Abstract
The breast cancer stem cell (BCSC) hypotheses suggest that breast cancer is derived from a single tumor-initiating cell with stem-like properties, but the source of these cells is unclear. We previously observed that induction of an immune response against an epithelial breast cancer led in vivo to the T cell-dependent outgrowth of a tumor, the cells of which had undergone epithelial to mesenchymal transition (EMT). The resulting mesenchymal tumor cells had a CD24−/loCD44+ phenotype, consistent with BCSCs. In the present study, we found that EMT was induced by CD8 T cells and the resulting tumors had characteristics of BCSCs, including potent tumorigenicity, ability to re-establish an epithelial tumor, and enhanced resistance to drugs and radiation. In contrast to the hierarchal cancer stem cell hypothesis which suggests that breast cancer arises from the transformation of a resident tissue stem cell, our results show that EMT can produce the BCSC phenotype. These findings have several important implications related to disease progression and relapse.
Keywords: EMT, breast cancer stem cells, generation in vivo, chemoresistance, radioresistance
Introduction
A current goal among breast cancer scientists worldwide is to reduce the risk of breast cancer recurrence following therapy (1). This risk, which remains high for at least one decade, suggests that treated patients still maintain a small population of tumorigenic, albeit dormant, cancer stem cells. Although previously described (2), it was not until 2003 that interest in breast cancer stem cells (BCSCs) was renewed (3). In that study, identification of the CD24−/loCD44+ tumorigenic breast cancer cells provided a valuable marker framework for testing hypotheses on BCSC involvement in the clinical course of breast cancer and its pathogenesis.
A prevalent BCSC hypothesis proposes that breast tumors are initiated and maintained by a small fraction of tumorigenic cells (4). What remains unclear about the BCSC hypothesis is the origin of the tumorigenic cells. One hypothesis is that transformed resident tissue stem cells undergo asymmetrical division, occasionally renewing a copy of themselves but most often generating proliferative epithelial daughter progeny with limited tumorigenicity. An alternative hypothesis is that BCSCs are derived from transformed, differentiated epithelial cells by acquisition of stem cell attributes. Determining the origin and biology of BCSCs is important for the development of therapies to reduce the risk of breast cancer relapse.
In prior work, we found that immunoediting of breast tumors in the neu-transgenic (neu-tg) mouse resulted in neu antigen-loss variant tumors (5–8). Our first observation was that the variants had a different proteomic profile and reduced inflammatory and danger signals (7). Subsequent analyses revealed that antigen-loss tumor cells had undergone T cell-dependent epithelial to mesenchymal transition (EMT) (5, 6). EMT, a de-differentiation program that converts epithelial cells into a mesenchymal phenotype, is involved in embryogenesis and used pathologically during cancer progression (9). Since our studies showed that mesenchymal antigen-loss variants were CD24−/lo, we assessed whether these antigen-loss tumor cells had characteristics of CD24−/lo BCSCs observed by Al-Hajj et al. (3, 6). Furthermore, we aimed to determine what T cell subset was involved in the induction of EMT. Herein, we show that EMT induction in vivo involves CD8 T cells and generates BCSCs.
Materials and Methods
Animals
Female neu-transgenic (neu-tg) mice on the FVB background and FVB/N mice were maintained as a colony in accordance with institutional policy.
Cell lines
The epithelial cell line (E), was derived as previously described (10). Four cell lines (M1, M2, M3 and M4) were obtained from relapsed tumors that underwent EMT following injection of E tumor cells into parental non-transgenic FVB/N mice (6). All cell lines were maintained in RPMI 1640 containing 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 1% sodium pyruvate, 2.5% HEPES and 2mM L-glutamine.
In vivo tumorigenicity assays
Neu-tg mice were inoculated, subcutaneously on the mid-dorsum unless otherwise specified, with doses of tumors cells ranging from 100 to 106 cells. Tumors were measured every other day with vernier calipers and volumes were calculated as the product of length × width × height × 0.5236.
Antibodies for in vivo cell depletion
Anti-CD4 (GK1.5) and anti-CD8 (53.6.72) monoclonal antibodies were prepared by the Mayo Antibody Core Facility (Rochester, MN). Mice were injected daily for 5 days with 100μg of antibody intravenously and E tumor cells were injected 48 hours later. For secondary depletion of CD8s, mice previously injected with E tumor cells received 5 more daily doses of anti-CD8 and were rechallenged with tumors.
Proliferation of M3 cells in response to CD8 T cell conditioned media
CD8 T cells were derived from lymph nodes and spleens of naïve FVB mice with a mouse CD8a+ T cell isolation kit per manufacturer’s recommendations using an autoMACS Separator (Miltenyi Biotec Inc Auburn, CA). Two million/well untouched CD8+ T cells were plated in 6 well plates. Unstimulated wells contained CD8s and media. CD8s were treated with 2 million irradiated splenocytes (3300 rads) and 250 μg of concanavalin A (ConA). Anti-CD3/CD28 received 400,000 mouse Dynabeads (Invitrogen, Carlsbad, CA) per well (~1 bead/5 T cells). 100 μl of 18 hour conditioned media were added to M cells previously seeded into 96-well plates. 3H thymidine was added, 0.273 nCi per well; 24 hours later cells were harvested and read on a TopCount NXT scintillation counter (Perkin Elmer, Waltham, MA).
Flow cytometry
Cells were incubated with primary antibodies at 4°C for 20–30 minutes, washed, followed by secondary antibody (if needed) for 20 minutes, washed and fixed with 0.5% formaldehyde. Samples were run on BD FACScan or FACSCalibur flow cytometers (BD Bioscience, San Jose, CA). Sorting of M3 cells was performed on a BD FACSVantage Cell Sorter and data was analyzed using BD CellQuest Pro software (ver. 4.0.2). Occasionally, results depicted the relative mean fluorescent intensity (rMFI) (ratio of specific marker intensity to the isotype or secondary antibody staining).
