Abstract
In addition to the rhodopsin crystal structure, high-resolution crystal structures of ligand-mediated G-protein-coupled receptors (GPCRs) have recently become available, and these have become attractive templates for developing homology models of several GPCRs of therapeutic interest. These crystal structures and the homology models derived from them have provided significant insights into ligand–receptor interactions. Moreover, several studies have demonstrated that the structural models are indeed suitable for virtual screening of compound databases to identify new ligands for various GPCRs. Recent examples of such virtual screening against GPCRs are discussed in this review.
Key words: GPCRs, homology modeling, virtual screening
INTRODUCTION
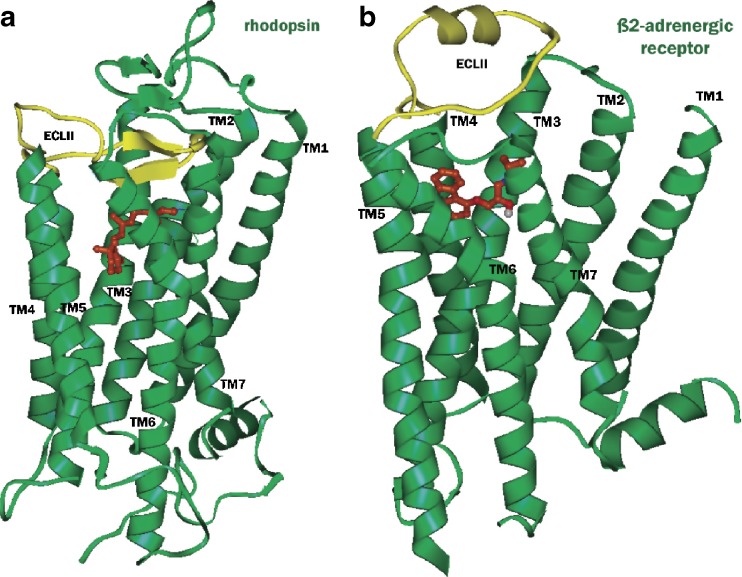
G-protein-coupled receptors (GPCRs) are the targets of 60–70% of the drugs in development today. In the past, drug discovery for GPCRs has primarily utilized ligand-based methods due to the absence of experimental structural information. However, the availability of the crystal structures of GPCRs coupled with advances in homology-based and ab initio modeling methods have led to an increase in the use of structure-based methods for drug discovery. The first mammalian GPCR for which experimental 3D structure was available was the bovine rhodopsin (1,2). Significant advances in protein expression and crystallization techniques have led to the determination of X-ray crystal structures of several additional GPCRs (3–9). The human β2-adrenergic receptor was the first non-rhodopsin GPCR to be cloned, and the X-ray structures of this receptor have provided the much needed templates for ligand-gated GPCRs. More recently, the X-ray structure of the β1-adrenergic receptor has also been published. A significant difference between rhodopsin containing the covalently bound retinal and the β2-adrenergic receptor containing the diffusible ligand carazolol is found in the second extracellular loop (ECL2, Fig. 1). The homology models developed from GPCR crystal structures have generally found widespread use in the analysis of mutational data, mapping ligand binding sites and docking of known ligands to gain insight into ligand–receptor interactions. In addition, results from several studies have demonstrated that homology models of GPCRs are also suitable for structure-based virtual screening for identification of novel agonist and antagonist ligands. Some of these efforts on structure-based virtual screening using homology models of GPCRs have been discussed in earlier reviews (10–14). Highlighted in this mini-review are recent examples of structure-based virtual screening studies on various GPCRs.
Fig. 1.
