb LA. Induction of microsatellite instability by oxidative DNA damage. Proc Natl Acad Sci U S A. 1998;95(21):12468–12473. doi: 10.1073/pnas.95.21.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 13.Gelfand CA, Plum GE, Grollman AP, Johnson F, Breslauer KJ. Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry. 1998;37(20):7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- 14.Ayadi L, Coulombeau C, Lavery R. Abasic sites in duplex DNA: molecular modeling of sequence-dependent effects on conformation. Biophys J. 1999;77(6):3218–3226. doi: 10.1016/S0006-3495(99)77152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat Res. 2006;166(5):693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- 16.Wilson SH, Sobol RW, Beard WA, Horton JK, Prasad R, Vande Berg BJ. DNA polymerase beta and mammalian base excision repair. Cold Spring Harb Symp Quant Biol. 2000;65:143–155. doi: 10.1101/sqb.2000.65.143. [DOI] [PubMed] [Google Scholar]
- 17.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 18.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci U S A. 1991;88(24):11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269(5224):699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 20.Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem. 1997;272(38):23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 21.Wiederhold L, Leppard JB, Kedar P, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15(2):209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Ye N, Holmquist GP, O’Connor TR. Heterogeneous repair of N-methylpurines at the nucleotide level in normal human cells. J Mol Biol. 1998;284(2):269–285. doi: 10.1006/jmbi.1998.2138. [DOI] [PubMed] [Google Scholar]
- 23.Xia L, Zheng L, Lee HW, et al. Human 3-methyladenine-DNA glycosylase: effect of sequence context on excision, association with PCNA, and stimulation by AP endonuclease. J Mol Biol. 2005;346(5):1259–1274. doi: 10.1016/j.jmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Eftedal I, Guddal PH, Slupphaug G, Volden G, Krokan HE. Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 1993;21(9):2095–2101. doi: 10.1093/nar/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seibert E, Ross JB, Osman R. Role of DNA flexibility in sequence-dependent activity of uracil DNA glycosylase. Biochemistry. 2002;41(36):10976–10984. doi: 10.1021/bi026121o. [DOI] [PubMed] [Google Scholar]
- 26.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401(6750):301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 27.Millar CB, Guy J, Sansom OJ, et al. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297(5580):403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 28.Sibghat U, Gallinari P, Xu YZ, et al. Base analog and neighboring base effects on substrate specificity of recombinant human G:T mismatch-specific thymine DNA-glycosylase. Biochemistry. 1996;35(39):12926–12932. doi: 10.1021/bi961022u. [DOI] [PubMed] [Google Scholar]
- 29.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc Natl Acad Sci U S A. 2008;105(26):8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278(50):49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 31.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36(37):11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 32.Beard WA, Shock DD, Yang XP, DeLauder SF, Wilson SH. Loss of DNA polymerase beta stacking interactions with templating purines, but not pyrimidines, alters catalytic efficiency and fidelity. J Biol Chem. 2002;277(10):8235–8242. doi: 10.1074/jbc.M107286200. [DOI] [PubMed] [Google Scholar]
- 33.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. “Action-at-a-distance” mutagenesis 8-oxo-7, 8-dihydro-2′-deoxyguanosine causes base substitution errors at neighboring template sites when copied by DNA polymerase beta. J Biol Chem. 1999;274(22):15920–15926. doi: 10.1074/jbc.274.22.15920. [DOI] [PubMed] [Google Scholar]
- 34.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3(8):998–1001. [PubMed] [Google Scholar]
- 35.Broderick P, Bagratuni T, Vijayakrishnan J, Lubbe S, Chandler I, Houlston RS. Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer. 2006;6:243. doi: 10.1186/1471-2407-6-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinmura K, Tao H, Goto M, et al. Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer. Carcinogenesis. 2004;25(12):2311–2317. doi: 10.1093/carcin/bgh267. [DOI] [PubMed] [Google Scholar]
- 37.Sugimura H, Kohno T, Wakai K, et al. hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1999;8(8):669–674. [PubMed] [Google Scholar]
- 38.Le Marchand L, Donlon T, Lum-Jones A, Seifried A, Wilkens LR. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11(4):409–412. [PubMed] [Google Scholar]
- 39.Xu J, Zheng SL, Turner A, et al. Associations between hOGG1 sequence variants and prostate cancer susceptibility. Cancer Res. 2002;62(8):2253–2257. [PubMed] [Google Scholar]
- 40.Elahi A, Zheng Z, Park J, Eyring K, McCaffrey T, Lazarus P. The human OGG1 DNA repair enzyme and its association with orolaryngeal cancer risk. Carcinogenesis. 2002;23(7):1229–1234. doi: 10.1093/carcin/23.7.1229. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Elahi A, Pow-Sang J, Lazarus P, Park J. Association between polymorphism of human oxoguanine glycosylase 1 and risk of prostate cancer. J Urol. 2003;170(6 Pt 1):2471–2474. doi: 10.1097/01.ju.0000087498.23008.bb. [DOI] [PubMed] [Google Scholar]
- 42.Park J, Chen L, Tockman MS, Elahi A, Lazarus P. The human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) DNA repair enzyme and its association with lung cancer risk. Pharmacogenetics. 2004;14(2):103–109. doi: 10.1097/00008571-200402000-00004. [DOI] [PubMed] [Google Scholar]
- 43.Zienolddiny S, Campa D, Lind H, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27(3):560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 44.Hill JW, Evans MK. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Res. 2006;34(5):1620–1632. doi: 10.1093/nar/gkl060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sweasy JB, Lang T, Starcevic D, et al. Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. Proc Natl Acad Sci U S A. 2005;102(40):14350–14355. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lang T, Maitra M, Starcevic D, Li SX, Sweasy JB. A DNA polymerase beta mutant from colon cancer cells induces mutations. Proc Natl Acad Sci U S A. 2004;101(16):6074–6079. doi: 10.1073/pnas.0308571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalal S, Hile S, Eckert KA, Sun KW, Starcevic D, Sweasy JB. Prostate-cancer-associated I260M variant of DNA polymerase beta is a sequence-specific mutator. Biochemistry. 2005;44(48):15664–15673. doi: 10.1021/bi051179z. [DOI] [PubMed] [Google Scholar]
- 48.Xie Y, Yang H, Cunanan C, et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64(9):3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 49.Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. Rna. 2008;14(7):1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34(9):2311–2321. [PubMed] [Google Scholar]
- 51.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103(48):18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fishel R, Lescoe MK, Rao MR, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75(5):1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]