Abstract
FDH (10-formyltetrahydrofolate dehydrogenase, Aldh1L1, EC 1.5.1.6) converts 10-formyltetrahydrofolate (10-formyl-THF) to tetrahydrofolate and CO2 in a NADP+-dependent reaction. It is a tetramer of four identical 902 amino acid residue subunits. The protein subunit is a product of a natural fusion of three unrelated genes and consists of three distinct domains. The N-terminal domain of FDH (residues 1–310) carries the folate binding site and shares sequence homology and structural topology with other enzymes utilizing 10-formyl-THF as a substrate. In vitro it functions as 10-formyl-THF hydrolase, and evidence indicate that this activity is a part of the overall FDH mechanism. The C-terminal domain of FDH (residues 400–902) originated from an aldehyde dehydrogenase-related gene and is capable of oxidation of short-chain aldehydes to corresponding acids. Similar to class 1 and 2 aldehyde dehydrogenases, this domain exists as a tetramer and defines the oligomeric structure of the full-length enzyme. The two catalytic domains are connected by an intermediate linker (residues 311–399), which is a structural and functional homolog of carrier proteins possessing a 4′-phosphopantetheine prosthetic group. In the FDH mechanism, the intermediate linker domain transfers a formyl, covalently attached to the sulfhydryl group of the phosphopantetheine arm, from the N-terminal domain to the C-terminal domain. The overall FDH mechanism is a coupling of two sequential reactions, a hydrolase and a formyl dehydrogenase, bridged by a substrate transfer step. In this mechanism, one domain provides the folate binding site and a hydrolase catalytic center to remove the formyl group from the folate substrate, another provides a transfer vehicle between catalytic centers and the third one contributes the dehydrogenase machinery further oxidizing formyl to CO2.
Keywords: folate metabolism, 10-formyltetrahydrofolate dehydrogenase, aldehyde dehydrogenase, functional domains, 4′-phosphopantetheine, enzyme mechanism
1. Introduction
1.1. Overview of folate metabolism
The present review is focused on the folate enzyme, 10-formyltetrahydrofolate dehydrogenase (FDH, Aldh1L1, EC 1.5.1.6). The crucial role of folate coenzymes in cellular metabolism is well established. They are key molecules in transferring one-carbon groups in numerous reactions [1] including de novo nucleotide biosynthesis [1–3] and the regeneration of methionine from homocysteine [1, 4, 5]. The latter process is connected to production of S-adenosylmethionine [1, 5, 6], a universal methyl donor in more than a hundred methylation reactions in the cell [7]. It is not surprising, therefore, that disruption of folate metabolism has dramatic consequences for the cell [8–10], ultimately reducing growth rate and impairing cell division [1, 11–13]. Of note, folate is a vitamin, meaning that humans are unable to synthesize it and strictly depend on diet to obtain this coenzyme [14]. For many years, folate metabolism attracted the attention of researchers due to strong correlations between folates status and several diseases. Epidemiological studies have linked low folate levels, or a low folate intake, with increased risk for several types of cancer [15], neural tube defects [16], and cardiovascular diseases [17]. At present, it is generally accepted that increased folate intake decreases the risk of these diseases in humans [15, 18].
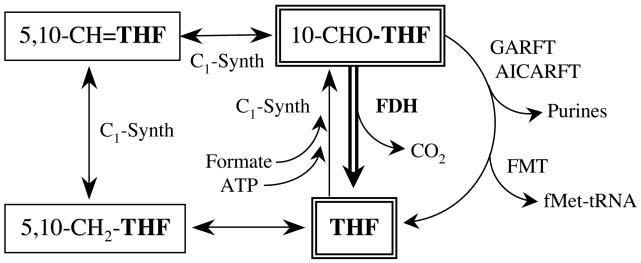
The intracellular folate pool consists of several major forms of folate that are interconvertible through multiple reactions catalyzed by more than a dozen enzymes [1, 19]. The FDH substrate, 10-formyltetrahydrofolate (10-formyl-THF), represents a major folate pool in the cell [20] and serves as a formyl donor in three biosynthetic reactions (Fig. 1) [1]. It is an essential substrate for two reactions of de novo purine biosynthesis, catalyzed by glycinamide ribonucleotide formyltransferase (GARFT, EC 2.1.2.2) and aminoimidazole-4-carboxamide ribonucleotide formyltransferase (EC 2.1.2.3) [2]. It is also required for the third reaction, formylation of methionyl-tRNA, that is an initiation step of translation in bacteria [21]. This reaction is catalyzed by methionyl-tRNA formyltransferase (FMT, EC 2.1.2.9). FMT is also present in higher organisms, including humans, where it might be required for translation initiation in mitochondria [21, 22]. While the importance of this reaction is not completely clear for mammalian cells [23], 10-formyl-THF-dependent reactions of purine biosynthesis are crucial for cellular proliferation. This was demonstrated in the experiments with inhibitors of GARFT [24], the first folate dependent enzyme in the de novo purine pathway: these inhibitors induce cytotoxicity by severe depletion of intracellular purine nucleotides [25–27]. In the cell, 10-formyl-THF is synthesized by one of two pathways (Fig. 1). The first includes two sequential reactions that convert 5,10-methylene-THF to 5,10-methenyl-THF and then to 10-formyl-THF [28]. The second is an ATP-dependent reaction that incorporates formate into the folate pool [28]. In the cytosol of mammalian liver, all three reactions are catalyzed by a trifunctional enzyme, C1-synthase [29].
Fig. 1.
Pathways of folate metabolism involving 10-formyl-THF. Abbreviations: THF, tetrahydrofolate; CH2, methylene; CH, methenyl. Enzymes involved in 10-formyl-THF metabolism are shown. C1-Synth, C1-synthase.
1.2. Disposition of FDH in cellular pathways
FDH converts 10-formyl-THF to THF and CO2 in a NADP+-dependent dehydrogenase reaction (Fig. 2). This pathway, in contrast to other 10-formyl-THF utilizing reactions, removes one-carbon groups from the folate pool instead of incorporating them into biosynthetic processes. FDH is an abundant enzyme in mammals [30]. Its levels are especially high in liver and kidney, the two principal sites of folate metabolism [30]. In rat liver, for example, it comprises about 1.2% of the total cytosolic protein [19] suggesting an essential function. The FDH gene is present in animals, including C. elegans and D. melanogaster, but is not found in prokaryotes, fungi and plants. The protein sequence is relatively conserved among different species (Table 1), including its catalytic residues (see below). It has been proposed that the FDH reaction is important for replenishing the pool of THF at the expense of 10-formyl-THF [31], thus allowing re-use of the coenzyme by other reactions [1]. The finding of a low THF level in mice lacking FDH supports the importance of the enzyme in THF regeneration [31]. These mice also showed a dramatically decreased reproductive efficiency [32], indicating that disruption of the FDH gene has significant physiological consequences. The FDH-catalyzed reaction also regulates de novo purine biosynthesis [30, 33] and formate degradation [34]. A recent study further implicated this reaction in regulation of methylation status of the cell [35]. Interestingly, although the precise biological role of FDH is not settled yet, the enzyme appears to be crucial regulator of cellular metabolism in general: it is strongly down regulated at certain physiological and pathological conditions [30, 36, 37], while its up-regulation can produce drastic antiproliferative effects [30].
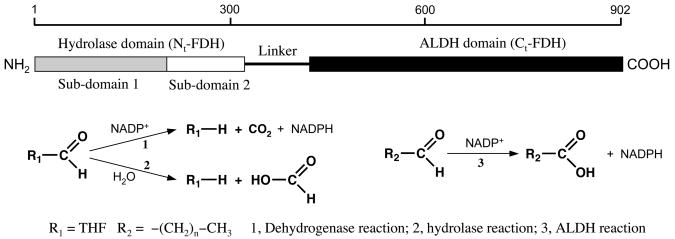
Fig. 2.
Schematic diagram of domain organization of the FDH subunit and the reactions catalyzed by the enzyme. Dehydrogenase reaction (reaction 1) requires the full-length enzyme; hydrolase reaction can be catalyzed by the full-length enzyme or by the hydrolase domain (residue 1–310); ALDH reaction can be catalyzed by the full-length enzyme or by the C-terminal ALDH domain (residue 420–902).
Table 1.
