Abstract
Alzheimer's disease is a progressive neurodegenerative disorder with high prevalence in old age. It is the most common cause of dementia, with a risk reaching 50% after the age of 85 years, and with the increasing age of the population it is one of the biggest healthcare challenges of the 21st century. Genetic variation is an important contributor to the risk for this disease, underlying an estimated heritability of about 70%. Alzheimer's genetics research in the 1990s was successful in identifying three genes accounting for most cases of early-onset disease with autosomal dominant inheritance, and one gene involved in the more common late-onset disease, which shows complex inheritance patterns. Despite the presence of significant remaining genetic contribution to the risk, the identification of genes since then has been elusive, reminiscent of most other complex disorders. In the past decade there have been significant efforts towards a systematic evaluation of the multiple genetic association studies for Alzheimer's disease, while the first genome-wide association studies are now being reported with promising results. As sample sizes grow through new collections and collaborative efforts, and as new technologies make it possible to test alternative hypotheses, it is expected that new genes involved in the disease will soon be identified and confirmed. The gene discoveries of the 1990s have taught us a lot about Alzheimer's disease pathogenesis, providing many therapeutic targets that are currently at various stages of testing for future clinical use. As new genes become known and the biological pathways leading to disease are further explored, the possibility of prevention and successful personalized treatment is becoming tangible, providing hope for the millions of patients with Alzheimer's disease and their caregivers.
Introduction: Alzheimer's genetics in the 20th century
Alzheimer's disease (AD), first described by Alois Alzheimer in 1907 [1], is a neurodegenerative disorder progressing from memory loss to profound dementia and death within an average of eight years. Although clinical diagnosis is reliable in more than 90% of cases, the definite diagnosis is assigned post mortem based on brain atrophy and the characteristic neuropathologic findings, which involve intracellular neurofibrillary tangles containing hyperphosphorylated tau protein and extracellular amyloid plaques containing deposits of beta amyloid (Aβ). AD is recognized today as the most common cause of dementia in the elderly.
In the 1930s a number of reports described familial cases of AD with multiple affected individuals in each generation, consistent with an autosomal dominant mode of inheritance [2-4]. Like Alzheimer's first patient, the age of onset of the familial cases was most often below 65 years, which led physicians to identify AD as pre-senile dementia, distinguishing it from senile dementia. By 1980, however, it had become evident that the two types of dementia are essentially identical, named by Terry and Davies [5] as dementia of the Alzheimer type.
Not surprisingly, the genes involved in the familial, auto-somal dominant AD (FAD) were quickly discovered during the golden years of gene mapping in the 1990s. Despite initial confusion and controversy due to the genetic heterogeneity of AD [6,7], extending even to the small subset of FAD, a combination of functional evidence, linkage analyses and sequence comparisons led to the identification of the three genes accounting for most FAD cases: APP [8], PSEN1 [9] and PSEN2 [10].
The majority (95%) of AD cases, however, are of later onset and do not follow Mendelian inheritance, despite showing significant heritability [11,12]. Like most other psychiatric disorders, the genetics of the late-onset disease appear to be complex. AD is the first complex disorder for which a gene was identified through an association with a DNA variant, a variant in the APOE gene whose effect has been consistently observed since. Apolipoprotein E was known to have three common isoforms, ε2, ε3 and ε4, that are determined by two coding single nucleotide polymorphisms (SNPs). After observing high-avidity binding of Aβ to APOE, and encouraged by previous linkage reports around the APOE locus on chromosome 19, Strittmatter et al. [13] examined the three isoforms in cases and controls and found a significant excess of ε4 in the cases. Today, after many follow-up studies, it has been determined in populations of European descent that ε3ε4 heterozygotes have a two- to three-fold higher risk of developing AD compared to ε3ε3 homozygotes. The increase in risk for ε4ε4 homozygotes is more than twice that of the ε3ε4 heterozygotes, while ε2 heterozygotes have a reduced risk.
The significance of the identification of APOE as a risk factor for AD stretched beyond the obvious importance for AD research. It was a proof of principle for the 'common disease - common variant' hypothesis that was at the time becoming increasingly popular. This result encouraged investigators to perform many candidate gene association studies for AD and other disorders; unfortunately, most did not enjoy the same degree of consistent replication.