Crystal structures of retinal-bound rhodopsin (a, PDB id 1U19) and carazolol-bound β2-adrenergic receptor (b, PDB id 2RH1). The ligands retinal and carazolol are shown in ball and stick representation with carbon atoms colored brown, while the extracellular loops 2 (ECL2) are shown in yellow. In rhodopsin, ECL2 and the N terminus form a lid over the ligand-binding pocket, whereas in β2-adrenergic receptor the ECL2 contains an extrahelical segment and is more exposed to the solvent, giving open access to the ligand-binding pocket
β2-ADRENERGIC RECEPTOR
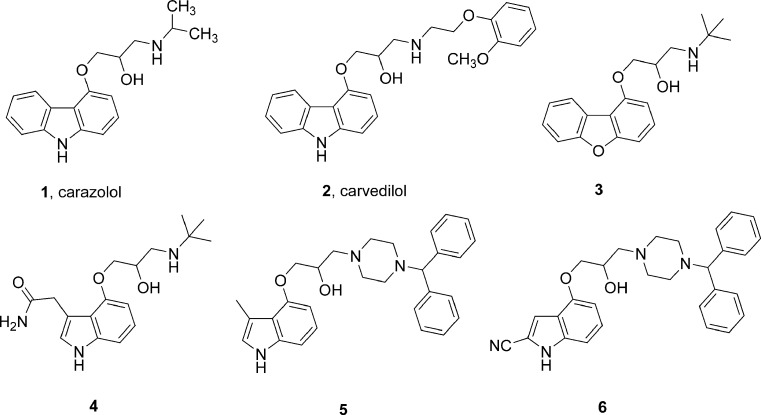
Topiol and Sabio have explored the potential utility of the recently reported X-ray structure of β2-adrenergic receptor for computer-aided drug design (15,16). In an initial validation, they used Glide-XP and GOLD-GoldScore docking tools to dock the S isomer of carazolol (1, Fig. 2) into the ligand binding site of the receptor and found that both the protocols were able to produce binding poses similar to that found in its co-crystal structure. In addition, the docking of six known beta blockers using Glide-XP gave binding poses that were consistent with expected interactions with several key residues. Following this validation, they performed virtual screening of an in-house database of approximately 400,000 compounds using Glide. The effectiveness of the protocol in docking and ranking was demonstrated by its ability to retrieve both carazolol and carvedilol (2) in the top-scoring 30 compounds. Among the top ranked 100 compounds, there were 11 compounds which were either known beta blockers or closely related analogues. The screening protocol was applied to the high-throughput docking of a database of approximately four million compounds. A comparison of the overlay of the top-scoring compounds from this screen was found to be similar to that obtained in the 400,000 compound screen. The top-scoring ligands consisted of structural classes that are both diverse and different from traditional beta blockers. Evaluation of 56 compounds from the top-scoring hits from the in-house database and 94 compounds from the commercial database led to the identification of a total of 30 compounds with affinities in the nanomolar to low micromolar range. Some of the carazolol-related high affinity ligands discovered in this study are shown in Fig. 2 (3–6).
Fig. 2.
Structures of carazolol, carvedilol, and related ligands predicted to bind like carazolol to β2 adrenergic receptor
In a recent study, de Graaf and Rognan (17) modified the β2-adrenergic receptor crystal structure to model an early conformational state of the activated state of the receptor to obtain an “agonist-customized” structure. These modifications included changing the rotameric states of two TM5 serines important for agonist binding (Ser5.43, Ser5.46), followed by energy minimizations with the agonist ligand in the active site. The crystal structure as well as the modified structure were then used to virtually screen a database of compounds consisting of 13 known β2-adrenergic receptor antagonists or inverse agonists, 13 known partial or full agonists, and 980 diverse drug-like compounds using the docking programs Surflex and Gold. Docked poses were ranked using built-in scoring functions and protein–ligand interaction fingerprint scoring (IFP). An analysis of the results revealed that docking into the crystal structure followed by IFP scoring yielded both agonist/antagonist ligands among the top-ranking sets. On the other hand, the use of the agonist-customized receptor structure for screening of the same 1,006 compounds selectively retrieved partial/full agonists. Their study also found that the use of the topological scoring function IFP was essential to properly rank the docking poses and achieve acceptable enrichments for partial and full agonists.
α1A-ADRENERGIC AND RELATED RECEPTORS
Evers et al. (11) performed a comparative evaluation of different virtual screening approaches for their effectiveness in identifying known antagonists of the biogenic amine-binding GPCRs, adrenergic α1A, 5HT2A, dopamine D2, and muscarinic M1 receptors. They generated a homology model of α1A receptor using rhodopsin crystal structure incorporating mutagenesis and ligand binding data from literature. This model was then used as template to generate models of 5HT2A, dopamine D2, and muscarinic M1 receptors. These receptors were screened against a database consisting of 950 inactive compounds and 42–48 known antagonists of each target receptor assembled from the MDDR database. For docking, GOLD and FlexX-Pharm were used and the poses were scored with GoldScore and FlexX-Score. Additionally, the poses were rescored using seven other scoring functions (D_Score, G_Score, ChemScore, PMF, F_Score, DrugScore, Xscore). The performance of virtual screening protocols was evaluated on the basis of their ability to retrieve known ligands. The results obtained showed that the performance of the scoring functions varied among the four target GPCRs. The results from this structure-based virtual screening were compared with ligand-based screening methods and found that some ligand-based methods showed surprisingly high enrichment factors. Nevertheless, the high hit rates (up to 60% among the top-ranked 1% of screened database) in the docking and ranking method indicate that the structure-based virtual screening using homology models of GPCRs can be a useful approach for finding new leads when little or no information about active ligands is available.