Protein sequence similarity among FDHs from different species.
| H. sapiens | R. norvegicus | M. musculus | C. elegans | D. melanogaster | |
|---|---|---|---|---|---|
| H. sapiens | 100 | 92 | 92 | 58 | 59 |
| R. norvegicus | 92 | 100 | 97 | 58 | 57 |
| M. musculus | 92 | 97 | 100 | 59 | 58 |
| C. elegans | 58 | 58 | 59 | 100 | 54 |
| D. melanogaster | 59 | 57 | 58 | 54 | 100 |
In vitro FDH catalyzes conversion of 10-formyl-THF to THF in two reactions: the NADP+-dependent dehydrogenase reaction with the formyl group ending up as CO2 and the NADP+-independent hydrolase reaction, in which formyl group is released as formate (Fig. 1). The enzymatic activity that deformylates 10-formyl-THF and yields free formate was first reported by Osborne et al. [38] in a preparation from beef liver. It has been also observed that the formation of THF was accelerated by addition of NADP+ [39] although this particular reaction does not require the participation of nicotinamide dinucleotide coenzymes. Kutzbach and Stokstad later demonstrated the NADP+-dependent oxidative deformylation of 10-formyl-THF (dehydrogenase reaction) using a partially purified enzyme [40, 41]. In this reaction the formyl group of 10-formyl-THF is converted to CO2 rather than to formate. More detailed characterization of the enzyme has confirmed that both activities, the hydrolase and the dehydrogenase, are catalyzed by the same enzyme [42, 43]. It has been also observed that the rate of the hydrolase reaction is 21% of the rate of the dehydrogenase reaction [42]. Despite these earlier findings, the hydrolase reaction was the subject of controversy for years. While Rios-Orlandi et al. showed that both dehydrogenase and hydrolase activities can take place simultaneously [44], Case et al. later reported that the two activities resided in two different but closely related enzymes, from cytosolic and mitochondrial fractions, respectively [45]. This controversy was resolved in favor of the original observation of an enzyme with dual catalytic activity after expression of recombinant rat liver FDH [46]. These studies have demonstrated that the recombinant enzyme indeed carries out both reactions, thus proving that both activities are associated with the product of a single gene, i.e. reside in one polypeptide [46].
2. Domain organization of FDH
2.1. Overview
Earlier studies have demonstrated that the enzyme is a tetramer of similar or identical subunits, each with a molecular mass of about 100 kDa [42]. Cloning of rat FDH cDNA by Cook et al. [47] confirmed that the protein is comprised of identical 902 amino acid residue subunits. Analysis of the FDH amino acid sequence performed in this study revealed the unexpected chimeric nature of the enzyme. Three distinct domains were discerned (Fig. 2): an N-terminal folate-binding domain (Nt-FDH, residues 1–310), an intermediate domain of unknown nature (residues 311–419) and an aldehyde dehydrogenase-like C-terminal domain (Ct-FDH, residue 420–902) [47]. The boundaries between the FDH domains are arbitrary and were first assigned based on the sequence similarity to other proteins. Thus, the N-terminal domain revealed identity with two enzymes utilizing 10-fFDH as a formyl donor in biosynthetic reactions: GARFT [48, 49] and FMT [50]. The C-terminal domain was found to be similar to aldehyde dehydrogenase family members (reviewed in [51, 52]). The discovery that FDH has high similarity with aldehyde dehydrogenases initiated a search for an aldehyde dehydrogenase activity of the enzyme. Unsurprisingly, studies have demonstrated that FDH possesses this activity towards short chain aldehydes, converting them to corresponding acids (Fig. 2) [47]. A later study employing proteolytic digestion of purified rabbit FDH has demonstrated that the 32 kDa N-terminal domain carries the folate binding site and the hydrolase activity while the 53 kDa C-terminal domain carries the NADP+ binding site and catalyses aldehyde dehydrogenase activity [53]. These findings have been further confirmed in experiments with expression of different recombinant constructs of rat FDH [54–56].
The intermediate domain was assigned a linker function between the two domains based on the absence of identity to either of the group of proteins [47]. Moreover, at that time, the intermediate domain did not reveal any sequence similarity to known proteins. Interestingly, in the absence of the intermediate domain, the two catalytic domains are not capable of producing 10-formyl-THF dehydrogenase activity [53, 54]. Therefore, the intermediate domain evidently couples the N-terminal and C-terminal domains into a functional enzyme.
2.2. Structure and function of the N-terminal domain of FDH
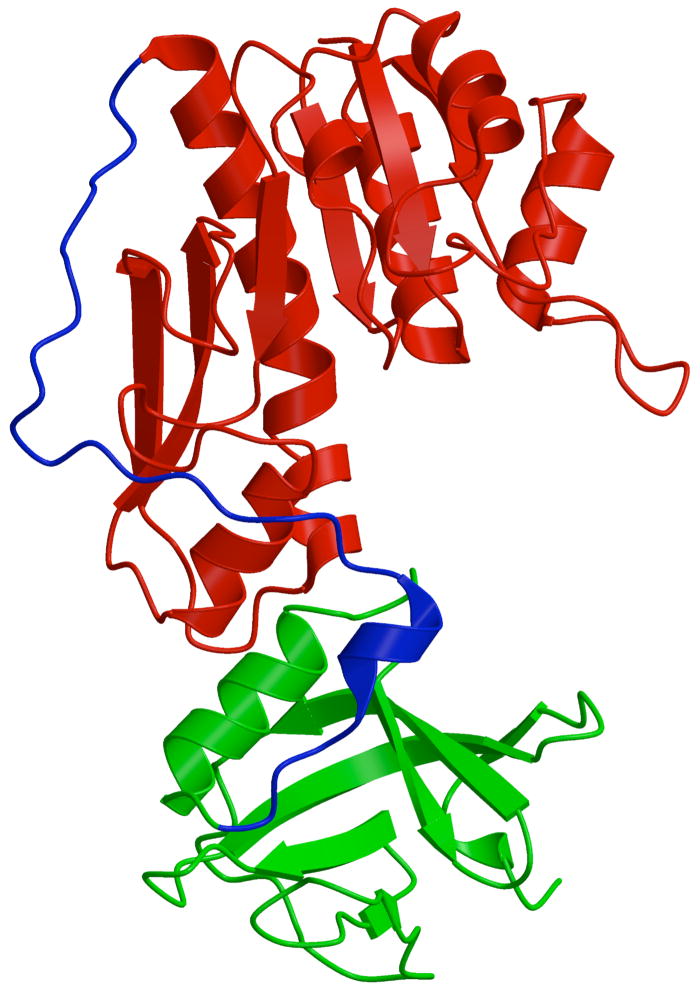
Nt-FDH (residues 1–310) can be expressed separately as a functional enzyme that allowed its detailed characterization [54]. The folate-binding site and the hydrolase activity reside in this domain, and two other domains of FDH do not contribute to folate binding or hydrolase reaction [53–55]. By itself, Nt-FDH exists as a monomer and is not involved in FDH tetramer formation. The crystal structures of the N-terminal domain of rat and human enzyme have been solved [57, 58]. The sequence similarity between Nt-FDH of the two species is 93% that obviously defines very close superimposition of their structures. The Nt-FDH can be divided into two sub-domains, N-terminal (subdomain 1) and C-terminal (sub-domain 2) connected by a long stretch of polypeptide chain, which generally lacks ordered secondary structure (Fig. 3). Sub-domain 1 contains a Rossmann fold [59] while sub-domain 2 represents a slightly open β-barrel that is similar in topology to the OB fold [60].
Fig. 3.
Overall structure of the N-terminal domain of FDH (Nt-FDH). Red, the folate-binding sub-domain; green, the sub-domain similar to tRNA binding domain of FMT; blue, the linker connecting two sub-domains.
The overall sequence identity between three 10-formyl-THF utilizing enzymes, Nt-FDH, GARFT and FMT, does not exceed 32%, but there is a region of about 70 amino acid residues in the middle of their sequences where identity increases to about 50% [61]. This region was originally predicted to be a folate binding site [47] that was later confirmed by crystal structure analysis [58, 62, 63]. Overall, sub-domain 1 of Nt-FDH shows a fold similar with both FMT and GART and superimposes closely in all three enzymes; with the closest overlap being the central seven-stranded sheet comprising the folate-binding site [57]. Two catalytically important and strictly conserved residues, aspartate 142 and histidine 106, have been identified in the active sites of FDH [61, 64]. These residues are also essential for catalysis in GARFT and FMT [65, 66]. They reside in the hydrophobic cleft that serves as a folate-binding pocket. The respective aspartates and histidines overlap very closely in superimposed structures of the three enzymes implying similarity of catalytic mechanisms. In the proposed mechanism of the hydrolase catalysis, Asp142 has two functions [57]: it activates a water molecule facilitating its nucleophilic attack on the carbonyl carbon of 10-formyl-THF and then it participate in the intermediate stabilization by compensating for a positive charge on quaternary amine at N10 of 10-formyl-THF. His106 in this mechanism participates in the stabilization of the other part of the intermediate, a negatively charged oxygen of a tetrahedral carbon [57].
Role of the C-terminal sub-domain of Nt-FDH
Compared to GARFT, Nt-FDH and FMT both have an additional sub-domain, which is very similar in topology in the two proteins. In FMT, it is responsible for binding methionyl-tRNA [67, 68]. A nucleic acid binding activity for FDH has not been reported, but several other folate-metabolizing enzymes do act as autoregulatory translational repressors by binding to mRNA [69, 70]. Unexpectedly, it has been revealed that this domain is important for the hydrolase catalysis: deletion experiments show that at least 300 residues are required for the functional hydrolase [57], although the folate-binding site and the hydrolase catalytic center are both located within the first 200 residues of the protein. Interestingly, sub-domain 2 of FMT is capable of substituting for the corresponding domain of Nt-FDH in the hydrolase catalysis [56]. The chimera did not reveal a detectable level of 10-formyl-THF dehydrogenase activity, though. This, however, was rather a result of distortion of the interface between the hydrolase and aldehyde dehydrogenase domains of FDH caused by differently positioned carboxyl-terminus of sub-domain 2 of FMT that oriented the aldehyde dehydrogenase domain of FDH away from the hydrolase catalytic center.