Alzheimer's genetics: recent progress
Since the discovery of an association of APOE with AD in 1993, there have been numerous publications of genetic association studies reporting mixed results and plagued by the lack of consistent replications. Initially, most studies examined one polymorphism per gene, often without evidence that the polymorphism had any functional significance. At that time (in the mid- to late nineties) this was considered acceptable practice because little genetic variation was known and dbSNP, the database of DNA polymorphisms [14], did not become available until 1998. It later became clear that a more dense survey of variation in and around a gene was necessary to be confident that it has been adequately tested for association. It also became clear that it is not necessary to genotype all SNPs in a gene because, as a result of linkage disequilibrium (LD), the number of haplo-types observed in the population is lower than the possible combinations of alleles [15]. As the International HapMap Project [16] started to make data available for characterizing LD, investigators started performing more comprehensive association analyses, genotyping enough SNPs to capture and investigate all known common variations in each gene. This, however, came with the price of increased multiple comparisons, further complicating the interpretation of results. Power was reduced due to statistical corrections, while the significance of studies that identified associations for the same gene but for different SNPs was hard to interpret. In an effort to put order into these results, Bertram et al. [17] catalogued all published association studies for AD, in a public database called AlzGene [18]. In December 2005 the AlzGene database contained 802 different polymorphisms in 277 genes, while meta-analyses pointed to a little over a dozen of these genes as the most reliable associations [17]. By January 2009 the contents of the database had doubled to 557 genes and 1,852 polymorphisms. This large assembly of studies is a useful tool for AD genetics investigators around the world and it has become increasingly valuable as it incorporates more recent results from genome-wide association studies (GWAS). However, it is important to remember the inherent biases of candidate gene studies, which comprise the majority of the AlzGene database entries. Most of the reported genes were chosen because of their known functions, linkage evidence or both. In addition, it is impossible to estimate publication bias and this is itself influenced by prior linkage and functional evidence. Thus, inferences about biological pathways or further support of linkage regions based on most of these data should be avoided, with the exception perhaps of results from GWAS.
Linkage studies on late-onset AD have suffered similar limitations to those observed in association studies. Multiple genome-wide studies have been performed, often including overlapping samples, yet they have not been consistent in the genomic regions they identify and they rarely replicate each other's results. Only a few genomic regions including chromosomes 9, 10 and 12 have been more consistently identified by linkage [19]; however, the presence of risk alleles within these regions remains unproven. The success of GWAS has led investigators today to pay less attention to linkage in complex disorder, leading to a current lack of new linkage results. It must be noted that under allelic heterogeneity, linkage would be successful where association tests would fail to give a positive result. In view of FAD being caused by more than 150 different mutations in the PSEN1 gene, it should be no surprise if the risk for late-onset AD is increased by more than ten different variants in a gene. Today it is sensible to follow the GWAS approach to find the first risk genes after APOE, because laboratory and analytical methods for identifying multiple rare disease-causing alleles across many genes are still in their infancy. New and emerging analytical methods and second-generation sequencing technologies will soon make it possible to explore this hypothesis in projects that will probably be guided by prior linkage and GWAS findings.
Genome-wide association studies for Alzheimer's disease risk loci
Through January 2009 there have been four reports of GWAS for AD that analyzed individual genotypes, and a report of a study that genotyped pooled samples. An additional study has used a pooling strategy to investigate potentially functional SNPs across the genome. Not surprisingly, all studies identified the APOE association, proving the power of the approach under the 'common disease - common variant' hypothesis. None of these studies exceeded a sample size of 2,500 (cases and controls) and they can therefore be considered of modest size for the purpose of a GWAS. The studies have shown no overlap in terms of the identified loci other than APOE, and most findings have yet to be replicated by independent groups. Nevertheless, they have each reported interesting results.
The study by Grupe et al. [20] in April 2007 used a pooling strategy and multiple follow-up sample sets to examine more than 17,000 gene-based putative functional SNPs across the genome. Although only SNPs around APOE reached study-wide significance, many others provided weaker evidence of association, overlapping significantly with known linkage regions.