DOPAMINE D2 RECEPTOR
Kortagere and Welsh (18) used a combined ligand-based and receptor structure-based method to demonstrate the potential of such an approach to identify new GPCR ligands. Ligand-based methods were applied to build GPCR-biased small molecule libraries which were then screened against rhodopsin and the rhodopsin-based dopamine D2 receptor model by structure-based methods. A random dataset of ~300,000 compounds was screened as follows. First, shape signatures encoding shape characteristics and electrostatic potential property were computed. Shape signatures were compared between database compounds and known ligands with confirmed activities against rhodopsin and dopamine D2 receptors. Second, 3D pharmacophore models capturing essential protein–ligand interactions between retinal and rhodopsin and between dopamine and the D2 receptor were developed. The compound library was then screened using these shape signature-based and 3D pharmacophore-based models. These led to the identification of 110 compounds as potential ligands of rhodopsin and 183 compounds for dopamine D2 receptor. These were then docked into the active sites of the rhodopsin and the dopamine D2 receptor. The docking was performed using GOLD and the docked poses were scored using GoldScore as well as a customized scoring function. This stepwise approach was shown to efficiently identify retinal-related compounds and dopamine-related compounds that are known binders of these two receptors. In addition, their study also led to the identification of 34 new compounds predicted to have high affinity for the D2 receptor. The experimental evaluation of these ligands against dopamine D2 receptor is, however, yet to be done.
HISTAMINE H4 RECEPTOR
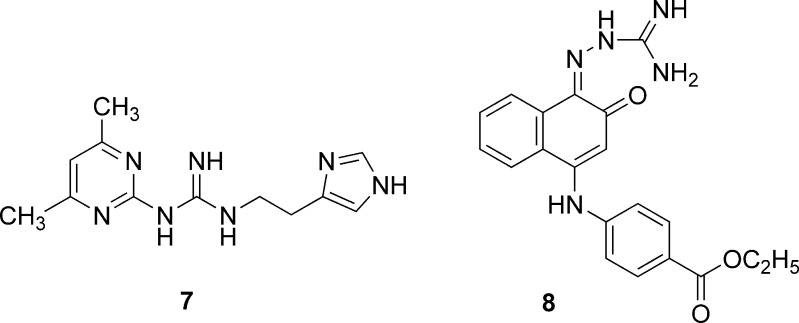
Recently, Kiss et al. (19) reported a successful large-scale virtual screening on a ligand-supported homology model of the human histamine H4 receptor (hH4R) that was developed and validated earlier (20). They compiled a compound database comprising more than 8.7 million structures representing more than five million unique compounds. These compounds were docked by FlexX and the docked ligands were ranked using ChemScore that had previously been shown to perform best in enrichment tests. The structures were inspected visually to identify the best 2,000 compounds that met both of the following criteria: (1) the entire ligand has to be positioned within the binding site cavity and (2) the ligand has to have acceptable protonation and tautomeric states. The presence of additional interaction(s) with key residues, Asp94 or Glu82, was considered as a favorable feature in the selection of 128 compounds of which 66 could be purchased from vendors. An additional set of 189 compounds was selected through an analysis of the top-scoring 45,000 ligands (top 0.5% of the ranked database) and their close analogues. Of the total 255 compounds that were evaluated in radioligand displacement assays, 16 compounds emerged as hits that displayed significant radioligand displacement at a concentration of 5 μM. These hits could be classified into compounds containing two guanidinium units (five compounds), compounds containing one guanidinium group (two compounds), and nine singletons. Concentration–response determinations on six compounds that displayed >60% ligand displacement at 5 μM led to the identification of 7 and 8 (Fig. 3) as ligands with binding Ki values of 85 and 219 nM, respectively.
Fig. 3.