Flow cytometry reagents
Antibodies and reagents from BD Pharmigen (San Diego, CA) included anti-CD24 FITC (M1/9), anti-CD44 FITC, PE-Cy5 (IM7), anti-Sca1 FITC (D7), anti-Annexin V APC and 7-AAD. Antibodies from eBioscience (San Diego, CA) were anti-CD24 PE (30-F1), anti-CD34 FITC (RAM34), anti-CD133/Prominin I FITC (13A4). Rabbit anti-claudin 3 (Z23.JM) was from Zymed/Invitrogen. The mouse monoclonal antibody against rat neu (7.16.4) was previously described (6). Secondary antibodies from Jackson ImmunoResearch Laboratories (West Grove, PA) were FITC goat anti-rabbit IgG and FITC anti-mouse IgG. Anti-mouse IgG2a/2b FITC (R2–40) was from BD Pharmigen. Isotype antibodies were PE Rat IgG2b (eBioscience), FITC Rat IgG2b (BD Pharmingen) and Pe-Cy5 Rat IgG2b (195-1, BD Pharmingen). BD Cytofix/Cytoperm Fixation/Permeabilization solution kit (BD Pharmingen) was used for Claudin 3 staining.
In vitro coculture of E cells with primed CD8s
FVB mice were injected with 5 million E tumor cells and spleens removed 7 days later to isolate primed CD8 T cells as described above. In 6 well plates, 2.5 × 105 E cells and 2 × 106 CD8 T cells were added per well. CD8 T cells were irradiated to 3300 rads. Cells were co-cultured for 24 hours, T cells washed off, and tumor cells stained for neu and CD24 and analyzed by flow cytometry.
PCR analysis
RNA was isolated using RNeasy (Qiagen, Valencia, CA). PCR primers, selected with MacVector software (MacVector, Inc., Cary, NC) were from Invitrogen. RT-PCR utilized SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen) using 100 or 200 ng of RNA in a Bio-Rad MyCycler. Samples were electrophoresed on 1% agarose gels, imaged on a Gel Doc XR (Bio-Rad, Hercules, CA), and band intensities quantified with Quantity One software (ver. 4.5.2). DNA repair gene expression analysis was done using DNA Damage Signaling RT2 Profiler PCR Array qPCR kit (SuperArray Bioscience, Frederick, MD) according to the manufacturer’s protocol and run on a Mx3005P thermal cycler (Stratagene, La Jolla, CA). Expression was normalized to housekeeping genes and expressed as fold relative to the E cell line.
Cell Photography
40X images of cell lines were taken using an Axiovert 200M inverted microscope and AxioCam HRm camera interfaced with Axiovision software (Carl Zeiss, Thornwood, NY). 20X images of spheroids were obtained with a Leica DC 200 microscope (Leica Microsystems, Wetzlar, Germany) and Fujifilm FinePix 6800 Zoom camera (Fujifilm, Tokyo, Japan).
Histology
M3 and E tumors from neu-tg mice were frozen in O.C.T. Compound (Tissue-Tek, Sakura Finetek USA, Torrance, CA), cryo sectioned and stained with hematoxylin and eosin. 20X images were gathered using a NanoZoomer Digital Pathology (Bacus Laboratories, Inc.) slide scanner and captured using WebSlide Enterprise software. Staining and imaging of frozen tumor sections was performed by the Tissue and Cell Molecular Analysis of the Mayo Clinic Cancer Center using standard techniques.
Western blotting
Protein was isolated from whole cell lysates using cell extraction buffer (Invitrogen) and concentration determined using the BCA assay (Pierce/Thermo Fisher Scientific Inc., Rockford, IL). Eighty to 150 μg of protein was subjected to SDS-PAGE electrophoresis and transferred to PVDF membranes which were blocked for 1 hour with skim milk (3% w/v of PBS with Tween 20: PBS-T) and incubated overnight (4°C) with monoclonal antibodies against BCRP (BXP-21) or PGP (C219) from Calbiochem (Gibbstown, NJ) diluted 1:50 and 1:500, respectively. Membranes were washed and incubated with alkaline phosphatase-conjugated anti-mouse IgG (Chemicon/Millipore, Billerica, MA) for 1 hour (1:8000 dilution) followed by detection with enhanced chemiluminescence (GE Healthcare). Anti-β-actin was employed as loading control (AC-74, Sigma Aldrich, St. Louis, MO).
Chemotherapy assays
Mitoxantrone dihydrochloride was from Sigma Aldrich, BCNU from the Chemical Synthesis Branch, NCI (Bethesda, MD), and Etoposide from Biomol International (Plymouth Meeting, PA). MTT cellular proliferation assays were employed (11) for mitoxantrone chemosensitivity. 5000 E and M cells were plated in 96-well plates in complete medium overnight and thereafter in serum free medium. Cell lines were exposed to increasing concentrations (0, 2.5, 5, 10 and 20 μM) of mitoxantrone (diluted in 95% ethanol) for 72 hours. Media was aspirated from wells and 15 μL/well MTT reagent (3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyltetrazolium bromide, Sigma Aldrich) was added and incubated for 4 h at 37°C followed by 100 μL of stop solution. The following day plate optical densities were measured at 562 nm in an ELISA SpectraMax 190 reader (Molecular Devices, Sunnyvale, California). Percent inhibition of growth was measured as a percent of control (no mitoxantrone). For apoptotic assays, cells were plated and treated with 2 μM etoposide for 24 hours followed by staining with Annexin V and 7-AAD. The percentage of apoptotic cells was determined by flow cytometry as described (12).
Clonogenic assays
500 cells were plated in P35 dishes and treated 24 hours later with irradiation or BCNU. Irradiation was performed with a 137Cs laboratory irradiator to generate a dose curve of 0, 4, 8, 16 and 32 Gy or cells were exposed to increasing doses (0, 2.5, 5, 10 and 20 μM) of BCNU. After 8 days, cells were washed, stained with crystal violet, and colonies counted.
MMP activity assay
The EnzoLyte 520 Generic MMP Fluorometric Assay Kit (AnaSpec, San Jose, CA) was used according to manufacturer’s directions. Cell culture supernatants were incubated with 1mM APMA for 24 hours at 37°C to activate pro-MMPs. Fluorescence was measured at 490/520nm Ex/Em using a Victor V plate reader (Perkin Elmer, Waltham, MA).
RNA microarray
Affymetrix microarray analysis was done by the Mayo Advanced Genomic Technology Center (Rochester, MN) as previously described (6) using GeneChip Mouse Genome 430 2.0 arrays. The expression analysis was performed with Affymetrix Genechip Operating Software with a scaling target signal of 500 and normalization of 1. Microarray data is posted on Gene Expression Omnibus as GSE13259.