In the same month, Coon et al. [21] reported results on half a million SNPs across the genome genotyped using the Affymetrix platform on over 1,000 histopathologically verified AD cases and controls. The main conclusion of that report was that APOE is the only major susceptibility gene, yet in a follow-up paper, Reiman et al. [22] stratified the cases by APOE genotype and detected a strong association with SNPs in the GAB2 gene, altered GAB2 transcript levels in vulnerable neurons, and an effect of GAB2 levels on tau phosphorylation. The association has since been tested in other samples, and although the results are conflicting [23-26], the occurrence of more than one independent replication [24,26] of an association that originated from an unbiased GWAS is encouraging.
In September 2008, Abraham et al. [27] reported on a study of over 1,000 pooled cases and 1,200 pooled controls genotyped on approximately 550,000 SNPs on the Illumina platform. The authors observed only one strong signal over the APOE gene. Follow-up of weaker signals by individual genotyping identified the LRAT gene, whose product plays a prominent role in the vitamin A cascade, as another potential association with AD.
In November 2008, Bertram et al. [28] reported on a GWAS analyzing 500,000 SNPs in a sample of 410 families and using multiple other samples for replications. Using family-based tests and incorporating age of onset information, they identified a SNP on chromosome 14q31 that, like the APOE variants, appears to be a modifier of age of onset, while other SNPs with weaker signals were also reported. No annotated gene was found in that genomic region in the University of California Santa Cruz (UCSC) database, with the exception of a computationally predicted gene and three expressed sequence tags (ESTs). This study also found some support for the GAB2 gene and one of the SNPs reported by the Grupe et al. study [20], while failing to support many other previous associations.
In January 2009, Beecham et al. [29] reported a GWAS on the Illumina platform analyzing approximately 550,000 SNPs in nearly 500 AD cases and 500 controls. Other than APOE, the study identified a SNP on chromosome 12q13 that met their criterion for genome-wide significance, a false-discovery rate of less than 0.20. This association was replicated in an independent sample, and it lies in a genomic region that has been previously reported to show significant genetic linkage to the disease [30-33]. Perhaps most interestingly, the authors reported that out of the 19 best distinct signals, 12 were found in regions with prior linkage evidence, consistent with the expectation of enrichment for true signals [34].
Carrasquillo et al. [35] have recently published a GWAS for AD on 844 cases and 1,255 controls, using an Illumina platform to assay approximately 314,000 SNPs. Although in their stage 1 analysis only APOE SNPs reached genome-wide significance, a signal on chromosome X provided strong evidence in the replication samples, giving a combined P-value as low as 3.9 × 10-12. This signal was within the PCDH11X gene, which encodes a protocadherin, a cell-cell adhesion molecule expressed in the brain.
It is still too early to draw definite conclusions from the results of the reported GWAS for AD. The sample sizes analyzed are considerably smaller than those of GWAS that have successfully identified genes for other complex disorders [36]. For example, a large GWAS of seven major diseases involving approximately 2,000 cases of each disease and approximately 3,000 controls successfully identified associations for five disorders but showed much weaker results for bipolar affective disorder and hypertension [37], indicating that this sample size is adequate for some but not all complex diseases. Some of the reported associations for AD will probably point to new genes as more investigators replicate them and further analyze the variants functionally; however, it appears that the GWAS on AD might still be underpowered and the effect sizes of the remaining AD-associated variants are as small as those seen in other GWAS. In other words, it becomes more and more clear that APOE is the exception rather than the rule, making AD an interesting case of a complex disorder that involves single genes with Mendelian transmission and extensive allelic heterogeneity, one gene with common allelic variation and a considerable effect on the risk and the age of onset, and multiple other variants with seemingly smaller effects. With a few genes already in hand, with the first GWAS opening the way for more to follow, and with new sequencing technologies emerging and promising to test the possibility of multiple rare variants, these are exciting times for AD genetics research.