Structures of human histamine H4 receptor identified by virtual screening
KAPPA OPIOID RECEPTOR
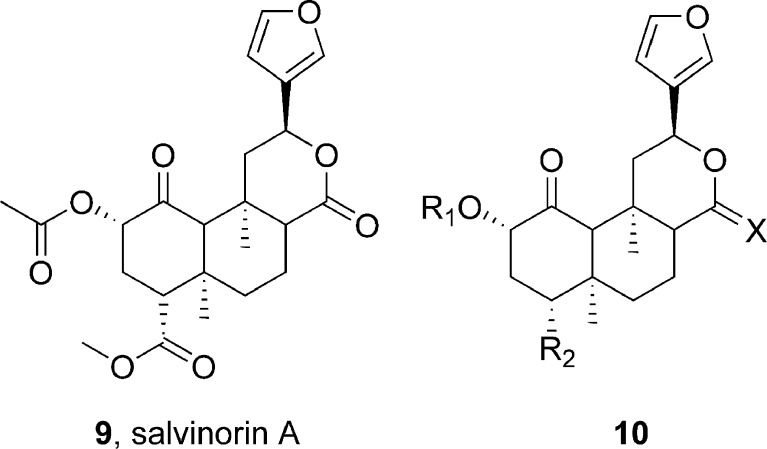
Singh et al. (21) have reported on their effort to develop a robust virtual screening model for the kappa opioid receptor (KOP) using a combined pharmacophore modeling and structure-based docking approach. The pharmacophore model was generated using the kappa agonist salvinorin A (9) and 14 chemically closely related analogues represented by 10 (Fig. 4) using Catalyst/HypoGen module. The best model that was generated consisted of two hydrogen bond acceptor and three hydrophobic features. The reliability of the pharmacophore model was tested in several validation procedures. In addition, the human kappa opioid receptor (hKOP) was modeled using the 2.2Å X-ray structure of bovine rhodopsin as the template. Multiple sequence alignments were generated from human kappa, delta, mu, and nociceptin receptors, and the models were generated and refined using Modeler/Discover of INSIGHTII. An agonist-bound state of the model was then generated by manually placing salvinorin A within the putative binding pocket followed by molecular dynamics simulation and minimizations. Although salvinorin A lacks a protonatable nitrogen to guide the placement of the ligand using the generally accepted key salt bridge interaction with conserved Asp138 in TM3, information gathered from site-directed mutagenesis studies was utilized to dock the ligand at the putative binding pocket. The quality of the refined models was evaluated using PROCHECK and PROSTAT. The model thus developed was consistent with the predictions from the pharmacophore model and correlated well with known SAR and mutational data. The docking protocol was then applied to 14 other ligands, generating ten solutions per ligand. For a majority of the ligands, top-scoring poses were found to display relevant binding interactions. A robust correlation (r2 = 0.80) was observed between the experimental binding affinity (pKi) and the GOLD score. Moreover, a quantitative comparison of the predicted activities from the pharmacophore model with the GOLD scores from docking initially gave a correlation of 0.67 (0.85 after removal of one outlier compound). This study indicates that a hybrid approach using models in which ligand-based and target-based information has been synergistically integrated could improve the effectiveness and performance of database searching for drug discovery through virtual screening.
Fig. 4.
Structures of salvinorin A and related compounds used in pharmacophore modeling and docking at kappa opioid receptor
MELANIN-CONCENTRATING HORMONE RECEPTOR 1
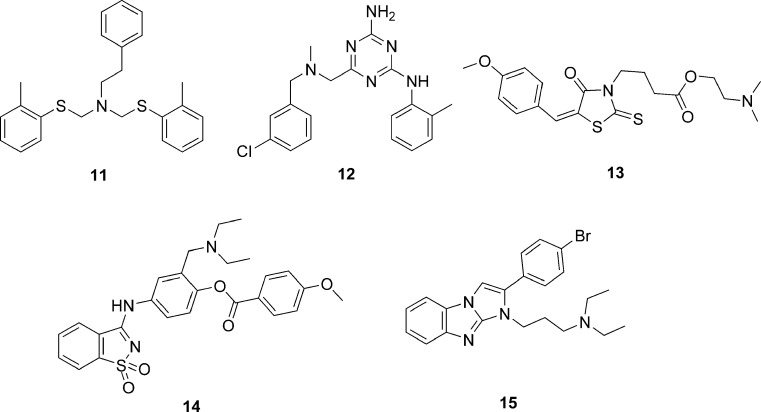
Cavasotto et al. (22) successfully used a ligand-steered homology modeling and virtual screening approach to identify novel classes of antagonist ligands of melanin-concentrating hormone receptor 1 (MCH-R1). An initial homology model of MCH-R1 was built using the rhodopsin template. Four known ligands of MCH-R1 were placed into the binding site of this model, and an ensemble of 200 structures for each ligand was generated by randomizing the position and orientation of the ligands followed by a multistep energy minimization. These models were then subjected to a flexible-ligand-flexible-receptor Monte Carlo docking and were analyzed by estimation of ligand–receptor interaction energy, binding pocket clustering, and visual inspection to select a final set of eight models. These final models were validated through a small-scale virtual screening with 11 known MCH-R1 antagonist and 5,497 decoy compounds. Analysis of the ability of the models to recover known ligands and the diversity observed among the top-ranking compounds led to the selection of the model that was finally used for large-scale virtual screening of 187,084 non-redundant compounds using the ICM virtual screening module. The docking of the entire compound set was performed three times and the best scoring pose for each compound was kept. The docked compounds were examined for the absence of docked ligand–receptor clashes and for the presence of a hydrogen bond between the ligand and the key aspartate residue in TM3. Compounds meeting these requirements were clustered according to chemical similarity, and the top-scoring compound from each cluster was chosen to obtain a set of 281 compounds. Of this final set, samples of 129 compounds were experimentally tested in competitive radioligand binding assay. Of the evaluated compounds, five compounds 11–15 (Fig. 5) displayed Ki values in the range of 7–12 μM. These results demonstrate that explicit ligand information can be applied to shape and optimize the binding site to reduce uncertainties in homology-modeled structures.