Spheroids
Cells from monolayer cultures were harvested and resuspended at a density of 4 × 104 viable cells per milliliter in serum-free MEBM (Cambrex Bioscience Walkersville Inc, MD) supplemented with 2% B27 (Invitrogen), 20ng/mL EGF (BD Biosciences), 100 μL mercaptoethanol, 4μg/mL insulin (Sigma), 1 ng/mL hydrocortisone, 1% penicillin/streptomycin, 0.25 μg/mL amphotericin B, 20 μg/mL gentamicin sulfate and seeded into 10 cm dishes coated with 1% agarose. Cultures were fed weekly.
Statistical analyses
Statistical analysis was performed using Prism 4 for Windows (version 4.03, GraphPad Software, San Diego, CA). Data were analyzed using one-way ANOVA when comparing E with each of the M cell lines with the Bonferroni multiple comparison test (Kruskal-Wallis as non-parametric) or one-tailed Student’s t-tests. Statistical significance was set at p≤0.05. The number of replicates is indicated with each figure.
Results
Induction of EMT in vivo requires CD8 T cells
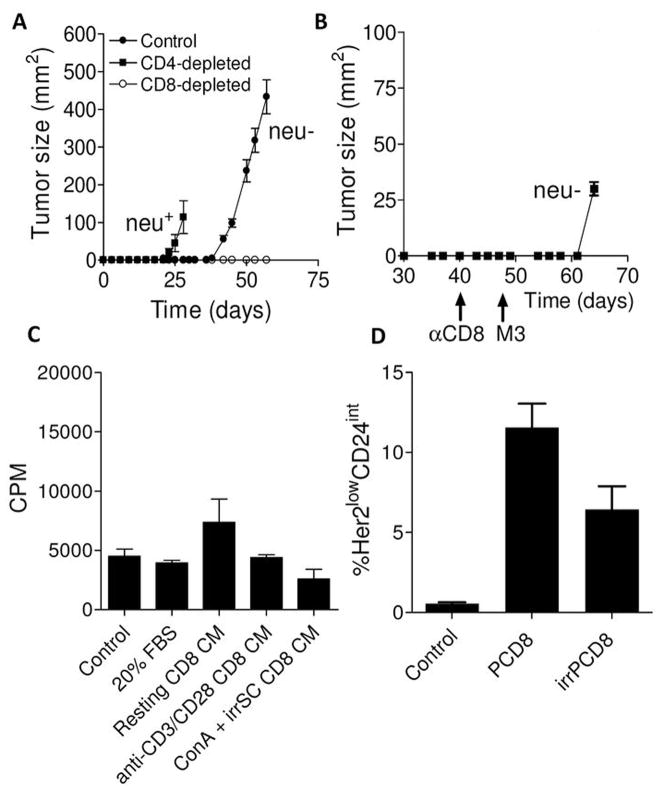
Immune rejection of neu+ epithelial (E) breast tumors in the FVB/N mouse, where neu is a foreign antigen, results in T cell- and IFN-γ-dependent induction of neu antigen-loss (neu−) variant tumors that have undergone EMT (5, 6). To determine which T cell subset was responsible, we depleted CD4 or CD8 T cells from FVB/N mice prior to challenge with neu+ E tumor cells. Control mice developed tumors around 40 days while those depleted of CD4 T cells developed tumors around 20 days (Figure 1A) (6). Consistent with prior work, control mice developed neu− variants while those depleted of CD4 T cells had neu+ tumors (not shown). Mice depleted of CD8 T cells, however, remained disease-free (Figure 1A). Long-term observation (>100 days) suggested complete tumor rejection (not shown). Four possible reasons could explain why tumors did not grow in CD8 T cell-depleted mice. The first is that CD8 T cell depletion permitted a more robust immune response against neu− EMT variants. To test this, CD8 T cell-depleted mice that rejected tumor were rechallenged with neu− EMT variants. If robust immunity against neu− EMT variants existed, these mice should reject tumor, which we did not observe (Figure 1B). The second possible reason is that CD8 T cells preferentially support enhanced growth of the neu− EMT variants contained as a small population within the E cell line. To test this, conditioned culture medium from CD8 T cells was applied to neu− EMT variants in vitro. Neu− EMT variant tumor cells exposed to conditioned medium did not grow any better than those incubated with control medium (p>0.05), ruling out the likelihood that growth of these cells was enhanced by growth factors derived from CD8 T cells (Figure 1C). A third possibility is that CD8 T cells promote the malignant transformation of stromal cells that associate with the nascent tumor. This is ruled out because these mesenchymal tumors contain the neu oncogene, which is not in the FVB/N genome (5). The only remaining possibility is that CD8 T cells directly induce neu− EMT variants. These findings, along with prior work, demonstrate that the generation of neu− EMT variants in the FVB/N mice is an active rather than a selective process (5). To further support this, we found that E cells cultured with tumor-primed CD8 T cells lost neu and CD24 expression (p=0.0009) (Figure 1D). This was partially ablated by prior irradiation of the CD8 T cells, which is known to blunt T cell proliferation and cytokine production. Naïve CD8 T cells failed to generate antigen-negative tumor cells (not shown).
Figure 1. Induction of EMT in vivo requires CD8 T cells.
Panel A shows tumor growth in control, CD4-depleted, and CD8-depleted mice. Each point is the mean (s.e.m.) of measurements from 3 tumors. A repeat experiment yielded identical results. Panel B, tumor growth (mean and s.e.m., n=3) in CD8-depleted mice that had been injected with parental E cells on day 0 and rechallenged at day 47 with antigen-loss variant tumor cells (M3). Panel C, shown are the proliferation rates (counts per minutes incorporation of thymidine) of antigen-negative tumor cells incubated with RPMI-1640 + 10% FBS (control medium), medium with 20% FBS, conditioned medium from resting CD8 T cells (resting CD8 CM), conditioned medium from anti-CD3/anti-CD28 stimulated T cells, or conditioned medium from ConA-stimulated T cells cocultured with irradiated splenocytes. Each bar is the mean (s.e.m., n=3). Panel D, shown are the percentages of tumor cells which lost neu and had reduced CD24 (epithelial marker) expression following exposure of epithelial (E) tumor cells to control (regular) medium, tumor-primed CD8 T cells (PCD8), or irradiated tumor-primed CD8 T cells (irPCD8). Each bar is the mean (s.e.m.) of 3 separate samples.