2.3. Structure and function of the C-terminal domain of FDH
Overall structure
The C-terminal domain of FDH (residues 420–902) is up to 50% homologous to enzymes from a family of aldehyde dehydrogenases [47, 55] suggesting that this part of the FDH molecule is derived from an aldehyde dehydrogenase-related gene [51, 52]. The identity between FDH and aldehyde dehydrogenases is spread through the entire sequence and includes all of the regions conserved in the aldehyde dehydrogenase family [47, 51, 52, 55]. All aldehyde dehydrogenases have a strictly conserved cysteine in the active site [51], which is a key catalytic residue serving as nucleophile [71, 72]. Using site-directed mutagenesis, it has been shown that the corresponding cysteine of FDH, Cys707, is directly involved in the dehydrogenase catalysis [73]: it is crucial in both dehydrogenase reactions utilizing either 10-formyl-THF (full-length enzyme catalysis) or aldehydes [73]. The C-terminal domain can be expressed separately as a functional aldehyde dehydrogenase [55] proving that ALDH activity is carried out by this domain and does not require participation of the two other domains of FDH. Similar to other class 1 and class 2 ALDH, the C-terminal domain exists as a tetramer [53, 55], indicating that this domain is responsible for the oligomerization of subunits of FDH. However, it is unable to oxidize 10-formyl-THF, due to its inability to bind the folate substrate: this domain has neither a folate-binding site nor participates in formation of such a site [53–55].
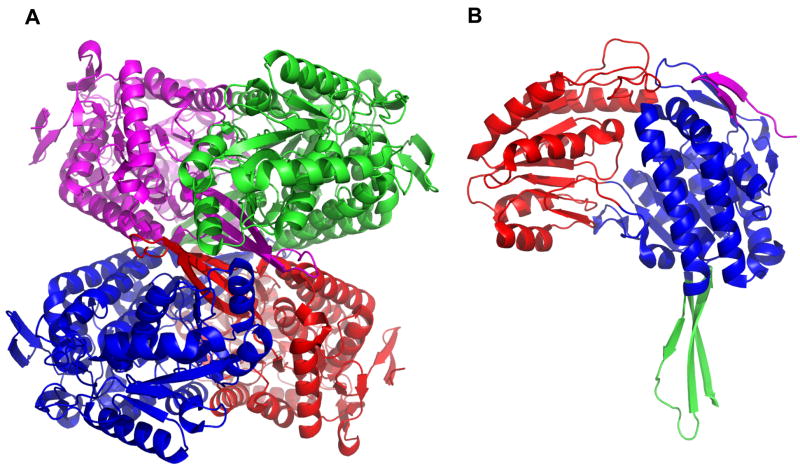
Since 1997, when the first two crystal structures of aldehyde dehydrogenases were solved [74, 75], a large number of aldehyde dehydrogenases structures have been reported. These studies permitted the identification of residues that are important for aldehyde dehydrogenase function. It was not surprising to find that most of the functional residues are present in the sequence of FDH. The recently solved crystal structure of Ct-FDH [76] confirmed a fold typical for class 1 and class 2 aldehyde dehydrogenases [75, 77] with four identical subunits arranged as a dimer of dimers [75, 77, 78] (Fig. 4A). Similar to other members of the family, the Ct-FDH subunit is comprised of three sub-domains (Fig. 4B): a NADP-binding sub-domain (residues 405–539, 564–675, and 867–893), a catalytic sub-domain (residues 676–866), and an oligomerization sub-domain (residues 540–563 and 894–902). The NADP-binding and catalytic domains are α/β in structure, whereas the oligomerization domain contains a three-stranded antiparallel β-sheet. While residues 423–902 of Ct-FDH can be superimposed closely into structures of other class 1 and 2 ALDHs, the rest of its amino-terminus is different from ALDH fold: it forms a β-hairpin, packed against the main body of the molecule (Fig. 4B). This region is not required for ALDH catalytic activity of Ct-FDH [55] and is likely a part of the interdomain interface in full-length FDH.
Fig. 4.
(A) Ribbon diagram of the Ct-FDH tetramer. The four subunits are in different colors. (B) Ribbon representation of the Ct-FDH subunit: the NADP-binding domain (blue); the catalytic domain (red); the oligomerization domain (green); a hairpin representing a part of the Ct-FDH interface with the rest of FDH is shown in magenta.
Active site and coenzyme specificity
The active site of Ct-FDH is similar to those of ALDH including the two residues essential for catalysis, Cys707 and Glu673 [75, 77]. The substrate entrance tunnel is situated between the coenzyme-binding and the catalytic domains of the protein. The amino acid composition of the tunnel is closer to that of the ALDH1 group, which is more specific towards larger substrates [77, 79]. Interestingly, while the overall mode of coenzyme binding is highly similar between Ct-FDH and class 1 and 2 ALDH, which are NAD+-specific, Ct-FDH, as well as the full-length enzyme, demonstrates a strong preference for NADP+: Km for NAD+ is about three order of magnitude higher that Km for NADP+ [55]. The structural basis for the shift in coenzyme specificity has been highlighted in our recent study [76]. Interestingly, while bacterial NADP+-specific ALDHs have been reported [80–82], and class 3 ALDH can use both NAD+ and NADP+ in vitro [83], it appears that FDH is the only example of strictly NADP+-specific ALDH in mammals. Comparison of two crystal structures of NADP+-specific bacterial ALDH, from Streptococcus mutans [81] and from Vibrio harveyi [82], with the structure of Ct-FDH showed that, in the latter, coordination of the 2′-phosphate of the coenzyme involves different residues, indicating a different mode for accommodating the extra phosphate.
Coenzyme-induced rearrangements in the active site and coenzyme mobility
As in other ALDH enzymes [74, 75], the coenzyme molecule is bound in Ct-FDH through a non-classical Rossmann fold. The nicotinamide moiety of the bound NADP+ is in the so-called hydride transfer, or extended conformation (the nicotinamide ring lies close to the cysteine nucleophile) and is buried in a pocket that approaches the active site from a direction opposite to the substrate entrance tunnel. Overall, binding of the cofactor does not result in large-scale rearrangements of the protein structure and the major differences between the apoenzyme and the NADP+-bound enzyme are observed in the active site. In the apoenzyme, the carboxylate group of Glu673 is within hydrogen bonding distance of the sulfhydryl of Cys707, whereas in the complex with NADP+, this side chain has rotated away from the cysteine and become hydrogen bonded to the amide group of the nicotinamide. In the complex with NADPH, Glu673 occupies the same position as in the apoenzyme, thus restoring the contact with Cys707. The nicotinamide moiety of the reduced coenzyme in this structure is distorted and displaced away from the cysteine and the catalytic center, similar to so-called hydrolysis position [74, 75, 77, 84, 85]. The two conformations of the coenzyme observed in the Ct-FDH structures may correlate with two states of the reaction, acylation and deacylation, as was proposed for other ALDHs [75, 77, 85–87]. In support of this hypothesis, the transition from well-ordered to distorted conformation of the nicotinamide portion of NADP+, bound to Ct-FDH, was directly observed after soaking crystals of binary in propanal, that apparently converted an oxidized coenzyme to its reduced form [76].
Unusual feature of Ct-FDH structure: a covalent bond between catalytic cysteine and nicotinamide
While the flexibility of the coenzyme bound to ALDH is a hallmark of this enzyme family and correlates with enzymatic activity [88], the Ct-FDH structure revealed an unusual mode of NADP+ binding. Similar to some other ALDH [86], two conformations of catalytic Cys707 were observed in crystal structures of both apoenzyme and its binary complex with NADP+ [76]. Surprisingly, in one of these conformations the distance between the sulfur of the cysteine and C4 atom of the nicotinamide ring is 1.6 Å, indicating a covalent bond between the two atoms. Furthermore, the electron density of the nicotinamide ring is distorted, which is consistent with the tetrahedral geometry of a covalent bond. The presence of such a bond was confirmed by absorption spectroscopy that revealed the appearance of an additional peak in the spectrum of NADP+ bound to Ct-FDH. The observed covalent bond is of a temporary (transient) nature and is easily reversible: NADP+ readily dissociates from the complex with Ct-FDH [76].