Fig. 5.
Structures of antagonist ligands of melanin-concentrating hormone receptor 1 identified by virtual screening
CANNABINOID RECEPTOR 2
Chen et al. (23) generated a model of the antagonist bound state of cannabinoid receptor 2 (CB2) by flexible docking of SR144528 at the ligand binding site of rhodopsin-based CB2 model using FlexiDock program. The model was further refined by MD/MM simulations with the INSIGHTII/Discover program. The placement of the antagonist ligand and refinement were aided by mutagenesis data implicating Ser161 and Ser165 as key residues for ligand binding. The model was validated by docking a test set containing 1,000 compounds consisting of 967 inactive NCI compounds and 33 other cannabinoid ligands, of which 15 were considered as active CB2 antagonists. The efficiency of single and combination of five different scoring functions in retrieving the known active ligands within the top 10% and 15% of hits was evaluated. The results from this validation study indicated that the developed virtual screening protocol is capable of identifying known CB2 ligands from randomly selected molecules.
THYROTROPIN-RELEASING HORMONE RECEPTOR
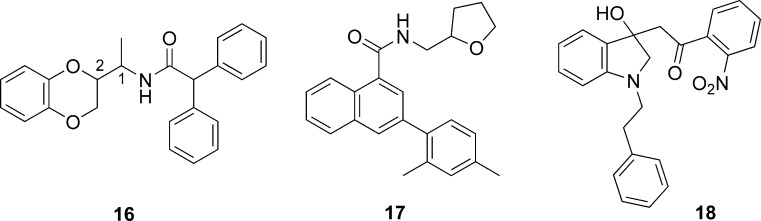
Recently, Engel et al. (24) applied a virtual screening approach to identify novel small molecule ligands of the thyrotropin-releasing hormone receptor 1 and 2 (TRH-R1 and TRH-R2) using a methodology that takes into consideration an expanded pharmacophore definition and protein flexibility. They defined a receptor-based pharmacophore model through a GRID analysis to identify potential fields of interaction with hydrogen donor, hydrogen acceptor, and hydrophobic molecular probes using rhodopsin-based homology model of thyrotropin-releasing hormone receptor. In this analysis, the side chains were allowed to be flexible to extend the potential interaction area. The pharmacophore model was used to screen one million commercially available drug-like compounds from ZINC database with Flex Search protocol (Unity) to obtain a subset of 100,000 compounds. A diversity subset of 10,000 compounds was selected and docked using FlexE against five alternative conformations of the binding pocket in order to mimic protein flexibility. Of the 1,000 compounds with best docking scores, 100 were selected using MACCS structural keys fingerprints as diverse representatives for experimental evaluation. The evaluation of these and related analogues for agonist or antagonist activity led to the identification of several unique scaffolds represented by 16–18 (Fig. 6) as antagonists. Among these, of particular interest was compound 16 possessing the 2,3-dihydro-1,4-benzodioxin scaffold which was present as the core structure among several highly ranked compounds from the virtual screen. The commercial sample of 16 containing all four stereoisomers displayed affinity against both TRH-R1 and TRH-R2 receptors. Docking of the four stereoisomers against TRH-R1 and scoring using LiaisonScore predicted that the C1-R, C2-S isomer will be the most potent ligand. This prediction was confirmed by experimental evaluation which revealed that this isomer binds to TRH-R1 with a binding Ki of 0.29 μM and shows nearly 13-fold selectivity over TRH-R2. The predicted binding modality of these antagonists was further verified through comprehensive mutational analysis of the key residues at the binding pocket of TRH receptors.
Fig. 6.