Stable mesenchymal cell lines were established
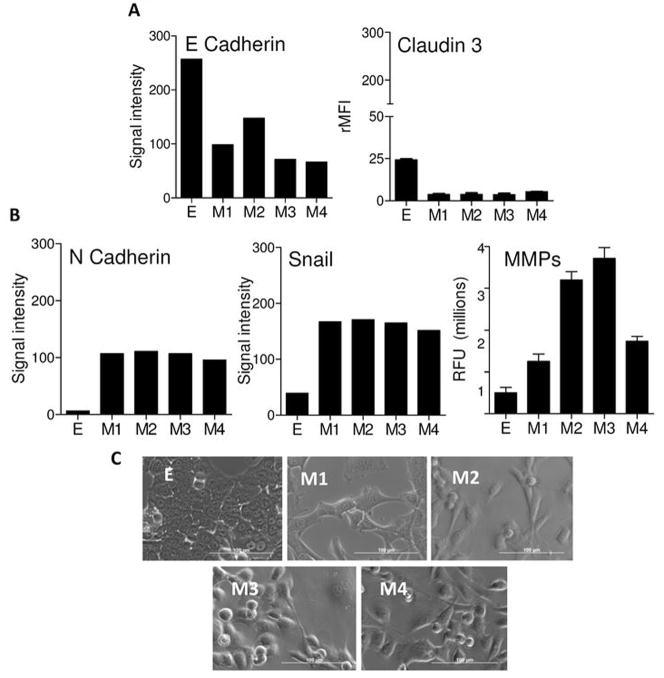
Four neu− EMT variant cell lines (M1, M2, M3, and M4) were established from 4 different tumors. All were neu− as assessed by RT-PCR (not shown). Consistent with having undergone EMT, the M cell lines had significantly reduced levels of the epithelial markers E-cadherin and claudin 3 (p=0.0001) (Figure 2A). Moreover, mesenchymal markers N-cadherin and Snail were upregulated in M cell lines (Figure 2B). Increased mesenchymal function, as measured by matrix metalloproteinase (MMP) activity, was increased significantly (p<0.01) in all lines, except M1 relative to the parental E cell line (Figure 2B). Visual inspection (Figure 2C) showed that M cell lines had a scattered, spindle-shaped appearance while the E cell line had strong cellular junctions with a cobblestone appearance. FISH karyotyping revealed that each M cell line contained, like the parental E cells, a single integration site for rat neu within chromosome 3 (Supplementary Figure 1).
Figure 2. Stable cell lines from tumor cells that underwent EMT were established.
Panel A shows expression levels of E-cadherin and claudin 3 in E and M cell lines. The E-cadherin experiments were repeated independently 3 times with similar results. Claudin 3 was measured using flow cytometry and each bar is the mean (s.e.m.) of 2 separate experiments. Panel B shows expression of N-cadherin, and Snail, and matrix metalloproteinase (MMP) activity for all cell lines. The bars represent the signal intensity derived using RT-PCR. RT-PCR results are representative of 2 experiments that gave nearly identical results. For MMP activity each bar is the mean (s.e.m.) of 3 samples. Panel C shows light microscope pictures (40X) of the cell lines. Bar=100 μm.
Mesenchymal cells are CD24−/loCD44+ and highly tumorigenic
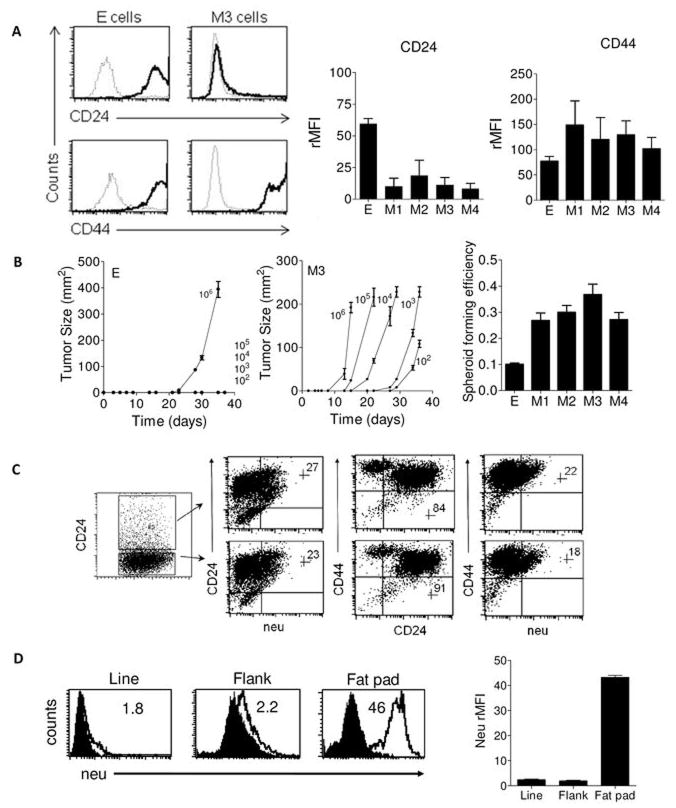
Human BCSCs have been distinguished from non-tumorigenic cells by the cell surface marker profile CD24−/loCD44+ (3). Since our prior work showed that CD24 was down-regulated in neu− EMT variant tumors, we hypothesized that M cells may have a BCSC profile. As shown in Figure 3A, the M cell lines were largely CD24−/lo and CD44+ in contrast to the CD24+CD44+ E cells. CD24 expression was significantly lower in all M cells relative to the E cells (p=0.004). CD34, a sialomucin expressed on mesenchymal stem cells (13), was not significantly elevated (p=0.63) in M cell lines relative to E cells (Supplementary Figure 2). Consistent with prior work, the M cells had low and intermediate expression of stem cell antigen-1 (Sca-1), although elevated relative to E cells it was not statistically significant (p=0.23) (Supplementary Figure 2). CD133, a marker of hematopoietic stem cells, was also examined given its association with human cancer stem cells (CSCs) (14). Staining revealed similarly low expression in both tumor types (p=0.74) (Supplementary Figure 2).
Figure 3. Mesenchymal cells are CD24−/loCD44+ and highly tumorigenic.