Contribution of AD genetics to treatment and diagnosis
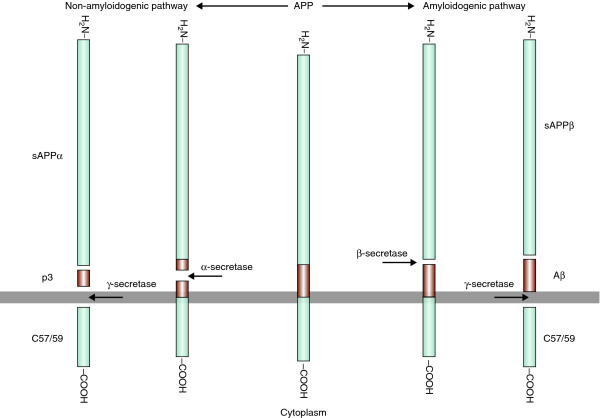
It is now more than 15 years since the discovery of the first genes involved in AD, and while we are still on a quest for additional genes, we have learned a lot about the disease. The processing of APP through cleavage by γ-secretase, an enzymatic complex whose catalytic subunit is formed by the presenilins [38], is considered by many as a key in the disease process. It leads to generation of the amyloidogenic peptide Aβ (Figure 1) and its aggregation into fibrils and toxic oligomeric forms, the earliest effectors of synaptic compromise [39], followed by neurodegeneration. The direct involvement of the products of at least three out of the four known genes in this hypothesis is no coincidence and significantly strengthens the confidence that this is a promising target for treatment. These genes have greatly enhanced our knowledge of the pathway leading to the production of amyloid (Figure 1), which has in turn provided targets for intervention.
Figure 1.
The amyloidogenic and non-amyloidogenic cleavage pathway of APP. Mutations in APP, PSEN1 or PSEN2 cause early-onset AD. PSEN1 and 2 are components of γ-secretase. The amyloidogenic peptide Aβ is shown in orange. reliable associations from GWAS, the best targets and strategies to intervene will become clearer.
Treatments targeting APP processing include those diverting cleavage toward the non-amyloidogenic α-secretase pathway (α-secretase enhancers) and those inhibiting the amyloido-genic pathway of beta and gamma secretase (β- or γ-secretase inhibitors). Pharmacological agents that enhance α-secretase activity include, among others, non-steroidal anti-inflammatory drugs (NSAIDs), statins and estrogens through activation of protein kinase C [40]. These agents have been tested with varying results. The effectiveness of estrogens has been suggested by many in vitro and in vivo studies; however, data from the Women's Health Initiative Memory Study [41], a large randomized controlled trial, did not show a consistent positive effect. The possibility that there is a critical period for neuroprotection [42] and that genetic variation or other predisposing factors might modify the effects of estrogens requires further examination [43]. The use of NSAIDs has seen support from epidemiological studies as likely to reduce the risk for the disease; however, the results of clinical investigations so far have not been encouraging [44,45]. The use of statins for AD prevention has shown conflicting results. Initial cross-sectional studies showed risk reductions that were better than 50% (for a review see Rockwood et al [46]). Clinical trials and cohort studies, however, failed to show the protective effect that has been consistently observed in cross-sectional studies [46].
The possibility of indication bias in cross-sectional studies (that is, people with AD are less likely to receive statin treatment) cannot be ruled out, although it has been accounted for by some studies [46]. The debate and interest in statins remains open, as support from prospective clinical trials is clearly necessary before they can be considered a preventive measure for AD [47]. The recognition that ADAM10 from the ADAM family of metalloproteases exhibits α-secretase activity [48] and is regulated by retinoic acid [49] has led to the inclusion of retinoic acid in the list of potential therapeutic agents [50]. Retinoic acid has shown promising results in an AD mouse model [51] but its potential as a therapeutic agent for AD in humans has not yet been examined.
Agents that inhibit the amyloidogenic pathway include β-and γ-secretase inhibitors. Beta-secretase inhibitors have only recently been developed [52], and initial tests on transgenic mice are positive, showing decreased Aβ production [53]. Gamma-secretase inhibitors have also shown positive results in laboratory animal models and, administered in low doses, they are safe in humans and reduce plasma Aβ [54]. A major limitation in the use of these inhibitors is that APP is not the only substrate of γ-secretase. Other substrates include NOTCH, ERBB4 and many other type I membrane protein stubs [54]; therefore, significant inhibition of the enzyme could lead to serious side-effects. This limitation might be bypassed to some extent as more selective agents are developed.