Structures of thyrotropin-releasing hormone receptor 1 antagonists identified by virtual screening
FREE FATTY ACID RECEPTOR 1
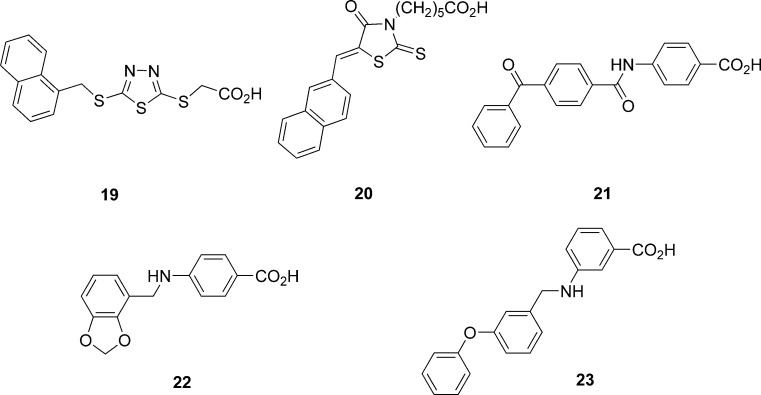
In an attempt to identify new ligands that activate or inhibit the free fatty acid receptor 1 (FFAR1), also known as GPR40, Tikhonova et al. (25) performed virtual screening starting with an initial library of 2.6 million drug-like compounds from the ZINC database. Sequentially, they performed a 2D similarity search, diversity subset selection, pharmacophore search, and docking against a structural model of FFAR1. In the first step, 2D similarity searches with two known high potency agonists were performed to obtain a set of 704,772 unique compounds using molecular operating environment. A diversity subset of 70,447 compounds (10%) was assembled using DiverseSolutions software. A 3D pharmacophore was defined on the basis of the docked conformations of the two known agonists of FFAR1. This pharmacophore was then used to search the 70,447 compound set to identify 1,581 compounds as hits. In parallel, the same database was subjected to high-throughput docking using Glide to obtain a set of 3,131 compounds with GlideScores lower than −8 kcal/mol. The 3D pharmacophore hits and docking hits contained 183 compounds that were common in both sets. The final subset selection was based on structural diversity calculated using BIT_MACCS structural keys and visual inspection. Of the final set of 52 compounds, 32 came from the 183 common hits and ten each from the unique compounds from docking and 3D pharmacophore search. Experimental evaluation of the 52 compounds led to the identification of six active compounds, two full agonists, three partial agonists, and one antagonist. The five ligands 19–23 (Fig. 7) identified by docking studies were either agonist or partial agonists but not antagonists, suggesting that the model was indeed biased toward agonist recognition. The experimental results also indicated that compounds identified by both 3D pharmacophore search and by docking had a better chance of emerging as hits than those retrieved by only one of the two techniques.
Fig. 7.
Structures of agonist or partial agonist ligands of the free fatty acid receptor 1 identified by virtual screening
CORTICOTROPIN-RELEASING FACTOR RECEPTOR 1 AND GLUCAGON RECEPTOR
The construction of reliable 3D models of the TM domain of class B and class C GPCRs suitable for structure-based virtual screening is a challenge, as these receptors share little primary structural homology with class A GPCRs. Recently, de Graaf et al. (26) have explored the application of structure-based virtual screening methods to the class B GPCRs, corticotropin-releasing factor receptor 1 (CRFR1) and glucagon receptor. They built three alternative CRFR1 models and performed retrospective virtual screening using 13 known CRFR1 antagonists and 987 decoys. The docking was performed with Gold and ranked with IFP. The relative performance of the three models was assessed through enrichment analyses. The best model was demonstrated to retrieve nine of the 13 known ligands in the top 1% of the screened set. Using the CRFR1 model as the template, they generated two models of the glucagon receptor and used them for prospective virtual screening of 144,000 compounds. From the two screens, 25 compounds were selected for experimental evaluation that led to the identification of six ligands as confirmed hits.
METABOTROPIC GLUTAMATE RECEPTOR 5
The metabotropic glutamate receptors (mGluRs) belong to the class C GPCRs. Recently, Radestock et al. (27) applied ligand-supported homology modeling of the allosteric ligand binding site of mGluR5, taking into consideration mutagenesis data and structure–activity relationship information from known ligands. The mGluR5 model was developed using a modified version of ligand-supported homology approach (MOBILE) of Evers et al. (11). A total of 159 known negative allosteric modulators were selected as reference ligands. Each of these reference ligands was docked into the ligand binding site using FlexX1.2, and the best poses were selected through visual inspection. The interaction fingerprints were then generated using these selected poses. For evaluation purposes, ten sets of 1,822 compounds each containing 11 known negative allosteric modulators and 1,791 presumably inactive compounds from NCI database were assembled. These were docked and ranked using interaction fingerprint-based similarity (IFS) scoring scheme. The enrichment rates obtained using the IFS-based ranking were significantly higher than those using other conventional scoring functions (Dock-score, PMF-Score, Gold-Score, ChemScore, and FlexX-score). This retrospective virtual screening validation study indicates that the described protocol could potentially be applied to successful virtual screening for ligands for the mGluR family.