Panel A shows a representative histogram demonstrating that mesenchymal (M) tumor cells are CD24−/lo CD44+. Dark and light tracings represent specific and isotype fluorescence, respectively. Panel A also shows graphs that summarize CD24 and CD44 expression in the E and M cells. Shown are the average (± s.e.m.) relative mean fluorescent intensities (rMFI) calculated from 3 experiments. Panel B shows tumor growth curves of the E (left) and M3 (right) cell lines injected into neu-tg mice. Each data point is the mean (s.e.m.) tumor size calculated from three mice. Numbers indicate the dose of cells injected. On the right, images of the tumors (20X) and spheroids (20X) corresponding to E and M3 cell lines. The far right graph shows the mean (± s.e.m, n=5 independent experiments) spheroid forming efficiency of each line. Panel C (left) is a summary of dot plots from CD24, CD44, and neu expression in tumors derived from sorted CD24−CD44+ M3 mesenchymal cells. This experiment is representative of 4 similar experiments done with each of the mesenchymal cells lines. Quadrants were established using M3 cell line cells (not shown). Inset numbers indicate percentage of gated cells in upper right quadrant. Panel D shows histograms of neu expression in cells from a mesenchymal cell line (Line), flank tumors (Flank), and mammary fat pad tumors (Fat pad) are shown. Inset numbers represent the relative mean fluorescent intensity. Nearly identical results were seen with 4 different samples of each cell type which is shown averaged in accompanying graph.
A key property of CSCs is their ability to seed tumors at very low numbers. In vivo studies showed that M tumor cells were far more tumorigenic than E cells (Figure 3B, Table). M cells formed tumors in 100% of mice at doses of ≥1000 cells, and M3 formed tumors with as few as 100 cells. E cells only formed tumors when 106 cells were injected (Figure 3B, Table). Despite large differences in tumorigenicity, both cell types were able to form spheroids in vitro under conditions that prevented attachment (Supplementary Figure 3). However, the M cells generated spheroids with greater efficiency than E cells (p<0.0001)(Figure 3B).
Table 1. Santisteban Table.
Frequency of tumor development following injection of E and M cell lines
| Dose of cells injected* | |||||
|---|---|---|---|---|---|
| Cell line | 106 | 105 | 104 | 103 | 102 |
| E | 6/6† | 0/6 | 0/6 | 0/6 | 0/6 |
| M1 | n.d. | n.d. | 3/3 | 3/3 | 0/3 |
| M2 | n.d. | n.d. | 3/3 | 3/3 | 0/3 |
| M3 | 6/6 | 6/6 | 6/6 | 6/6 | 3/6 |
| M4 | n.d. | n.d. | 3/3 | 3/3 | 0/3 |
Tumors were injected subcutaneously;
Numerator represents number of mice developing tumor; Denominator is number of mice injected; n.d.: not done.
Mesenchymal tumor cells generate neu+CD24hiCD44+ tumors in neu-tg mice
Another unique feature of BCSCs is an ability to give rise to epithelial progeny that constitute the bulk of a tumor. To examine whether the mesenchymal cells could reconstitute an epithelial tumor, M tumor cells were injected into the neu-tg mouse, which carries the neu transgene and will not reject neu+ tumors. Since some M cells showed a minor population with moderate CD24 expression, we sorted CD24− tumor cells from CD24lo/int tumor cells. Both fractions formed tumors upon injection of 105, 104 or 103 cells with nearly equal efficacy (not shown). M cells were able to reconstitute tumors that regained CD24 expression and maintained high CD44 expression levels (Figure 3C). Histologic analysis of tumors from E and M3 cells show no remarkable differences (Supplementary Figure 4). However, neu expression was poorly upregulated, a finding consistent with our prior study showing that the neu-transgene MMTV promoter in the neu− EMT variant tumors is heavily methylated (5). Indeed, neu expression could be regained by injection of M tumor cells into the mammary fat pad, a site of preferential activation of the MMTV promoter. In fact, M1 cells regained complete neu expression in the mammary fat pad but not in the flank (p= 0.002) (Figures 3C). CD24 and neu expression were also induced, albeit moderately, in M1 cells grown under spheroid forming conditions (Supplementary Figure 5).
Mesenchymal cell lines have elevated expression of drug pumps and DNA repair enzymes and are resistant to cytotoxic agents
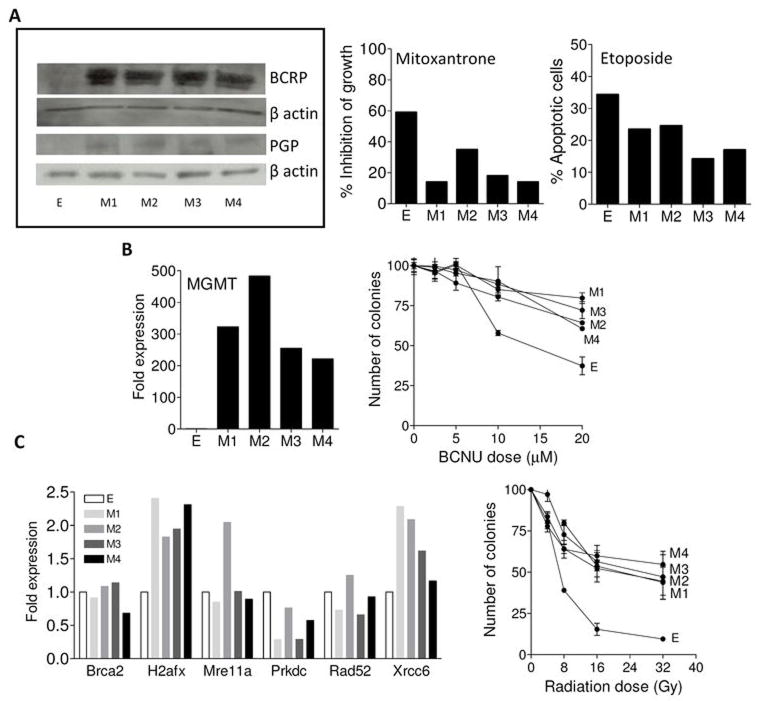
Another hallmark of BCSCs is their ability to resist environmental insults. Both mesenchymal cells and BCSCs have been linked to resistance to drugs and radiation in recent studies (15). Cytotoxic resistance is often due to elevated expression of drug pumps. Two pumps associated with resistance in breast cancer are BCRP and PGP (16). We observed higher expression of both pumps in mesenchymal cells relative to parental cells (Figure 4A). Enhanced chemoresistance of M cells to mitoxantrone or etoposide (substrates for BCRP and PGP, respectively) suggests that these pumps are active (Figure 4A).
Figure 4. Mesenchymal cell lines have elevated expression of drug pumps, DNA repair enzymes and are resistant to environmental insults.
