Abstract
Previous studies have shown that administration of fibroblast growth factor-19 (FGF-19) reverses diabetes, hepatic steatosis, hyperlipidemia, and adipose accretion in animal models of obesity. To investigate the mechanism for this effect, we determined whether FGF-19 modulated hepatic fatty acid synthesis, a key process controlling glucose tolerance and triacylglycerol accumulation in liver, blood, and adipose tissue. Incubating primary hepatocyte cultures with recombinant FGF-19 suppressed the ability of insulin to stimulate fatty acid synthesis. This effect was associated with a reduction in the expression of lipogenic enzymes. FGF-19 also suppressed the insulin-induced expression of sterol regulatory element-binding protein-1c (SREBP-1c), a key transcriptional activator of lipogenic genes. FGF-19 inhibition of lipogenic enzyme expression was not mediated by alterations in the activity of the insulin signal transduction pathway or changes in the activity of ERK, p38 MAPK, and AMP-activated protein kinase (AMPK). In contrast, FGF-19 increased the activity of STAT3, an inhibitor of SREBP-1c expression and decreased the expression of peroxisome proliferator-activated receptor-γ coactivator-1β (PGC-1β), an activator of SREBP-1c activity. FGF-19 also increased the expression of small heterodimer partner (SHP), a transcriptional repressor that inhibits lipogenic enzyme expression via a SREBP-1c-independent mechanism. Inhibition of SREBP-1c activity by changes in STAT3 and PGC-1β activity and inhibition of gene transcription by an elevation in SHP expression can explain the inhibition of lipogenesis caused by FGF-19. In summary, the inhibitory effect of FGF-19 on insulin activation of hepatic fatty acid synthesis constitutes a mechanism that would explain the beneficial effect of FGF-19 on metabolic syndrome.
Metabolic syndrome is a state of metabolic dysregulation that is characterized by obesity, hepatic steatosis, hyperlipidemia, atherosclerosis, and glucose intolerance (1). A key mechanism contributing to the development of metabolic syndrome is an elevation in the rate of hepatic fatty acid synthesis (2, 3). Hepatic fatty acid synthesis drives the synthesis of triacylglycerols that accumulate in the liver, blood, and adipose tissue. An elevation in hepatic fatty acid synthesis also promotes glucose intolerance, as accumulation of fatty acid metabolites in the liver suppresses the ability of insulin activate glycogen synthesis and inhibit gluconeogenesis (3). Accordingly, one approach to treating metabolic syndrome has been to manipulate the activity of signal transduction pathways that modulate hepatic fatty acid synthesis. For example, the beneficial effect of metformin on glucose tolerance in diabetic animals is mediated by a decrease in the rate of hepatic fatty acid synthesis (4). Metformin suppresses fatty acid synthesis by inhibiting the activity of acetyl-CoA carboxylase-α (ACCα)2 and decreasing the expression of sterol regulatory element-binding protein-1c (SREBP-1c), a key transcriptional activator of lipogenic genes. Metformin also increases the rate of hepatic fatty acid oxidation, an effect that contributes to the improvement in glucose tolerance. Metformin-induced changes in hepatic fatty acid synthesis and fatty acid oxidation are mediated by an activation of AMP-activated protein kinase (AMPK). As metformin administration causes undesirable side effects, the identification of new signaling pathways that modulate hepatic fatty acid metabolism may lead to the development of more effective therapies for treating metabolic syndrome.
Fibroblast growth factor-19 (FGF-19) was originally identified as a signal promoting the development of the inner ear in chick embryos (5). Subsequent studies have shown that FGF-19 and its mouse ortholog, FGF-15, also function in adult animals. For example, FGF-19/FGF-15 expressed in the small intestine acts as an enterohepatic hormone, mediating the inhibitory effects of intestinal bile acids on expression of hepatic cholesterol 7α-hydroxylase (CYP7A1), a key regulatory step in the bile acid synthesis pathway (6, 7). FGF-19 also regulates carbohydrate and lipid metabolism in adult animals. Administration of recombinant human FGF-19 or transgenic expression of the human FGF-19 gene in obese/diabetic mice causes an increase in energy expenditure and a decrease in adipose tissue stores (8, 9). Treatment of obese/diabetic mice with FGF-19 also reduces serum and liver triacylglycerol levels and enhances glucose tolerance. These observations indicate that activation of FGF-19 signaling is a potential approach to the treatment of metabolic syndrome. Currently, there is no information on how FGF-19 enhances glucose tolerance and inhibits triacylglycerol accumulation in the liver, adipose tissue, and blood. In the present study, we show that FGF-19 suppresses the ability of insulin to stimulate the rate of fatty acid synthesis and the expression of lipogenic enzymes in primary cultures of hepatocytes. We propose that alterations in hepatic fatty acid synthesis play a role in mediating the beneficial effects of FGF-19 on diseases associated with metabolic syndrome.