At first glance, such a covalent bond appears to be highly unusual. In general, however, nucleophiles, including the -S− anion, can add reversibly to the para position (C4) of nicotinamide to form adducts, producing structures reminiscent of those of the reduced coenzymes [89, 90]. Accordingly, covalent adducts of the coenzyme in other enzymes have also been reported [91–94], including demonstration of a covalent attachment of NADH to a catalytic cysteine in GAPDH [95]. Interestingly, prediction of such a bond by molecular dynamics simulations in ALDH3 [96] indicates that its formation is thermodynamically favorable. With this regard, re-examination of some existing structures of ALDH, as well as generation of high-resolution structures, could provide additional examples of the covalent adduct. Indeed, in two cases, such a bond has been already detected [76, 97]: in the structure of the allosteric nonphosphorylating GAPDH from Thermoproteus tenax [87] and in ALDH2 [85]. It is not clear at present whether the covalent bond is a part of ALDH mechanism. In support of this possibility, chemical activation of the nucleophilic cysteine in non-phosphorylating GAPDH upon coenzyme binding was observed [98].
There are several reasons why the equilibrium of covalent/dissociated states might exist. (1) The interaction may serve to maintain the cysteine in an activated state after its proton has been abstracted by Glu673 and the glutamate has been displaced by NADP+. (2) It may also help activate the C4 of the nicotinamide for hydride transfer. (3) It may hold the coenzyme in the hydride transfer position prior to substrate binding preventing it from occupying the hydrolysis position. It has been also proposed that the covalent bond may protect the cysteine from oxidation during the reaction [97]. Formation of a covalent bond between enzyme and coenzyme might also be a mechanism of activation of the esterase reaction by NAD+/NADP+ [99]. ALDHs display an esterase activity that is enhanced by a nicotinamide coenzyme even though it is not a participant in the esterase reaction [100]. FDH also displays this type of activity [101]. Thus, stabilization of a deprotonated cysteine by a positively charged coenzyme might facilitate a nucleophilic attack on the substrate. We have also hypothesized [76] that “half-of-the-site” reactivity of some ALDH [102–104] could be explained in the context of a covalent bond between enzyme and coenzyme: equilibrium between covalent and non-covalent states of NADP+, each with approximately 50% occupancy, could result in only half of the active sites being active at a specific time-point.
2.4. Role of the intermediate domain of FDH
Until recently, the origin of the intermediate domain (residues 311–399) and its precise function remained unknown. Indeed, it does not have sufficient sequence identity to known proteins to allow its assignment to a specific protein family. Early studies have demonstrated that this domain is important for catalysis: in its absence the mixture of two separately expressed catalytic domains of FDH, the N- and C-terminal, did not produce 10-formyl-THF dehydrogenase activity (34). It has been proposed that this domain is not directly involved in the enzymatic mechanism but serves as a linker, which brings two catalytic domains in close proximity to each other to allow catalysis (35). Therefore, it was surprising to find out that this domain is a structural and functional homolog of carrier proteins. These proteins are components of multi-enzyme complexes involved in fatty acid, polyketide, and non-ribosomal peptide biosynthesis (reviewed in [105, 106]). Their characteristic feature is the 4′-phosphopantetheine (4′-PP) moiety, covalently attached to a conserved serine residue through a phosphodiester bond, functioning as a moving arm that holds a growing chain of fatty acids, polyketides, or peptides [107]. Another enzyme with a 4′-PP modification is α-aminoadipate semialdehyde reductase, a component of lysine biosynthesis specific to fungi [108, 109]. In higher organisms (including humans) the reversal of this pathway has been suggested to serves as a lysine degradation route [110].
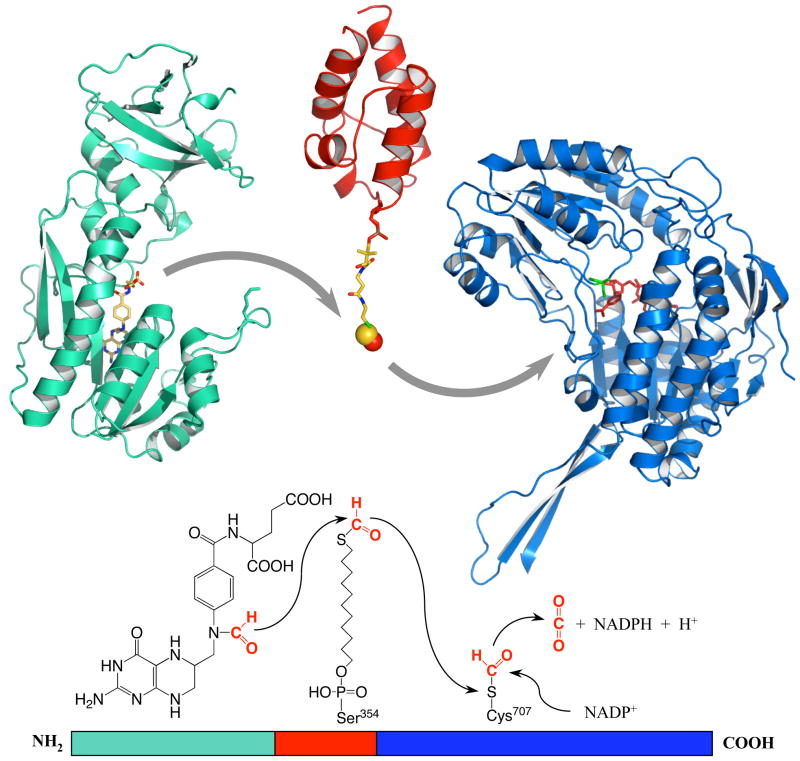
All carrier proteins are similar in structure representing a three- or four-helix bundle connected by loops with 4′-PP arm covalently attached to a conserved serine located in one of these loops. The NMR structure of the intermediate domain of human FDH, recently solved by the RIKEN Structural Genomics/Proteomics Initiative (Protein Data Bank # 2CQ8), has demonstrated that the fold is similar to other carrier proteins (Fig. 5). While this structure does not have a prosthetic group, the alignment with other carrier proteins has identified Ser354 as the residue carrying this modification, a suggestion confirmed by site-directed mutagenesis experiments [111]. These studies add an additional layer of complexity to the FDH functional organization by highlighting the requirement for a post-translational modification. The presence of the 4′-PP group, which extends about 20 Å from the peptide part of the intermediate domain, provides a mechanism to access the catalytic centers of FDH, a feature especially crucial for the ALDH center which is situated at the end of 12 Å deep tunnel. How the 4′-PP arm transfers formyl between domains is not clear, but conformational changes within the intermediate domain [112], as well as the catalytic domains [57], are likely to assist in this process.
Fig. 5.
FDH mechanism. FDH subunit (a single polypeptide) consists of three domains: N-terminal (green), intermediate (red) and C-terminal (blue). FDH converts its substrate, 10-formyl-THF, to THF and CO2 in three steps: (i) N-terminal domain hydrolytically cleaves the bond between formyl group and THF; (ii) 4′-PP prosthetic group (shown in stick mode), covalently bound to the intermediate domain at Ser354, transfers formyl (red-yellow space fill) from N-terminal to C-terminal domain; (iii) formyl is oxidized in the C-terminal domain to CO2 by function of catalytic nucleophile Cys707 with participation of NADP+. For simplicity, Ct-FDH is shown as a monomer. Bound folate substrate and NADP+ molecule are shown in N- and C-terminal domains, correspondingly.
3. Mechanism of FDH catalysis
The site-directed mutagenesis studies provided evidence that the hydrolase reaction and aldehyde dehydrogenase reaction are parts of the overall dehydrogenase catalytic mechanism of FDH towards 10-formyl-THF. Thus, replacement of either of the two conserved catalytic residues in the hydrolase active center, His106 and Asp142, resulted in mutant enzymes lacking both hydrolase and dehydrogenase activities and suggested a tight connection between the two mechanisms [61, 64]. Likewise, mutation of catalytically important Cys707 [73] or Glu673 [76] in the aldehyde dehydrogenase C-terminal domain abolished both dehydrogenase activities but the hydrolase activity remained. The likely explanation of these results is that the 10-formyl-THF dehydrogenase reaction is not a separate reaction but a combination of two steps, the hydrolase reaction and aldehyde dehydrogenase reaction and that the 10-formyl-THF hydrolysis step should always precede the dehydrogenase catalysis. In this mechanism, uncoupling the Ct-FDH domain would stop the overall reaction after the hydrolase step releasing formate [112]. While it is not clear whether hydrolase and ALDH reactions take place in vivo, they are a reflection of the fact that the catalytic domains of FDH can function as separate enzymes [54, 55].
Thus, to convert 10-formyl-THF to THF and CO2 FDH employs a complex mechanism, which includes three general sequential steps, two catalytic and one transfer as schematically shown in Fig. 5. Each step is accomplished by a separate domain of the enzyme. In the first step, which takes place in the N-terminal hydrolase domain, the formyl group is removed by a hydrolytic cleavage from 10-formylTHF. In the second step, generated formyl is transferred onto the sulfhydryl moiety of 4′-PP arm of the intermediate domain. The arm then swings from the hydrolase catalytic center into the ALDH catalytic center and donates the formyl to the ALDH active site Cys707. In the third step, carried out by the ALDH C-terminal domain, the formyl is oxidized to CO2 through an ALDH-like mechanism, which involves reduction of NADP+ into NADPH.