Panel A (left) shows western blot analysis of BCRP and PGP drug pumps in all cell lines. β actin was used as a loading control. Western blots are representative of three experiments for each protein. Panel A (middle) shows inhibition of growth (i.e. compared to control treated cells) of the tumor cells lines after in vitro treatment with mitoxantrone. Panel A (right) shows the apoptotic response to etoposide as assessed using flow cytometry. Each bar is expressed as the % apoptotic cells following treatment. A representative experiment is shown for MTT and apoptotic assays, both performed twice. Panel B (left) depicts the expression levels of MGMT in the tumor cell lines by real time PCR. Results are expressed as fold expression over E and represent one experiment. Panel B (right) shows the results of a clonogenic assay of the tumor cells following exposure to escalating doses of BCNU. Each data point is the mean (s.e.m.) of 3 replicates. Panel C (left) shows the results of real-time PCR analysis of the double stranded DNA repair genes. Results are expressed as fold expression over E. Panel C (right) shows the results of clonogenic assay after escalating doses of gamma irradiation. Each data point is the mean (± s.e.m.) of 3 replicate plates.
Our mesenchymal tumor cells also have elevated expression of key DNA repair enzymes. One enzyme involved in drug resistance in human cancers is O6-methylguanine-DNA methyltransferase (MGMT), a ubiquitous DNA repair protein that removes O6-alkyl-guanine lesions from damaged DNA and contributes to the resistance of brain cancers to α-chloro-nitrosourea (BCNU) (17). Figure 4B shows that M cells have high expression of MGMT (>200 fold higher relative to E cells) and are resistant to BCNU treatment (p=0.0002 at 20μM). Since human BCSCs are resistant to ionizing radiation (18), we evaluated this and expression of double-stranded DNA repair pathway components (i.e. Brca2, H2afx, Mre11a, Prkdc, Rad52, and Xrcc6). Relative to E cells, M tumor cell lines showed elevated expression of H2afx and Xrcc6 (Figure 4C). Activation of H2afx enhances DNA repair efficiency (19) and the KU-70 protein (encoded by Xrcc6) has a key role by recruiting the DNA-dependent protein kinase (20). In general, levels of double-stranded DNA repair genes were similar to E cells, except Prkdc, the gene encoding the catalytic subunit of the DNA-dependent protein kinase (DNA-PK), whose expression was reduced in two M cell lines. Nonetheless, this altered expression of the double-stranded DNA repair pathway was associated with strong resistance of M cells to γ-irradiation (p=0.003 at 16Gy) (Figure 4C). Additional irradiation experiments demonstrated E cells were more sensitive to irradiation as compared to M cells, when assessed by apoptosis assays (not shown).
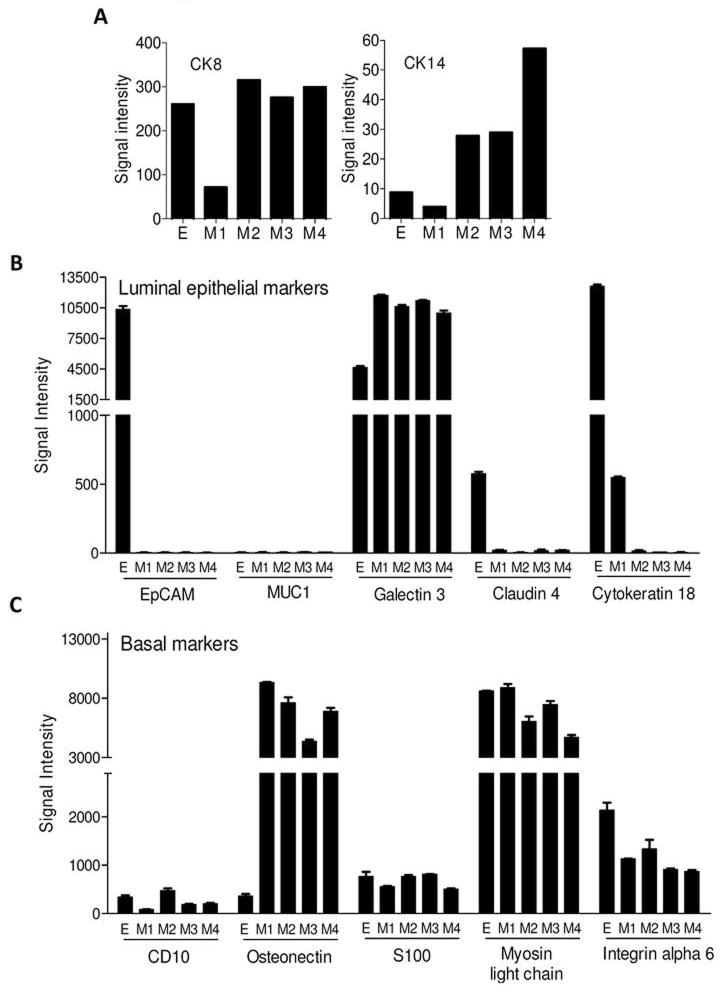
Gene expression of luminal or basal epithelial markers in mesenchymal cells suggests that EMT is incomplete
The normal mammary epithelium contains two layers, basal and luminal. These layers are distinguished by unique cytokeratins (Cks) (21). Ck5 and Ck14 are expressed by the basal layer and Ck8 and Ck18 by the luminal layer (21, 22). Since Ck expression is confined to epithelial cells, we investigated whether EMT was complete based on loss of Ck expression in our cell lines. E tumor cells have the characteristic signature of high Ck8 and low Ck14 expression consistent with their luminal differentiation (Figure 5A). There was no consistent pattern of modulation of Cks observed in the mesenchymal cells. While the M1 cell line showed loss of Ck8 and maintained low level Ck14 expression, M2, M3, and M4 cell lines maintained Ck8 and increased Ck14 expression. Other frequently cited markers of luminal (EpCAM, MUC1, galectin 3, claudin 4 and Ck18) and basal (CD10, osteonectin, S100, myosin light chain and integrin alpha 6) epithelial phenotypes were also assessed (Figures 5B–C). Of the luminal markers, EpCAM (p<0.0001) and claudin 4 (p<0.0001) were consistently downregulated in all M lines relative to the E lines and MUC1 was not expressed by any line. Like Ck8, Ck18 expression was inconsistent. Of the basal markers, only osteonectin was convincingly elevated (p<0.0001), an expected finding given its expression in mesenchymal stem cells (23) (Figure 5C). The other basal markers did not change and failed to distinguish E from M tumor cells. Collectively, the results show that EMT is incomplete.