EXPERIMENTAL PROCEDURES
Cell Culture and Analytical Procedures—Hepatocytes were isolated from male Sprague-Dawley rats (∼200 g) starved for 24 h as described by Stabile et al. (10). Cells (3 × 106) were plated on 60-mm collagen-coated dishes containing Waymouth's medium MD752/1 supplemented with 20 mm HEPES, pH 7.4, 0.5 mm serine, 0.5 mm alanine, penicillin (100 μg/ml), streptomycin (100 μg/ml), gentamicin (50 mg/ml), and 5% newborn calf serum. After 4 h of incubation, the medium was replaced with one of the same composition lacking newborn calf serum. A Matrigel overlay (0.3 mg/ml) was added at this time. After an additional 16 h of incubation, the medium was replaced with serum-free medium containing the treatments indicated in the figure legends 1-7. In experiments to examine the effects of FGF-19 on glucose regulation of gene expression, hepatocytes were incubated in serum-free RPMI medium containing 5 mm glucose or 25 mm glucose. Hepatocyte cultures were maintained in a humidified chamber at 37 °C in 5% CO2, 95% air.
Human HepG2 hepatoma cells were plated on 60-mm dishes containing Waymouth's medium MD752/1 supplemented with penicillin (100 μg/ml), streptomycin (100 μg/ml), and 10% fetal bovine serum. After the cells reached 80% confluence, the medium was changed to one of the same composition lacking fetal bovine serum. After 24 h of incubation, the medium was replaced with serum-free medium containing the treatments indicated in the figure legend 3. Recombinant human FGF-19 was obtained from R&D Systems. Bovine insulin was a gift from Lilly. T0-901317 was purchased from Cayman Chemical. 3,5,3′-Triiodothyronine (T3) was obtained from Sigma. cAMP levels in acid-soluble total cell extracts were measured using the cyclic AMP enzyme immunoassay kit from Cayman Chemical.
Measurement of Fatty Acid Synthesis—The rate of de novo fatty acid synthesis was measured in hepatocyte cultures using the tritiated water method (11). Cells were incubated with 0.2 mCi/ml 3H2O during the last 3 h of a 24-h treatment period, with or without insulin, in the absence or presence of FGF-19. After removal of the incubation medium, the cells were harvested in 8 n KOH and transferred to screw-cap tubes. An equal volume of ethanol was added, and the tubes were heated in a boiling water bath for 2 h. Nonsaponifiable lipids were extracted three times with petroleum ether and then discarded. The aqueous phase was acidified with an 0.5 volume of 12 n HCl, and saponifiable lipids were extracted three times with petroleum ether. The pooled petroleum extracts were washed once with 0.5% acetic acid and dried under a stream of N2. 3H radioactivity was determined by scintillation counting. Fatty acid synthesis rates were calculated as described previously (11).