The role of APOE in AD appears to be more complex than that of APP and the presenilins. Studies of its functional involvement have implicated the homeostasis of cholesterol and phospholipids, synaptic integrity, amyloid metabolism, phosphorylation of tau, accumulation of neurofibrillary tangles and neuronal survival [55]. Nevertheless, APOE is an important player in pharmacological intervention research. Many studies have suggested that the APOE genotype can influence the outcome of existing treatments [56], making it interesting from a pharmacogenetics perspective, while others have suggested that targeting the regulation of APOE expression is a potential treatment approach [55,57]. Interestingly, drugs that modify the expression of APOE include statins which, as discussed above, have already shown promise.
Many of the genes that have been implicated in AD - albeit not consistently - by association studies are involved in multiple aspects of the disease [58], including the generation of neurofibrillary tangles, amyloid aggregation, amyloid clearance, oxidative stress and hypoxia, inflammation and apoptotic cell death. Most of these processes are already targeted by therapeutic agents either directly or through effects of drugs chosen to target other disease mechanisms. When the validity and exact nature of the genetic associations is elucidated in the near future, together with new reliable associations from GWAS, the best targets and strategies to intervene will become clearer.
At the level of molecular genetic diagnosis and risk assessment, there is a sharp divide between early- and late-onset disease. In FAD, mutations in one of three genes (APP, PSEN1 and PSEN2) can be found in more than 80% of patients [59]. Although there is currently no effective cure or prevention strategy, knowledge of the risk, and prenatal diagnosis, might be desired and is possible. For late-onset AD, however, our current knowledge does not allow useful genetic testing. The increase in risk by APOE ε4 is of questionable use as most carriers will not become affected and half of the patients do not carry this allele. This is even more questionable for less established genetic associations. As we begin to discover more and more genes and genetic variants involved in AD and learn the details of their functions and interactions with each other and the environment, it is most likely that one day in the not so distant future accurate risk and age of onset prediction will become a reality. Such capability, combined with strategies for prevention and effective treatments, perhaps tailored to each patient through pharmacogenetics, could lead to solving this major public health problem.
Conclusion
Initial successes in relation to the genetics of AD were followed by frustration as investigators addressed the more common and genetically complex late-onset disease. The four AD genes identified in the 1990s provided important knowledge on the disease pathogenesis at the molecular level. While no new gene has been positively identified since then, there is a wealth of new data from potentially important genetic linkage and association studies. As powerful tools for high-throughput DNA analysis are becoming available, the first GWAS are emerging, and extensive DNA sequencing studies will probably follow. Those will soon lead to further breakthroughs, improving our understanding of genetic risk, and allowing better gene-based approaches to prevention and treatment.
Abbreviations
Aβ: amyloid beta; AD: Alzheimer's disease; EST: expressed sequence tag; FAD: familial Alzheimer's disease; GWAS: genome-wide association studies; LD: linkage disequilibrium; NSAID: non-steroidal anti-inflammatory drug; SNP: single nucleotide polymorphism; UCSC: University of California Santa Cruz.
Competing interests
The author declares that he has no competing interests.
Author's information
DA has been researching the genetic causes of AD since 2001 through linkage, association and gene expression studies.
Acknowledgements
The author thanks Dr David Valle and Megan Szymanski for critical review of the manuscript. DA is supported for this work by funding provided from the National Institute of Aging (grant R01AG022099 to DA).