Figure 5. Gene expression of luminal or basal epithelial markers in mesenchymal cells suggests that EMT is incomplete.
Panel A shows the RT-PCR results depicting expression of CK8 (left) and CK14 (right) in the tumor cell lines. Results are representative of 3 independent experiments. Panels B and C show RNA microarray analyses of common luminal epithelial and basal markers in the cell lines, respectively. Shown are the mean (s.e.m.) signal intensities of 2 microarray analyses for each cell line. Each bar represents the signal intensity for a unique probe.
Discussion
While great strides in breast cancer treatment have been made, women treated for the disease remain at high risk for recurrence. Recent hypotheses suggest that recurrence (and possibly tumor initiation) is caused by a subset of tumor cells with stem cell qualities including self renewal, ability to differentiate and reconstitute a tumor, and resistance to chemotherapeutic drugs and radiation. While studies have confirmed the existence of this tumorigenic subset, their origin is unclear. Understanding the origin and biology of these cells may reveal strategies for targeting them therapeutically. In this study, we made three novel observations which provide an additional framework for BCSCs. These observations are that (1) EMT was induced in vivo by CD8 T cells, (2) EMT generates mesenchymal tumor cells with BCSC properties, and (3) EMT (and BCSC genesis) is not associated with complete loss of epithelial characteristics.
Although much evidence demonstrates the importance of EMT in embryogenesis, investigation of its role in cancer has been confined mainly to in vitro model systems. As a result, the relevance of natural EMT to in vivo malignancy is controversial. In the current study, we found that EMT was induced in vivo without prior manipulation of tumor cells. Our conclusion that the tumor cells had undergone EMT was based on a number of molecular and cellular changes including (1) loss of the epithelial markers CD24, E-cadherin, and claudin 3, (2) gain of validated mesenchymal markers N-cadherin and Snail, (3) gain of matrix metalloproteinase activity, and (4) acquisition of a scattered phenotype. Prior studies showed that mesenchymal tumors develop in the neu-tg mouse, but it remained unclear whether the appearance of the mesenchymal tumors was due to selection or induction (5, 6, 24). Our findings clarify this by showing CD8 T cell-dependent EMT induction in vivo.
Various immune effector cells are known to produce factors capable of inducing EMT, such as TGF-β and TNF-α (25, 26). Of those, the notable example is TGF-β. While CD4 T regulatory cells are the predominant source of TGF-β among T cells, newer studies show that chronically-stimulated effector CD8 T cells can also become regulatory and produce TGF-β (27). Another potential mediator is the recently discovered product of the FAM3C gene, interleukin-like EMT inducer (ILEI). Although little is known of ILEI, it appears to be highly expressed in lymphocytes associated with chronic inflammatory lesions (e.g. arthritis and tumors) (24). Bates and colleagues observed that TGF-β and TNF-α synergistically induced EMT in human colon cancer cells (25). The requirement of multiple stimuli for EMT is supported by our prior studies showing that engagement of the IFN-γ receptor is necessary for the generation of antigen-loss variants in vivo, but IFN-γ stimulation in vitro leads only to a modest loss of neu antigen (5). Whatever mechanism is operative, the induction of EMT and the generation of the BCSC phenotype were observed in vivo, suggesting that our results may be biologically relevant events. Our findings in mice are also consistent with prior studies which showed that CD8 T cell infiltration is associated with lymph node metastasis in breast cancer patients (28).
EMT was accompanied by the acquisition of BCSCs’ properties, including tumorigenicity, resistance to environmental insults, ability to re-differentiate into an epithelial tumor, and ability to form spheroids. These findings are consistent with those of Mani and colleagues who recently showed that forced expression of EMT-associated molecules such as Snail and Twist or treatment with TGF-β resulted in cells with a CSC phenotype (29). Similarly, Morel et al. with the same cell lines demonstrated that adding active Ras generated a population of CD24−CD44+ cells that underwent EMT and had BCSC attributes (30). One key characteristic of CSCs called into question by our studies is the need for self renewal, a property that ensures preservation of CSCs in the tumor. Our hypothesis offers an alternative to self renewal, suggesting that a tumor cell with cancer stem cell properties could be generated from differentiated epithelial tumor cells through the aberrant use of EMT. This premise of de-differentiation is supported by the recently described generation of pluripotent stem cells from seemingly terminally-differentiated somatic cells (31, 32). Inducing a CSC from differentiated daughter cells could likely explain the shortcomings of the hierarchal self-renewing BCSC hypothesis. That hypothesis does not easily explain why relapse is closely associated with metastasis and invasion, features not readily linked to normal mammary stem cells (33, 34). Based on the current study and hallmarks of these prior studies, a likely alternative explanation for relapse and disease progression is that a few of the tumor cells within the primary tumor separate from the primary lesion using EMT and are not eliminated by conventional therapies, due in part to physical separation but primarily to the inherent drug and radio resistance of the mesenchymal tumor cells. Such a hypothesis is supported by Moody et al. which showed that the EMT inducer Snail is linked to relapse in human breast cancer (24). EMT might also explain why relapse in breast cancer is linked with a wound response signature (35). Mesenchymal tumor cells may have several characteristics in common with normal healthy mesenchymal stem cells, which have roles in the wound repair and healing response (36).
Stemness has been associated with the CD24+ phenotype in murine mammary models including CD24+CD29hi (37) and CD24+Sca1− (21). In contrast to these data, we have found stemness associated with the CD24−/lo cells. Although not directly addressed by our study, there are at least two possibilities to explain these discrepancies. First there may be multiple types of cells, generated during malignant transformation and progression that have BCSC-like properties. Secondly, markers used to identify BCSC traits may be variable and reflect the context or microenvironment from which the cells were derived as has been described by Bissell and others (38).
Although our results demonstrate that the tumor cells underwent EMT, as assessed using well-established markers, gene transcription analysis suggests that EMT was incomplete because luminal and basal epithelial associated genes remained expressed. These results are consistent with observations that EMT is associated with some but not all genetic changes ordinarily associated with the stromal or mesenchymal phenotype (39). In other words, EMT may be incomplete or aberrant. Our results are also consistent with a recent study in human breast cancer that revealed that a small fraction (<10%) of the disseminated Ck expressing tumor cells demonstrate the CD24−/loCD44+ BCSC phenotype (40). The implications of our findings could be important for identifying evidence of EMT in breast cancer lesions with strict marker paradigms that include the absence of Cks and E-cadherin expression.