The sequence of events proposed above reflects the overall mechanism of FDH but its details await future investigations. For example, studies of the hydrolase catalysis of FDH indicated that participation of a sulfhydryl moiety is essential to complete the reaction: in vitro it requires the presence of millimolar concentrations of 2-mercaptoethanol [46, 101]. A molecule of 2-mercaptoethanol bridging two catalytic residues, His106 and Asp142, was seen in the crystal structure of rat Nt-FDH [57]. It occupies the same spatial position as the expected position of the formyl group of 10-formyl-THF bound in the active site [62], thus mimicking the formyl moiety. It has been proposed therefore that, in the absence of the rest of FDH molecule, THF and formate remain bound in the active site after the hydrolase reaction but displacement of formate by 2-mercaptoethanol brings the reaction to completion. In light of the function of the intermediate domain it is more likely, however, that 2-mercaptoethanol replaces a functionality lost in the absence of the intermediate domain thus forming a thiol ester with formate, which is then rapidly hydrolyzed yielding formic acid. Of note, 10-formyl-THF hydrolase, which shares some sequence similarity with Nt-FDH, GARFT and FMT, exists in bacteria [113, 114]. The fact that this enzyme does not require participation of a thiol for catalysis implies a feature of Nt-FDH mechanism that prevents hydrolase reaction if an assisted discharge of formyl is not available.
While the ALDH catalysis of FDH is similar to a common ALDH mechanism, the exact mechanism of oxidation of formyl to CO2 as a part of overall 10-formyl-THF dehydrogenase reaction is not clear. With this regard it should be noted that the oxidation states of a substrate and a product of ALDH reaction (an aldehyde and an acid oxidation level, correspondingly) are different from those of 10-formyl-THF dehydrogenase reaction (an acid and CO2 oxidation level, correspondingly). Therefore, oxidation of formyl should proceed through a different intermediate that, in turn, might require an altered ALDH mechanism. Curiously, an NAD+-dependent dehydrogenase oxidizing formate to CO2, which exists in bacteria, yeast and plants, employs a mechanism different from that of ALDH: it directly transfers a hydride ion from the substrate to NAD+ without stages of acid-base catalysis [115].
4. Conclusion
Domain organization is an intrinsic element of protein structure and the majority of proteins consist of distinctive domains that can act independently or cooperatively to achieve a unique function [116, 117]. The interaction between functional modules within a single polypeptide determines mechanisms by which multidomain proteins operate. Often evolution combines enzymes from the same metabolic pathway in one polypeptide thus compartmentalizing the synthesis of the final product and allowing channeling intermediates between reactions within the pathway [49, 118–121]. In such multi-functional fusions each module serves its own catalytic purpose. In the case of FDH, however, the combination of three genes creates an end product with new enzymatic activity.
Acknowledgments
I would like to thank the following people who helped me in preparation of this review: Christopher Davies, Henry Donato, Natalia I. Krupenko, Graham Solomons and Yaroslav Tsybovsky. This work was supported by National Institute of Health grants DK54388 and CA95030.
Abbreviations
- 4′-PP
4′-phosphopantetheine
- ALDH
aldehyde dehydrogenase
- Ct-FDH
C-terminal domain of FDH
- FDH
10-formyltetrahydrofolate dehydrogenase
- FMT
methionyl-tRNA formyltransferase
- GARFT
glycinamid rybonucleotide formyltransferase
- Nt-FDH
N-terminal domain of FDH
- THF
tetrahydrofolate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- 1.Wagner C. Biochemical role of folate in cellular metabolism. In: Bailey LB, editor. Folate in Health and Disease. Marcel Dekker, Inc.; New York: 1995. pp. 23–42. [Google Scholar]
- 2.Benkovic SJ. The transformylase enzymes in de novo purine biosynthesis. Trends Biochem Sci. 1984;9:320–322. [Google Scholar]
- 3.Assaraf YG. Molecular basis of antifolate resistance. Cancer Metastasis Rev. 2007;26(1):153–181. doi: 10.1007/s10555-007-9049-z. [DOI] [PubMed] [Google Scholar]
- 4.Stanger O. Physiology of folic acid in health and disease. Curr Drug Metab. 2002;3(2):211–223. doi: 10.2174/1389200024605163. [DOI] [PubMed] [Google Scholar]
- 5.Blom HJ, Shaw GM, den Heijer M, Finnell RH. Neural tube defects and folate: case far from closed. Nat Rev Neurosci. 2006;7(9):724–731. doi: 10.1038/nrn1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stover PJ. Physiology of folate and vitamin B12 in health and disease. Nutr Rev. 2004;62(6 Pt 2):S3–12. doi: 10.1111/j.1753-4887.2004.tb00070.x. discussion S13. [DOI] [PubMed] [Google Scholar]
- 7.Glynn SA, Albanes D. Folate and cancer: A review of the literature. Nutrition and Cancer. 1994;22:101–119. doi: 10.1080/01635589409514336. [DOI] [PubMed] [Google Scholar]
- 8.Jhaveri MS, Wagner C, Trepel JB. Impact of extracellular folate levels on global gene expression. Mol Pharmacol. 2001;60(6):1288–1295. doi: 10.1124/mol.60.6.1288. [DOI] [PubMed] [Google Scholar]
- 9.Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci U S A. 1997;94(7):3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruman II, Kumaravel TS, Lohani A, Pedersen WA, Cutler RG, Kruman Y, Haughey N, Lee J, Evans M, Mattson MP. Folic acid deficiency and homocysteine impair DNA repair in hippocampal neurons and sensitize them to amyloid toxicity in experimental models of Alzheimer’s disease. J Neurosci. 2002;22(5):1752–1762. doi: 10.1523/JNEUROSCI.22-05-01752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borman LS, Branda RF. Nutritional folate deficiency in Chinese hamster ovary cells. I. Characterization of the pleiotropic response and its modulation by nucleic acid precursors. J Cell Physiol. 1989;140(2):335–343. doi: 10.1002/jcp.1041400220. [DOI] [PubMed] [Google Scholar]
- 12.Huang RF, Ho YH, Lin HL, Wei JS, Liu TZ. Folate deficiency induces a cell cycle-specific apoptosis in HepG2 cells. J Nutr. 1999;129(1):25–31. doi: 10.1093/jn/129.1.25. [DOI] [PubMed] [Google Scholar]
- 13.Chu E, Allegra CJ. Antifolates. In: Chabner BA, Longo DL, editors. Cancer Chemotherapy and Biotherapy: Principles and Practice. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 109–148. [Google Scholar]
- 14.Cooper BA. Folate nutrition in man and animals. In: Blakley RL, Whitehead VM, editors. Folates and pterins. John Wiley & Sons, Inc; New York: 1986. pp. 49–74. [Google Scholar]
- 15.Rock CL, Lampe JW, Patterson RE. Nutrition, genetics, and risks of cancer. Annu Rev Public Health. 2000;21:47–64. doi: 10.1146/annurev.publhealth.21.1.47. [DOI] [PubMed] [Google Scholar]
- 16.Fleming A. The role of folate in the prevention of neural tube defects: human and animal studies. Nutr Rev. 2001;59(8 Pt 2):S13–20. doi: 10.1111/j.1753-4887.2001.tb05497.x. discussion S21–13. [DOI] [PubMed] [Google Scholar]
- 17.Moat SJ, Lang D, McDowell IF, Clarke ZL, Madhavan AK, Lewis MJ, Goodfellow J. Folate, homocysteine, endothelial function and cardiovascular disease. J Nutr Biochem. 2004;15(2):64–79. doi: 10.1016/j.jnutbio.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Choi SW, Mason JB. Folate and carcinogenesis: an integrated scheme. J Nutr. 2000;130(2):129–132. doi: 10.1093/jn/130.2.129. [DOI] [PubMed] [Google Scholar]
- 19.Kisliuk RL. Folate biochemistry in relation to antifolate selectivity. In: Jackman AL, editor. Antifolate drugs in cancer therapy. Humana Press; Totowa, New Jersey: 1999. pp. 13–36. [Google Scholar]
- 20.Horne DW, Patterson D, Cook RJ. Effect of nitrous oxide inactivation of vitamin B12-dependent methionine synthetase on the subcellular distribution of folate coenzymes in rat liver. Arch Biochem Biophys. 1989;270(2):729–733. doi: 10.1016/0003-9861(89)90556-0. [DOI] [PubMed] [Google Scholar]