References
- Alzheimer A. Ueber eine eigenartige Erkrankung der Himrinde. Allg Z Psychiat Med. 1907;64:146–148. [Google Scholar]
- Lowenberg K, Waggoner RW. Familial organic psychosis (Alzheimer's type). Arch Neurol Psychiatry. 1934;31:737–754. [Google Scholar]
- McMenemey WH, Worster-Drought C, Flind J, Williams HG. Familial presenile dementia: report of a case with clinical and pathological features of Alzheimer's disease. J Neurol Psychopathol. 1939;2:293–302. doi: 10.1136/jnnp.2.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottky J. Ueber praesenile Verbloedungen. Z Ges Neurol Psychiat. 1932;140:333–397. [Google Scholar]
- Terry RD, Davies P. Dementia of the Alzheimer type. Annu Rev Neurosci. 1980;3:77–95. doi: 10.1146/annurev.ne.03.030180.000453. [DOI] [PubMed] [Google Scholar]
- Tanzi R, Gaston S, Bush A, Romano D, Pettingell W, Peppercorn J, Paradis M, Gurubhagavatula S, Jenkins B, Wasco W. Genetic heterogeneity of gene defects responsible for familial Alzheimer disease. Genetica. 1993;91:255–263. doi: 10.1007/BF01436002. [DOI] [PubMed] [Google Scholar]
- Haines JL. The genetics of Alzheimer disease - a teasing problem. Am J Hum Genet. 1991;48:1021–1025. [PMC free article] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance M, Roses AD, Williamson R, Rossor M, Owen MJ, Hardy J. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin JF, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, Da Silva HAR, Haines JL, Pericak-Vance M, Tanzi R, Roses AD. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E, Wijsman EM, Nemens E, Anderson L, Goddard KA, Weber JL, Bird TD, Schellenberg GD. A familial Alzheimer's disease locus on chromosome 1. Science. 1995;269:970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- Bergem AL, Engedal K, Kringlen E. The role of heredity in late-onset Alzheimer disease and vascular dementia. A twin study. Arch Gen Psychiatry. 1997;54:264–270. doi: 10.1001/archpsyc.1997.01830150090013. [DOI] [PubMed] [Google Scholar]
- Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, Posner SF, Viitanen M, Winblad B, Ahlbom A. Heritability for Alzheimer's disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52:M117–125. doi: 10.1093/gerona/52a.2.m117. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, Nguyen BT, Norris MC, Sheehan JB, Shen N, Stern D, Stokowski RP, Thomas DJ, Trulson MO, Vyas KR, Frazer KA, Fodor SP, Cox DR. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294:1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- The AlzGene database. http://www.alzgene.org
- Myers AJ, Goate AM. The genetics of late-onset Alzheimer's disease. Curr Opin Neurol. 2001;14:433–440. doi: 10.1097/00019052-200108000-00002. [DOI] [PubMed] [Google Scholar]
- Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, Jehu L, Segurado R, Stone D, Schadt E, Karnoub M, Nowotny P, Tacey K, Catanese J, Sninsky J, Brayne C, Rubinsztein D, Gill M, Lawlor B, Lovestone S, Holmans P, O'Donovan M, Morris JC, Thal L, Goate A, Owen MJ, Williams J. Evidence for novel susceptibility genes for late-onset Alzheimer's disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16:865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Walker DG, Ravid R, Heward CB, Rogers J, Papassotiropoulos A, Reiman EM, Hardy J, Stephan DA. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, Craig DW, Coon KD, Liang WS, Herbert RH, Beach T, Rohrer KC, Zhao AS, Leung D, Bryden L, Marlowe L, Kaleem M, Mastroeni D, Grover A, Heward CB, Ravid R, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis J, Hannequin D, Pasquier F, Bentham P, Brice A, Leber I, Frebourg T, Deleuze JF, Cousin E, Thaker U, Amouyel P, Mann D, Lendon C, Campion D, Lambert JC. Association study of the GAB2 gene with the risk of developing Alzheimer's disease. Neurobiol Dis. 2008;30:103–106. doi: 10.1016/j.nbd.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Sleegers K, Bettens K, Brouwers N, Engelborghs S, van Miegroet H, De Deyn PP, Van Broeckhoven C. Common variation in GRB-associated Binding Protein 2 (GAB2) and increased risk for Alzheimer dementia. Hum Mutat. 2009;30:E338–344. doi: 10.1002/humu.20909. [DOI] [PubMed] [Google Scholar]