Supplementary Material
Acknowledgments
This work was supported by generous gifts from Martha and Bruce Atwater (KLK, LH), the Mayo Breast SPORE, P50-CA116201 (JI, KLK), K01-CA100764 (KLK), and R01-CA113861 (KLK, SF). We thank the Cytogenetics Shared Resource at Mayo Clinic.
References
- 1.Group EBCTC. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Pierce GB, Nakane PK, Martinez-Hernandez A, Ward JM. Ultrastructural comparison of differentiation of stem cells of murine adenocarcinomas of colon and breast with their normal counterparts. J Natl Cancer Inst. 1977;58:1329–45. doi: 10.1093/jnci/58.5.1329. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho RW, Wang X, Diehn M, et al. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells. 2008;26:364–71. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- 5.Kmieciak M, Knutson KL, Dumur CI, Manjili MH. HER-2/neu antigen loss and relapse of mouse mammary carcinoma are actively induced by T cell-mediated anti-tumor responses. Eur J Immunol. 2007;37:675–85. doi: 10.1002/eji.200636639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knutson KL, Lu H, Stone B, et al. Immunoediting of cancers may lead to epithelial to mesenchymal transition. J Immunol. 2006;177:1526–33. doi: 10.4049/jimmunol.177.3.1526. [DOI] [PubMed] [Google Scholar]
- 7.Manjili MH, Arnouk H, Knutson KL, et al. Emergence of immune escape variant of mammary tumors that has distinct proteomic profile and a reduced ability to induce “danger signals”. Breast Cancer Res Treat. 2006;96:233–41. doi: 10.1007/s10549-005-9044-4. [DOI] [PubMed] [Google Scholar]
- 8.Worschech A, Kmieciak M, Knutson KL, et al. Signatures associated with rejection or recurrence in HER-2/neu-positive mammary tumors. Cancer Res. 2008;68:2436–46. doi: 10.1158/0008-5472.CAN-07-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 10.Knutson KL, Almand B, Dang Y, Disis ML. Neu antigen-negative variants can be generated after neu-specific antibody therapy in neu transgenic mice. Cancer Res. 2004;64:1146–51. doi: 10.1158/0008-5472.can-03-0173. [DOI] [PubMed] [Google Scholar]
- 11.Parrizas M, Saltiel AR, LeRoith D. Insulin-like growth factor 1 inhibits apoptosis using the phosphatidylinositol 3′-kinase and mitogen-activated protein kinase pathways. J Biol Chem. 1997;272:154–61. doi: 10.1074/jbc.272.1.154. [DOI] [PubMed] [Google Scholar]
- 12.Herault O, Colombat P, Domenech J, et al. A rapid single-laser flow cytometric method for discrimination of early apoptotic cells in a heterogenous cell population. Br J Haematol. 1999;104:530–7. doi: 10.1046/j.1365-2141.1999.01203.x. [DOI] [PubMed] [Google Scholar]
- 13.Nadri S, Soleimani M. Isolation murine mesenchymal stem cells by positive selection. In Vitro Cell Dev Biol Anim. 2007;43:276–82. doi: 10.1007/s11626-007-9041-5. [DOI] [PubMed] [Google Scholar]
- 14.Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004;117:3539–45. doi: 10.1242/jcs.01222. [DOI] [PubMed] [Google Scholar]
- 15.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 16.Hait WN, Yang JM. Clinical management of recurrent breast cancer: development of multidrug resistance (MDR) and strategies to circumvent it. Semin Oncol. 2005;32:S16–21. doi: 10.1053/j.seminoncol.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Gerson SL. Targeted modulation of MGMT: clinical implications. Clin Cancer Res. 2006;12:328–31. doi: 10.1158/1078-0432.CCR-05-2543. [DOI] [PubMed] [Google Scholar]
- 18.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–85. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 19.Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: histone modifications and the DNA damage response. Cell. 2005;121:973–6. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol Rev. 2004;200:132–41. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu JC, Deng T, Lehal RS, Kim J, Zacksenhaus E. Identification of tumorsphere- and tumor-initiating cells in HER2/Neu-induced mammary tumors. Cancer Res. 2007;67:8671–81. doi: 10.1158/0008-5472.CAN-07-1486. [DOI] [PubMed] [Google Scholar]
- 22.Deugnier MA, Faraldo MM, Janji B, Rousselle P, Thiery JP, Glukhova MA. EGF controls the in vivo developmental potential of a mammary epithelial cell line possessing progenitor properties. J Cell Biol. 2002;159:453–63. doi: 10.1083/jcb.200207138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva WA, Jr, Covas DT, Panepucci RA, et al. The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells. 2003;21:661–9. doi: 10.1634/stemcells.21-6-661. [DOI] [PubMed] [Google Scholar]
- 24.Moody SE, Perez D, Pan TC, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Bates RC, Mercurio AM. Tumor necrosis factor-alpha stimulates the epithelial-to-mesenchymal transition of human colonic organoids. Mol Biol Cell. 2003;14:1790–800. doi: 10.1091/mbc.E02-09-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–74. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 27.Mahic M, Henjum K, Yaqub S, et al. Generation of highly suppressive adaptive CD8(+)CD25(+)FOXP3(+) regulatory T cells by continuous antigen stimulation. Eur J Immunol. 2008;38:640–6. doi: 10.1002/eji.200737529. [DOI] [PubMed] [Google Scholar]
- 28.Bilik R, Mor C, Hazaz B, Moroz C. Characterization of T-lymphocyte subpopulations infiltrating primary breast cancer. Cancer Immunol Immunother. 1989;28:143–7. doi: 10.1007/BF00199115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 32.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 33.Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–26. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 35.Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galie M, Konstantinidou G, Peroni D, et al. Mesenchymal stem cells share molecular signature with mesenchymal tumor cells and favor early tumor growth in syngeneic mice. Oncogene. 2008;27:2542–51. doi: 10.1038/sj.onc.1210920. [DOI] [PubMed] [Google Scholar]
- 37.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 38.Bissell MJ, Labarge MA. Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell. 2005;7:17–23. doi: 10.1016/j.ccr.2004.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turley EA, Veiseh M, Radisky DC, Bissell MJ. Mechanisms of disease: epithelial-mesenchymal transition--does cellular plasticity fuel neoplastic progression? Nat Clin Pract Oncol. 2008;5:280–90. doi: 10.1038/ncponc1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balic M, Lin H, Young L, et al. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res. 2006;12:5615–21. doi: 10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.