VIRTUAL SCREENING FOR LIGANDS THAT INHIBIT GPCR/G-PROTEIN INTERACTIONS
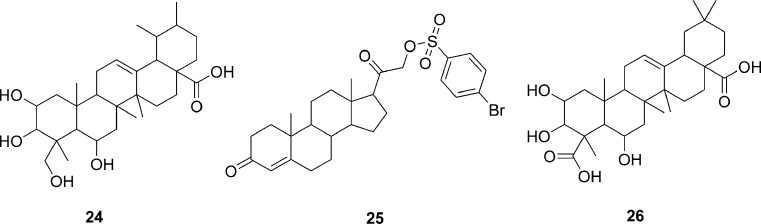
The development of homology models and virtual screening have primarily been focused on finding agonist or antagonist molecules that bind to the transmembrane domain closest to the extracellular side. In a recent effort, Taylor et al. (28) have investigated the possibility of targeting the interface between an activated GPCR and its G-protein to block signal transduction. The complex structure of photoactivated rhodopsin in MII state (R* state) bound to a segment of its G-protein, transducin (Gt) was modeled using known structure of rhodopsin in the R* state and TrNOE structure of Gt for the segment between residues 340 and 350. Rhodopsin intracellular loop structure in a conformation binding to Gt was previously modeled based on the best fit between loop conformations and Gt, as predicted through docking the Gt(340–350) segment. The NCI diversity set containing 1,990 compounds were then docked into this model and scored using X-Score, Autodock score, and CSCORE. A series of subset selections from the top-scoring ligands were performed to finally select nine compounds for experimental testing. These nine compounds were assayed to determine their ability to stabilize MII-photoactivated rhodopsin and inhibit the interaction between MII-photoactivated rhodopsin and Gt. In the MII stabilization assay, two of the tested compounds, 24 and 25 (Fig. 8), were found to be active, with an EC50 value of 120 and 45 μM, respectively. Further, the dose-dependent inhibition of activated rhodopsin–Gt showed that both 24 and 25 inhibited rhodopsin–Gt interaction with an IC50 value of 180 and 15 μM, respectively. An analysis of the docked conformations for 24 and 25 revealed several key interactions of these ligands with the intracellular loop structure. A search of the full NCI database of 140,000 for compounds similar to 24 or 25 and evaluation of seven compounds thus identified yielded an additional compound 26 that displayed an EC50 value of 350 μM and IC50 value of 2 mM for inhibition of activated rhodopsin–Gt interaction. Thus, this first virtual screening effort using modeled intracellular loop structures of an activated form of GPCR was successful in identifying compounds. These hits from the virtual screening were comparable in potency to those found in experimental high-throughput screening. This study indicates the potential of virtual screening approaches for discovery of small molecule inhibitors of signal transduction processes of GPCRs.
Fig. 8.
Structures of compounds identified as inhibitors of photoactivated rohodopsin and transducin by virtual screening
CONCLUSIONS
During the past few years, as highlighted in this review, there have been several reports on the application of homology models of GPCRs for structure-based virtual screening. While some of the efforts have focused on the validation of the models and virtual screening protocols, others have focused on the successful identification of novel agonist or antagonist ligands for various GPCRs. With the availability of the X-ray crystal structures of the β1- and β2-adrenergic receptors as additional templates for homology modeling, along with the advances in protein modeling/refinement techniques, docking/scoring methods, and increased computing power, structure-based virtual screening is becoming an increasingly useful approach for identifying novel ligands for therapeutically relevant GPCRs.
References
- 1.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 2.Teller D. C., Okada T., Behnke C. A., Palczewski K., Stenkamp R. E. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Kobilka B. K. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen S. G., Choi H. J., Rosenbaum D. M., Kobilka T. S., Thian F. S., Edwards P. C., Burghammer M., Ratnala V. R., Sanishvili R., Fischetti R. F., Schertler G. F., Weis W. I., Kobilka B. K. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 6.Hanson M. A., Cherezov V., Griffith M. T., Roth C. B., Jaakola V. P., Chien E. Y., Velasquez J., Kuhn P., Stevens R. C. A specific cholesterol binding site is established by the 2.8 a structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J. H., Scheerer P., Hofmann K. P., Choe H. W., Ernst O. P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 9.Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 10.Bissantz C., Bernard P., Hibert M., Rognan D. Protein-based virtual screening of chemical databases. II. Are homology models of G-protein coupled receptors suitable targets? Proteins. 2003;50:5–25. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 11.Evers A., Hessler G., Matter H., Klabunde T. Virtual screening of biogenic amine-binding G-protein coupled receptors: Comparative evaluation of protein- and ligand-based virtual screening protocols. J. Med. Chem. 2005;48:5448–5465. doi: 10.1021/jm050090o. [DOI] [PubMed] [Google Scholar]