- Miyashita A, Arai H, Asada T, Imagawa M, Shoji M, Higuchi S, Urakami K, Toyabe S, Akazawa K, Kanazawa I, Ihara Y, Kuwano R. GAB2 is not associated with late-onset Alzheimer's disease in Japanese. Eur J Hum Genet. 2008. in press . [DOI] [PMC free article] [PubMed]
- Ikram MA, Liu F, Oostra BA, Hofman A, van Duijn CM, Breteler MM. The GAB2 gene and the risk of Alzheimer's disease: replication and meta-analysis. Biol Psychiatry. 2008. in press . [DOI] [PubMed]
- Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L, Dowzell K, Cichon S, Hillmer AM, O'Donovan MC, Williams J, Owen MJ, Kirov G. A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Med Genomics. 2008;1:44. doi: 10.1186/1755-8794-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I, Jiang H, Laird N, Moscarillo T, Ohlsen KL, Elliott K, Wang X, Hu-Lince D, Ryder M, Murphy A, Wagner SL, Blacker D, Becker KD, Tanzi RE. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL, Pericak-Vance MA. Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet. 2009;84:35–43. doi: 10.1016/j.ajhg.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeux R, Lee JH, Romas SN, Mayo D, Santana V, Williamson J, Ciappa A, Rondon HZ, Estevez P, Lantigua R, Medrano M, Torres M, Stern Y, Tycko B, Knowles JA. Chromosome-12 mapping of late-onset Alzheimer disease among Caribbean Hispanics. Am J Hum Genet. 2002;70:237–243. doi: 10.1086/324773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Vance JM, Saunders AM, Roses AD, Haines JL. Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. JAMA. 1997;278:1237–1241. [PubMed] [Google Scholar]
- Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Knowles H, Morris JC, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, Rice F, Giles P, Tunstall N, Jones L, Lovestone S, Williams J, Owen MJ, Hardy J, Goate A. Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet. 2002;114:235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- Liang X, Schnetz-Boutaud N, Kenealy SJ, Jiang L, Bartlett J, Lynch B, Gaskell PC, Gwirtsman H, McFarland L, Bembe ML, Bronson P, Gilbert JR, Martin ER, Pericak-Vance MA, Haines JL. Covariate analysis of late-onset Alzheimer disease refines the chromosome 12 locus. Mol Psychiatry. 2006;11:280–285. doi: 10.1038/sj.mp.4001766. [DOI] [PubMed] [Google Scholar]
- Avramopoulos D, Zandi P, Gherman A, Fallin MD, Bassett SS. Testing groups of genomic locations for enrichment in disease loci using linkage scan data: a method for hypothesis testing. Hum Genomics. 2006;2:345–352. doi: 10.1186/1479-7364-2-6-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, Younkin SG, Younkin CS, Younkin LH, Bisceglio GD, Ertekin-Taner N, Crook JE, Dickson DW, Petersen RC, Graff-Radford NR, Younkin SG. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer's disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkan AL, Goate AM. Presenilin function and gamma-secretase activity. J Neurochem. 2005;93:769–792. doi: 10.1111/j.1471-4159.2005.03099.x. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Klyubin I, Shankar GM, Townsend M, Fadeeva JV, Betts V, Podlisny MB, Cleary JP, Ashe KH, Rowan MJ, Selkoe DJ. The role of cell-derived oligomers of Abeta in Alzheimer's disease and avenues for therapeutic intervention. Biochem Soc Trans. 2005;33:1087–1090. doi: 10.1042/BST20051087. [DOI] [PubMed] [Google Scholar]
- Bullock R. Future directions in the treatment of Alzheimer's disease. Expert Opin Investig Drugs. 2004;13:303–314. doi: 10.1517/13543784.13.4.303. [DOI] [PubMed] [Google Scholar]
- Resnick SM, Maki PM, Rapp SR, Espeland MA, Brunner R, Coker LH, Granek IA, Hogan P, Ockene JK, Shumaker SA. Effects of combination estrogen plus progestin hormone treatment on cognition and affect. J Clin Endocrinol Metab. 2006;91:1802–1810. doi: 10.1210/jc.2005-2097. [DOI] [PubMed] [Google Scholar]