- 21.Bianchetti R, Lucchini G, Crosti P, Tortora P. Dependence of mitochondrial protein synthesis initiation on formylation of the initiator methionyl-tRNAf. J Biol Chem. 1977;252(8):2519–2523. [PubMed] [Google Scholar]
- 22.Spencer AC, Spremulli LL. Interaction of mitochondrial initiation factor 2 with mitochondrial fMet-tRNA. Nucleic Acids Res. 2004;32(18):5464–5470. doi: 10.1093/nar/gkh886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibbetts AS, Oesterlin L, Chan SY, Kramer G, Hardesty B, Appling DR. Mammalian mitochondrial initiation factor 2 supports yeast mitochondrial translation without formylated initiator tRNA. J Biol Chem. 2003;278(34):31774–31780. doi: 10.1074/jbc.M304962200. [DOI] [PubMed] [Google Scholar]
- 24.Beardsley GP, Moroson BA, Taylor EC, Moran RG. A new folate antimetabolite, 5,10-dideaza-5,6,7,8-tetrahydrofolate is a potent inhibitor of de novo purine synthesis. J Biol Chem. 1989;264(1):328–333. [PubMed] [Google Scholar]
- 25.Pizzorno G, Moroson BA, Cashmore AR, Beardsley GP. (6R)-5,10-Dideaza-5,6,7,8-tetrahydrofolic acid effects on nucleotide metabolism in CCRF-CEM human T-lymphoblast leukemia cells. Cancer Res. 1991;51(9):2291–2295. [PubMed] [Google Scholar]
- 26.Pizzorno G, Davis SJ, Hartigan DJ, Russello O. Enhancement of antineoplastic activity of 5-fluorouracil in mice bearing colon 38 tumor by (6R)5,10-dideazatetrahydrofolic acid. Biochem Pharmacol. 1994;47(11):1981–1988. doi: 10.1016/0006-2952(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 27.Mendelsohn WLGJF, Walling JM. Preclinical and clinical evaluation of the glycinamide ribonucleotide formyltransferase inhibitors Lometrexol and LY309887. In: Jackman AL, editor. Antifolate drugs in cancer therapy. Humana Press; Totowa, New Jersey: 1999. pp. 261–280. [Google Scholar]
- 28.MacKenzie RE. Biogenesis and interconversion of substituted tetrahydrofolates. In: Blakley RL, Benkocic SJ, editors. Folates and Pterins. John Wiley & Sons, Inc.; New York: 1984. pp. 255–306. [Google Scholar]
- 29.Hum DW, Bell AW, Rozen R, MacKenzie RE. Primary structure of a human trifunctional enzyme. Isolation of a cDNA encoding methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase. J Biol Chem. 1988;263(31):15946–15950. [PubMed] [Google Scholar]
- 30.Krupenko SA, Oleinik NV. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth Differ. 2002;13(5):227–236. [PubMed] [Google Scholar]
- 31.Champion KM, Cook RJ, Tollaksen SL, Giometti CS. Identification of a heritable deficiency of the folate-dependent enzyme 10-formyltetrahydrofolate dehydrogenase in mice. Proc Natl Acad Sci U S A. 1994;91(24):11338–11342. doi: 10.1073/pnas.91.24.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giometti CS, Tollaksen SL, Grahn D. Altered protein expression detected in the F1 offspring of male mice exposed to fission neutrons. Mutat Res. 1994;320(1–2):75–85. doi: 10.1016/0165-1218(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 33.Oleinik NV, Krupenko NI, Priest DG, Krupenko SA. Cancer cells activate p53 in response to 10-formyltetrahydrofolate dehydrogenase expression. Biochem J. 2005;391(Pt 3):503–511. doi: 10.1042/BJ20050533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tephly TR. The toxicity of methanol. Life Sci. 1991;48(11):1031–1041. doi: 10.1016/0024-3205(91)90504-5. [DOI] [PubMed] [Google Scholar]
- 35.Anguera MC, Field MS, Perry C, Ghandour H, Chiang EP, Selhub J, Shane B, Stover PJ. Regulation of Folate-mediated One-carbon Metabolism by 10-Formyltetrahydrofolate Dehydrogenase. J Biol Chem. 2006;281(27):18335–18342. doi: 10.1074/jbc.M510623200. [DOI] [PubMed] [Google Scholar]
- 36.Epperson LE, Dahl TA, Martin SL. Quantitative analysis of liver protein expression during hibernation in the golden-mantled ground squirrel. Mol Cell Proteomics. 2004;3(9):920–933. doi: 10.1074/mcp.M400042-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Leonard JF, Courcol M, Mariet C, Charbonnier A, Boitier E, Duchesne M, Parker F, Genet B, Supatto F, Roberts R, Gautier JC. Proteomic characterization of the effects of clofibrate on protein expression in rat liver. Proteomics. 2006;6(6):1915–1933. doi: 10.1002/pmic.200500251. [DOI] [PubMed] [Google Scholar]
- 38.Osborn MJ, Hatefi Y, Kay LD, Huennekens FM. Evidence for the enzymic deacylation of N10-formyl tetrahydro-folic acid. Biochim Biophys Acta. 1957;26(1):208–210. doi: 10.1016/0006-3002(57)90077-x. [DOI] [PubMed] [Google Scholar]
- 39.Huennekens FM, Scrimgeour KG. Methods in Enzymology. 1963;6:673–675. [Google Scholar]
- 40.Kutzbach C, Stokstad EL. Partial purification of a 10-formyl-tetrahydrofolate: NADP oxidoreductase from mammalian liver. Biochem Biophys Res Commun. 1968;30(2):111–117. doi: 10.1016/0006-291x(68)90456-7. [DOI] [PubMed] [Google Scholar]
- 41.Kutzbach C, Stokstad ELR. 10-Formyl tetrahydrofolate: NADP oxidoreductase. Methods Enzymol. 1971;18B:793–798. doi: 10.1016/0006-291x(68)90456-7. [DOI] [PubMed] [Google Scholar]
- 42.Scrutton MC, Beis I. Inhibitory effects of histidine and their reversal. The roles of pyruvate carboxylase and N10-formyltetrahydrofolate dehydrogenase. Biochem J. 1979;177(3):833–846. doi: 10.1042/bj1770833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Min H, Shane B, Stokstad EL. Identification of 10-formyltetrahydrofolate dehydrogenase-hydrolase as a major folate binding protein in liver cytosol. Biochim Biophys Acta. 1988;967(3):348–353. doi: 10.1016/0304-4165(88)90097-9. [DOI] [PubMed] [Google Scholar]
- 44.Rios-Orlandi EM, Zarkadas CG, MacKenzie RE. Formyltetrahydrofolate dehydrogenase-hydrolase from pig liver: simultaneous assay of the activities. Biochim Biophys Acta. 1986;871(1):24–35. doi: 10.1016/0167-4838(86)90129-9. [DOI] [PubMed] [Google Scholar]
- 45.Case GL, Kaisaki PJ, Steele RD. Resolution of rat liver 10-formyltetrahydrofolate dehydrogenase/hydrolase activities. J Biol Chem. 1988;263(21):10204–10207. [PubMed] [Google Scholar]
- 46.Krupenko SA, Wagner C, Cook RJ. Recombinant 10-formyltetrahydrofolate dehydrogenase catalyses both dehydrogenase and hydrolase reactions utilizing the synthetic substrate 10-formyl-5,8-dideazafolate. Biochem J. 1995;306(Pt 3):651–655. doi: 10.1042/bj3060651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cook RJ, Lloyd RS, Wagner C. Isolation and characterization of cDNA clones for rat liver 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1991;266(8):4965–4973. [PubMed] [Google Scholar]
- 48.Smith JM, Daum HA., 3rd Identification and nucleotide sequence of a gene encoding 5′-phosphoribosylglycinamide transformylase in Escherichia coli K12. J Biol Chem. 1987;262(22):10565–10569. [PubMed] [Google Scholar]
- 49.Schild D, Brake AJ, Kiefer MC, Young D, Barr PJ. Cloning of three human multifunctional de novo purine biosynthetic genes by functional complementation of yeast mutations. Proc Natl Acad Sci U S A. 1990;87(8):2916–2920. doi: 10.1073/pnas.87.8.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guillon JM, Mechulam Y, Schmitter JM, Blanquet S, Fayat G. Disruption of the gene for Met-tRNA(fMet) formyltransferase severely impairs growth of Escherichia coli. J Bacteriol. 1992;174(13):4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hempel J, Nicholas H, Lindahl R. Aldehyde dehydrogenases: widespread structural and functional diversity within a shared framework. Protein Sci. 1993;2(11):1890–1900. doi: 10.1002/pro.5560021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perozich J, Nicholas H, Wang BC, Lindahl R, Hempel J. Relationships within the aldehyde dehydrogenase extended family. Protein Sci. 1999;8(1):137–146. doi: 10.1110/ps.8.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schirch D, Villar E, Maras B, Barra D, Schirch V. Domain structure and function of 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1994;269(40):24728–24735. [PubMed] [Google Scholar]
- 54.Krupenko SA, Wagner C, Cook RJ. Domain structure of rat 10-formyltetrahydrofolate dehydrogenase. Resolution of the amino-terminal domain as 10-formyltetrahydrofolate hydrolase. J Biol Chem. 1997;272(15):10273–10278. doi: 10.1074/jbc.272.15.10273. [DOI] [PubMed] [Google Scholar]