- 12.Schlyer S., Horuk R. I want a new drug: G-protein-coupled receptors in drug development. Drug Discov. Today. 2006;11:481–493. doi: 10.1016/j.drudis.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh S., Nie A., An J., Huang Z. Structure-based virtual screening of chemical libraries for drug discovery. Curr. Opin. Chem. Biol. 2006;10:194–202. doi: 10.1016/j.cbpa.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Klebe G. Virtual ligand screening: Strategies, perspectives and limitations. Drug Discov. Today. 2006;11:580–594. doi: 10.1016/j.drudis.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topiol S., Sabio M. Use of the X-ray structure of the beta2-adrenergic receptor for drug discovery. Bioorg. Med. Chem. Lett. 2008;18:1598–1602. doi: 10.1016/j.bmcl.2008.01.063. [DOI] [PubMed] [Google Scholar]
- 16.Sabio M., Jones K., Topiol S. Use of the X-ray structure of the beta2-adrenergic receptor for drug discovery. Part 2: Identification of active compounds. Bioorg. Med. Chem. Lett. 2008;18:5391–5395. doi: 10.1016/j.bmcl.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 17.de Graaf C., Rognan D. Selective structure-based virtual screening for full and partial agonists of the beta2 adrenergic receptor. J. Med. Chem. 2008;51:4978–4985. doi: 10.1021/jm800710x. [DOI] [PubMed] [Google Scholar]
- 18.Kortagere S., Welsh W. J. Development and application of hybrid structure based method for efficient screening of ligands binding to G-protein coupled receptors. J. Comput. Aided Mol. Des. 2006;20:789–802. doi: 10.1007/s10822-006-9077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiss R., Kiss B., Konczol A., Szalai F., Jelinek I., Laszlo V., Noszal B., Falus A., Keseru G. M. Discovery of novel human histamine H4 receptor ligands by large-scale structure-based virtual screening. J. Med. Chem. 2008;51:3145–3153. doi: 10.1021/jm7014777. [DOI] [PubMed] [Google Scholar]
- 20.Kiss R., Noszal B., Racz A., Falus A., Eros D., Keseru G. M. Binding mode analysis and enrichment studies on homology models of the human histamine H4 receptor. Eur. J. Med. Chem. 2008;43:1059–1070. doi: 10.1016/j.ejmech.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Singh N., Cheve G., Ferguson D. M., McCurdy C. R. A combined ligand-based and target-based drug design approach for G-protein coupled receptors: Application to salvinorin A, a selective kappa opioid receptor agonist. J. Comput. Aided Mol. Des. 2006;20:471–493. doi: 10.1007/s10822-006-9067-x. [DOI] [PubMed] [Google Scholar]
- 22.Cavasotto C. N., Orry A. J., Murgolo N. J., Czarniecki M. F., Kocsi S. A., Hawes B. E., O'Neill K. A., Hine H., Burton M. S., Voigt J. H., Abagyan R. A., Bayne M. L., Monsma F. J., Jr. Discovery of novel chemotypes to a G-protein-coupled receptor through ligand-steered homology modeling and structure-based virtual screening. J. Med. Chem. 2008;51:581–588. doi: 10.1021/jm070759m. [DOI] [PubMed] [Google Scholar]
- 23.Chen J. Z., Wang J., Xie X. Q. GPCR structure-based virtual screening approach for CB2 antagonist search. J. Chem. Inf. Model. 2007;47:1626–1637. doi: 10.1021/ci7000814. [DOI] [PubMed] [Google Scholar]
- 24.Engel S., Skoumbourdis A. P., Childress J., Neumann S., Deschamps J. R., Thomas C. J., Colson A. O., Costanzi S., Gershengorn M. C. A virtual screen for diverse ligands: Discovery of selective G protein-coupled receptor antagonists. J. Am. Chem. Soc. 2008;130:5115–5123. doi: 10.1021/ja077620l. [DOI] [PubMed] [Google Scholar]
- 25.Tikhonova I. G., Sum C. S., Neumann S., Engel S., Raaka B. M., Costanzi S., Gershengorn M. C. Discovery of novel agonists and antagonists of the free fatty acid receptor 1 (ffar1) using virtual screening. J. Med. Chem. 2008;51:625–633. doi: 10.1021/jm7012425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. de Graaf, F. Giordanetto, G. Abbas, C. G. Unson, and D. Rognan. Class B G-protein coupled receptors as targets for protein-based virtual screening. The 236th American Chemical Society National Meeting, Philadelphia, PA, Aug 17–21, MEDI-409 (2008).
- 27.Radestock S., Weil T., Renner S. Homology model-based virtual screening for GPCR ligands using docking and target-biased scoring. J. Chem. Inf. Model. 2008;48:1104–1117. doi: 10.1021/ci8000265. [DOI] [PubMed] [Google Scholar]
- 28.Taylor C. M., Barda Y., Kisselev O. G., Marshall G. R. Modulating G-protein coupled receptor/G-protein signal transduction by small molecules suggested by virtual screening. J. Med. Chem. 2008;51:5297–5303. doi: 10.1021/jm800326q. [DOI] [PMC free article] [PubMed] [Google Scholar]