- Zandi PP, Carlson MC, Plassman BL, Welsh-Bohmer KA, Mayer LS, Steffens DC, Breitner JC. Hormone replacement therapy and incidence of Alzheimer disease in older women: the Cache County Study. JAMA. 2002;288:2123–2129. doi: 10.1001/jama.288.17.2123. [DOI] [PubMed] [Google Scholar]
- Ryan J, Scali J, Carriere I, Ritchie K, Ancelin ML. Hormonal treatment, mild cognitive impairment and Alzheimer's disease. Int Psychogeriatr. 2008;20:47–56. doi: 10.1017/S1041610207006485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely CA, Town T, Zandi PP. NSAIDs for the chemoprevention of Alzheimer's disease. Subcell Biochem. 2007;42:229–248. doi: 10.1007/1-4020-5688-5_11. [DOI] [PubMed] [Google Scholar]
- McGeer PL, McGeer EG. NSAIDs and Alzheimer disease: epidemiological, animal model and clinical studies. Neurobiol Aging. 2007;28:639–647. doi: 10.1016/j.neurobiolaging.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Rockwood K. Epidemiological and clinical trials evidence about a preventive role for statins in Alzheimer's disease. Acta Neurol Scand Suppl. 2006;185:71–77. doi: 10.1111/j.1600-0404.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- Xiong GL, Benson A, Doraiswamy PM. Statins and cognition: what can we learn from existing randomized trials? CNS Spectr. 2005;10:867–874. doi: 10.1017/s1092852900019817. [DOI] [PubMed] [Google Scholar]
- Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci USA. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinzen C, Muller U, Endres K, Fahrenholz F, Postina R. Genomic structure and functional characterization of the human ADAM10 promoter. FASEB J. 2005;19:1522–1524. doi: 10.1096/fj.04-3619fje. [DOI] [PubMed] [Google Scholar]
- Fahrenholz F, Postina R. Alpha-secretase activation - an approach to Alzheimer's disease therapy. Neurodegener Dis. 2006;3:255–261. doi: 10.1159/000095264. [DOI] [PubMed] [Google Scholar]
- Ding Y, Qiao A, Wang Z, Goodwin JS, Lee ES, Block ML, Allsbrook M, McDonald MP, Fan GH. Retinoic acid attenuates beta-amyloid deposition and rescues memory deficits in an Alzheimer's disease transgenic mouse model. J Neurosci. 2008;28:11622–11634. doi: 10.1523/JNEUROSCI.3153-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AK, Kumaragurubaran N, Hong L, Koelsh G, Tang J. Memapsin 2 (beta-secretase) inhibitors: drug development. Curr Alzheimer Res. 2008;5:121–131. doi: 10.2174/156720508783954730. [DOI] [PubMed] [Google Scholar]
- Hussain I, Hawkins J, Harrison D, Hille C, Wayne G, Cutler L, Buck T, Walter D, Demont E, Howes C, Naylor A, Jeffrey P, Gonzalez MI, Dingwall C, Michel A, Redshaw S, Davis JB. Oral administration of a potent and selective non-peptidic BACE-1 inhibitor decreases beta-cleavage of amyloid precursor protein and amyloid-beta production in vivo. J Neurochem. 2007;100:802–809. doi: 10.1111/j.1471-4159.2006.04260.x. [DOI] [PubMed] [Google Scholar]
- Wolfe MS. Inhibition and modulation of gamma-secretase for Alzheimer's disease. Neurotherapeutics. 2008;5:391–398. doi: 10.1016/j.nurt.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier J. Apolipoprotein E represents a potent gene-based therapeutic target for the treatment of sporadic Alzheimer's disease. Alzheimers Dement. 2008;4:S91–97. doi: 10.1016/j.jalz.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Crentsil V. The pharmacogenomics of Alzheimer's disease. Ageing Res Rev. 2004;3:153–169. doi: 10.1016/j.arr.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Lahiri DK. Apolipoprotein E as a target for developing new therapeutics for Alzheimer's disease based on studies from protein, RNA, and regulatory region of the gene. J Mol Neurosci. 2004;23:225–233. doi: 10.1385/JMN:23:3:225. [DOI] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci. 2008;9:768–778. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- Raux G, Guyant-Marechal L, Martin C, Bou J, Penet C, Brice A, Hannequin D, Frebourg T, Campion D. Molecular diagnosis of autosomal dominant early onset Alzheimer's disease: an update. J Med Genet. 2005;42:793–795. doi: 10.1136/jmg.2005.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]