- 55.Krupenko SA, Wagner C, Cook RJ. Expression, purification, and properties of the aldehyde dehydrogenase homologous carboxyl-terminal domain of rat 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1997;272(15):10266–10272. doi: 10.1074/jbc.272.15.10266. [DOI] [PubMed] [Google Scholar]
- 56.Reuland SN, Vlasov AP, Krupenko SA. Modular organization of FDH: Exploring the basis of hydrolase catalysis. Protein Sci. 2006;15(5):1076–1084. doi: 10.1110/ps.052062806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chumanevich AA, Krupenko SA, Davies C. The crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase: mechanism of hydrolysis and its interplay with the dehydrogenase domain. J Biol Chem. 2004;279(14):14355–14364. doi: 10.1074/jbc.M313934200. [DOI] [PubMed] [Google Scholar]
- 58.Kursula P, Schuler H, Flodin S, Nilsson-Ehle P, Ogg DJ, Savitsky P, Nordlund P, Stenmark P. Structures of the hydrolase domain of human 10-formyltetrahydrofolate dehydrogenase and its complex with a substrate analogue. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 11):1294–1299. doi: 10.1107/S0907444906026849. [DOI] [PubMed] [Google Scholar]
- 59.Rossmann MG, Moras D, Olsen KW. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 60.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. Embo J. 1993;12(3):861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krupenko SA, Wagner C. Aspartate 142 is involved in both hydrolase and dehydrogenase catalytic centers of 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1999;274(50):35777–35784. doi: 10.1074/jbc.274.50.35777. [DOI] [PubMed] [Google Scholar]
- 62.Almassy RJ, Janson CA, Kan CC, Hostomska Z. Structures of apo and complexed Escherichia coli glycinamide ribonucleotide transformylase. Proc Natl Acad Sci U S A. 1992;89(13):6114–6118. doi: 10.1073/pnas.89.13.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klein C, Chen P, Arevalo JH, Stura EA, Marolewski A, Warren MS, Benkovic SJ, Wilson IA. Towards structure-based drug design: crystal structure of a multisubstrate adduct complex of glycinamide ribonucleotide transformylase at 1.96 A resolution. J Mol Biol. 1995;249(1):153–175. doi: 10.1006/jmbi.1995.0286. [DOI] [PubMed] [Google Scholar]
- 64.Krupenko SA, Vlasov AP, Wagner C. On the role of conserved histidine 106 in 10-formyltetrahydrofolate dehydrogenase catalysis: connection between hydrolase and dehydrogenase mechanisms. J Biol Chem. 2001;276(26):24030–24037. doi: 10.1074/jbc.M009257200. [DOI] [PubMed] [Google Scholar]
- 65.Warren MS, Marolewski AE, Benkovic SJ. A rapid screen of active site mutants in glycinamide ribonucleotide transformylase. Biochemistry. 1996;35(27):8855–8862. doi: 10.1021/bi9528715. [DOI] [PubMed] [Google Scholar]
- 66.Newton DT, Mangroo D. Mapping the active site of the Haemophilus influenzae methionyl-tRNA formyltransferase: residues important for catalysis and tRNA binding. Biochem J. 1999;339(Pt 1):63–69. [PMC free article] [PubMed] [Google Scholar]
- 67.Schmitt E, Blanquet S, Mechulam Y. Structure of crystalline Escherichia coli methionyl-tRNA(f)Met formyltransferase: comparison with glycinamide ribonucleotide formyltransferase. Embo J. 1996;15(17):4749–4758. [PMC free article] [PubMed] [Google Scholar]
- 68.Schmitt E, Panvert M, Blanquet S, Mechulam Y. Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet. Embo J. 1998;17(23):6819–6826. doi: 10.1093/emboj/17.23.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hentze MW. Enzymes as RNA-binding proteins: a role for (di)nucleotide-binding domains? Trends Biochem Sci. 1994;19(3):101–103. doi: 10.1016/0968-0004(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 70.Liu X, Reig B, Nasrallah IM, Stover PJ. Human cytoplasmic serine hydroxymethyltransferase is an mRNA binding protein. Biochemistry. 2000;39(38):11523–11531. doi: 10.1021/bi000665d. [DOI] [PubMed] [Google Scholar]
- 71.Weiner H, Farres J, Rout UJ, Wang X, Zheng CF. Site directed mutagenesis to probe for active site components of liver mitochondrial aldehyde dehydrogenase. Adv Exp Med Biol. 1995;372:1–7. doi: 10.1007/978-1-4615-1965-2_1. [DOI] [PubMed] [Google Scholar]
- 72.Farres J, Wang TT, Cunningham SJ, Weiner H. Investigation of the active site cysteine residue of rat liver mitochondrial aldehyde dehydrogenase by site-directed mutagenesis. Biochemistry. 1995;34(8):2592–2598. doi: 10.1021/bi00008a025. [DOI] [PubMed] [Google Scholar]
- 73.Krupenko SA, Wagner C, Cook RJ. Cysteine 707 is involved in the dehydrogenase activity site of rat 10-formyltetrahydrofolate dehydrogenase. J Biol Chem. 1995;270(2):519–522. doi: 10.1074/jbc.270.2.519. [DOI] [PubMed] [Google Scholar]
- 74.Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, Kuo I, Perozich J, Lindahl R, Hempel J, Wang BC. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4(4):317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 75.Steinmetz CG, Xie P, Weiner H, Hurley TD. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5(5):701–711. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- 76.Tsybovsky Y, Donato H, Krupenko NI, Davies C, Krupenko SA. Crystal structures of the carboxyl terminal domain of rat 10-formyltetrahydrofolate dehydrogenase: implications for the catalytic mechanism of aldehyde dehydrogenases. Biochemistry. 2007;46(11):2917–2929. doi: 10.1021/bi0619573. [DOI] [PubMed] [Google Scholar]
- 77.Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN. Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6(12):1541–1551. doi: 10.1016/s0969-2126(98)00152-x. [DOI] [PubMed] [Google Scholar]
- 78.Johansson K, El-Ahmad M, Ramaswamy S, Hjelmqvist L, Jornvall H, Eklund H. Structure of betaine aldehyde dehydrogenase at 2.1 A resolution. Protein Sci. 1998;7(10):2106–2117. doi: 10.1002/pro.5560071007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lamb AL, Newcomer ME. The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity. Biochemistry. 1999;38(19):6003–6011. doi: 10.1021/bi9900471. [DOI] [PubMed] [Google Scholar]
- 80.Habenicht A, Hellman U, Cerff R. Non-phosphorylating GAPDH of higher plants is a member of the aldehyde dehydrogenase superfamily with no sequence homology to phosphorylating GAPDH. J Mol Biol. 1994;237(1):165–171. doi: 10.1006/jmbi.1994.1217. [DOI] [PubMed] [Google Scholar]
- 81.Cobessi D, Tete-Favier F, Marchal S, Azza S, Branlant G, Aubry A. Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. J Mol Biol. 1999;290(1):161–173. doi: 10.1006/jmbi.1999.2853. [DOI] [PubMed] [Google Scholar]
- 82.Ahvazi B, Coulombe R, Delarge M, Vedadi M, Zhang L, Meighen E, Vrielink A. Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity. Biochem J. 2000;349(Pt 3):853–861. [PMC free article] [PubMed] [Google Scholar]
- 83.Perozich J, Kuo I, Lindahl R, Hempel J. Coenzyme specificity in aldehyde dehydrogenase. Chem Biol Interact. 2001;130–132(1–3):115–124. doi: 10.1016/s0009-2797(00)00227-1. [DOI] [PubMed] [Google Scholar]
- 84.Hurley TD, Perez-Miller S, Breen H. Order and disorder in mitochondrial aldehyde dehydrogenase. Chem Biol Interact. 2001;130–132(1–3):3–14. doi: 10.1016/s0009-2797(00)00217-9. [DOI] [PubMed] [Google Scholar]
- 85.Perez-Miller SJ, Hurley TD. Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase. Biochemistry. 2003;42(23):7100–7109. doi: 10.1021/bi034182w. [DOI] [PubMed] [Google Scholar]
- 86.Gruez A, Roig-Zamboni V, Grisel S, Salomoni A, Valencia C, Campanacci V, Tegoni M, Cambillau C. Crystal structure and kinetics identify Escherichia coli YdcW gene product as a medium-chain aldehyde dehydrogenase. J Mol Biol. 2004;343(1):29–41. doi: 10.1016/j.jmb.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 87.Pohl E, Brunner N, Wilmanns M, Hensel R. The crystal structure of the allosteric non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeum Thermoproteus tenax. J Biol Chem. 2002;277(22):19938–19945. doi: 10.1074/jbc.M112244200. [DOI] [PubMed] [Google Scholar]
- 88.Hammen PK, Allali-Hassani A, Hallenga K, Hurley TD, Weiner H. Multiple conformations of NAD and NADH when bound to human cytosolic and mitochondrial aldehyde dehydrogenase. Biochemistry. 2002;41(22):7156–7168. doi: 10.1021/bi012197t. [DOI] [PubMed] [Google Scholar]
- 89.Dittmer DC, Kolyer JM. Addition of compounds of thiol and 1-substituted nicotinamides. J Am Chem Soc. 1963;28:1720–1722. [Google Scholar]
- 90.Metzler DE. Biochemistry. The chemical reactions of living cells. Academic Press; New York: 1977. [Google Scholar]
- 91.Choe J, Guerra D, Michels PA, Hol WG. Leishmania mexicana glycerol-3-phosphate dehydrogenase showed conformational changes upon binding a bi-substrate adduct. J Mol Biol. 2003;329(2):335–349. doi: 10.1016/s0022-2836(03)00421-2. [DOI] [PubMed] [Google Scholar]
- 92.Benach J, Atrian S, Gonzalez-Duarte R, Ladenstein R. The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography. J Mol Biol. 1999;289(2):335–355. doi: 10.1006/jmbi.1999.2765. [DOI] [PubMed] [Google Scholar]
- 93.Rubach JK, Plapp BV. Amino acid residues in the nicotinamide binding site contribute to catalysis by horse liver alcohol dehydrogenase. Biochemistry. 2003;42(10):2907–2915. doi: 10.1021/bi0272656. [DOI] [PubMed] [Google Scholar]
- 94.Bossi RT, Aliverti A, Raimondi D, Fischer F, Zanetti G, Ferrari D, Tahallah N, Maier CS, Heck AJ, Rizzi M, Mattevi A. A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase. Biochemistry. 2002;41(28):8807–8818. doi: 10.1021/bi025858a. [DOI] [PubMed] [Google Scholar]
- 95.Mohr S, Stamler JS, Brune B. Posttranslational modification of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosylation and subsequent NADH attachment. J Biol Chem. 1996;271(8):4209–4214. doi: 10.1074/jbc.271.8.4209. [DOI] [PubMed] [Google Scholar]
- 96.Hempel J, Nicholas HB, Jr, Brown ST, Wymore T. Unexpected encounters in simulations of the ALDH mechanism. In: Weiner H, Maser E, Lindahl R, Plapp B, editors. Enzymology and Molecular Biology of Carbonyl Metabolism. Pardue University Press; 2007. pp. 9–13. [Google Scholar]
- 97.Wymore T, Deerfield DW, 2nd, Hempel J. Mechanistic implications of the cysteine-nicotinamide adduct in aldehyde dehydrogenase based on quantum mechanical/molecular mechanical simulations. Biochemistry. 2007;46(33):9495–9506. doi: 10.1021/bi700555g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marchal S, Branlant G. Evidence for the chemical activation of essential cys-302 upon cofactor binding to nonphosphorylating glyceraldehyde 3-phosphate dehydrogenase from Streptococcus mutans. Biochemistry. 1999;38(39):12950–12958. doi: 10.1021/bi990453k. [DOI] [PubMed] [Google Scholar]
- 99.Takahashi K, Weiner H. Nicotinamide adenine dinucleotide activation of the esterase reaction of horse liver aldehyde dehydrogenase. Biochemistry. 1981;20(10):2720–2726. doi: 10.1021/bi00513a003. [DOI] [PubMed] [Google Scholar]
- 100.Feldman RI, Weiner H. Horse liver aldehyde dehydrogenase. II. Kinetics and mechanistic implications of the dehydrogenase and esterase activity. J Biol Chem. 1972;247(1):267–272. [PubMed] [Google Scholar]
- 101.Cook RJ, Wagner C. Enzymatic activities of rat liver cytosol 10-formyltetrahydrofolate dehydrogenase. Arch Biochem Biophys. 1995;321(2):336–344. doi: 10.1006/abbi.1995.1403. [DOI] [PubMed] [Google Scholar]
- 102.Weiner H, Hu JH, Sanny CG. Rate-limiting steps for the esterase and dehydrogenase reaction catalyzed by horse liver aldehyde dehydrogenase. J Biol Chem. 1976;251(13):3853–3855. [PubMed] [Google Scholar]
- 103.Zhou J, Weiner H. Basis for half-of-the-site reactivity and the dominance of the K487 oriental subunit over the E487 subunit in heterotetrameric human liver mitochondrial aldehyde dehydrogenase. Biochemistry. 2000;39(39):12019–12024. doi: 10.1021/bi001221k. [DOI] [PubMed] [Google Scholar]
- 104.Weiner H, Wei B, Zhou J. Subunit communication in tetrameric class 2 human liver aldehyde dehydrogenase as the basis for half-of-the-site reactivity and the dominance of the oriental subunit in a heterotetramer. Chem Biol Interact. 2001;130–132(1–3):47–56. doi: 10.1016/s0009-2797(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 105.Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 106.Lai JR, Koglin A, Walsh CT. Carrier protein structure and recognition in polyketide and nonribosomal peptide biosynthesis. Biochemistry. 2006;45(50):14869–14879. doi: 10.1021/bi061979p. [DOI] [PubMed] [Google Scholar]
- 107.White SW, Zheng J, Zhang YM, Rock The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 108.Guo S, Evans SA, Wilkes MB, Bhattacharjee JK. Novel posttranslational activation of the LYS2-encoded alpha-aminoadipate reductase for biosynthesis of lysine and site-directed mutational analysis of conserved amino acid residues in the activation domain of Candida albicans. J Bacteriol. 2001;183(24):7120–7125. doi: 10.1128/JB.183.24.7120-7125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xu H, Andi B, Qian J, West AH, Cook PF. The alpha-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem Biophys. 2006;46(1):43–64. doi: 10.1385/CBB:46:1:43. [DOI] [PubMed] [Google Scholar]
- 110.Sacksteder KA, Biery BJ, Morrell JC, Goodman BK, Geisbrecht BV, Cox RP, Gould SJ, Geraghty MT. Identification of the alpha-aminoadipic semialdehyde synthase gene, which is defective in familial hyperlysinemia. Am J Hum Genet. 2000;66(6):1736–1743. doi: 10.1086/302919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Donato H, Krupenko NI, Tsybovsky Y, Krupenko SA. 10-formyltetrahydrofolate dehydrogenase requires a 4′-phosphopantetheine prosthetic group for catalysis. J Biol Chem. 2007;282(47):34159–34166. doi: 10.1074/jbc.M707627200. [DOI] [PubMed] [Google Scholar]
- 112.Reuland SN, Vlasov AP, Krupenko SA. Disruption of a calmodulin central helix-like region of 10-formyltetrahydrofolate dehydrogenase impairs its dehydrogenase activity by uncoupling the functional domains. J Biol Chem. 2003;278(25):22894–22900. doi: 10.1074/jbc.M302948200. [DOI] [PubMed] [Google Scholar]
- 113.Nagy PL, Marolewski A, Benkovic SJ, Zalkin H. Formyltetrahydrofolate hydrolase, a regulatory enzyme that functions to balance pools of tetrahydrofolate and one-carbon tetrahydrofolate adducts in Escherichia coli. J Bacteriol. 1995;177(5):1292–1298. doi: 10.1128/jb.177.5.1292-1298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nagy PL, McCorkle GM, Zalkin H. purU, a source of formate for purT-dependent phosphoribosyl-N-formylglycinamide synthesis. J Bacteriol. 1993;175(21):7066–7073. doi: 10.1128/jb.175.21.7066-7073.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tishkov VI, Popov VO. Catalytic mechanism and application of formate dehydrogenase. Biochemistry (Mosc) 2004;69(11):1252–1267. doi: 10.1007/s10541-005-0071-x. [DOI] [PubMed] [Google Scholar]
- 116.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14(2):208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 117.Bornberg-Bauer E, Beaussart F, Kummerfeld SK, Teichmann SA, Weiner J., 3rd The evolution of domain arrangements in proteins and interaction networks. Cell Mol Life Sci. 2005;62(4):435–445. doi: 10.1007/s00018-004-4416-1. [DOI] [PubMed] [Google Scholar]
- 118.Appling DR, Rabinowitz JC. Regulation of expression of the ADE3 gene for yeast C1-tetrahydrofolate synthase, a trifunctional enzyme involved in one-carbon metabolism. J Biol Chem. 1985;260(2):1248–1256. [PubMed] [Google Scholar]
- 119.Ivanetich KM, Santi DV. Bifunctional thymidylate synthase-dihydrofolate reductase in protozoa. Faseb J. 1990;4(6):1591–1597. doi: 10.1096/fasebj.4.6.2180768. [DOI] [PubMed] [Google Scholar]
- 120.Davidson JN, Chen KC, Jamison RS, Musmanno LA, Kern CB. The evolutionary history of the first three enzymes in pyrimidine biosynthesis. Bioessays. 1993;15(3):157–164. doi: 10.1002/bies.950150303. [DOI] [PubMed] [Google Scholar]
- 121.Martins A, Shuman S. Characterization of a baculovirus enzyme with RNA ligase, polynucleotide 5′-kinase, and polynucleotide 3′-phosphatase activities. J Biol Chem. 2004;279(18):18220–18231. doi: 10.1074/jbc.M313386200. [DOI] [PubMed] [Google Scholar]