Isolation of RNA and Quantitation of mRNA Levels—Medium was removed and total RNA extracted from hepatocytes by the guanidinium thiocyanate/phenol/chloroform method (12). The abundance of mRNA encoding glucokinase (GK), fatty-acid synthase (FAS), l-pyruvate kinase (l-PK), ATP-citrate lyase (ATP-CL), ACCα, ACCβ, stearoyl-CoA desaturase-1 (SCD1), spot 14 (S14), glucose-6-phosphate dehydrogenase (G6PD), malic enzyme (ME), ATP-binding cassette transporter A1 (ABCA1), SREBP-1c, CYP7A1, small heterodimer partner (SHP), liver X receptor-α (LXRα), peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α), PGC-1β, and FGF-19 was measured by quantitative real-time PCR analysis using the Qiagen Quantitect SYBR Green reverse transcription-PCR system. Samples of DNase I-treated RNA (100 ng) were analyzed in triplicate according to the manufacturer's instructions. PCR was performed in 96-well plates using a Bio-Rad iCycler iQ. The relative amount of mRNA was calculated using the comparative Ct method. Rat cyclophilin and human glyceraldehyde-3-phosphate dehydrogenase were used as reference genes. Amplification of specific transcripts was confirmed by analyzing the melting curve profile performed at the end of each run and determining the size of the PCR products using agarose electrophoresis and ethidium bromide staining. The primer set for each gene is shown in supplemental Table S1.
Western Analysis—For analysis of SREBP-1 and SHP, total cell extracts were prepared from hepatocytes as described by Hansmannel et al. (13). Equal amounts of denatured protein (20 μg/lane) were subjected to electrophoresis in 10% SDS-polyacrylamide gels and then transferred to polyvinylidene difluoride membranes (Immobilon-FL, Millipore) using an electroblotting apparatus (Bio-Rad). The blots were blocked in TBST (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 0.1% Tween) containing 5% nonfat dry milk for 1 h at room temperature and then incubated with mouse monoclonal antibody against SREBP-1 (IgG-2A4, American Type Culture Collection) diluted 1:2000 in TBST containing 5% bovine serum albumin. After incubation with primary antibody for 12 h at 4 °C, the blots were washed in TBST. Next, the blots were incubated with secondary antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch) diluted 1:5000 in TBST, 5% nonfat dry milk for 1 h at room temperature. After washing with TBST, antibody/protein complexes on blots were detected using enhanced chemiluminescence (Amersham Biosciences). Fluorescence on the blots was visualized using a Typhoon 9410 imager, and signals were quantified using ImageQuant software. For analysis of ACCα, FAS, ABCA1, extracellular signal-regulated kinases 1 and 2 (ERK1/2), p38 mitogen-activated protein kinase (p38 MAPK), AMPK, protein kinase B (Akt), FoxO1, glycogen synthase kinase 3 (GSK-3), protein kinase C ζ/λ (PKC ζ/λ), and STAT3, total cell extracts were prepared as described above for SREBP-1 except that the lysis buffer contained 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 200 mm NaF, 1 mm Na3VO4, 1 mm β-glycerophosphate, 2.5 mm sodium pyrophosphate, 1 mm phenylmethylsulfonyl fluoride, and a mixture of protease inhibitors (Complete, Roche Applied Science). Antibodies against phosphorylated p38 MAPK (Thr180/Tyr182), phosphorylated ERK1/2 (Thr202Tyr204), phosphorylated Akt (Ser473), phosphorylated FoxO1 (Ser256), phosphorylated GSK-3α/β (Ser21/9), phosphorylated AMPKα (Thr172), phosphorylated PKC ζ/λ (Thr410/403), phosphorylated STAT3 (Tyr705), phosphorylated STAT3 (Ser727), total p38 MAPK, total ERK1/2, total Akt, total FoxO1, total GSK-3, total AMPK, total STAT3, and ACCα were obtained from Cell Signaling Technology. The antibodies against PKC ζ/λ and SHP were obtained from Santa Cruz Biotechnology. The antibody against ABCA1 was obtained from Novus.
Immunoprecipitation Analysis—Total cell extracts (1 mg protein) were precleared with prewashed protein A-agarose beads and then incubated with 4 and 10 μg of antibody against insulin receptor substrate-1 (IRS-1) (Millipore, 06-248) for 16 h at 4 °C. Protein A-agarose beads were then added to the reactions, and the incubation was continued for an additional 1 h at 4 °C. After centrifugation, supernatants were removed, and the beads were washed three times in lysis buffer. Electrophoresis sample loading buffer (80 μl) was then added to the beads, and the samples were boiled for 5 min. Samples of the supernatant were subjected to Western analysis using antibodies against phosphotyrosine (Millipore, 4G10 Platinum) and IRS-1.
Measurement of Protein Kinase Activity—PKC ζ/λ was measured as described by Standaert et al. (14). Briefly, total cell extracts were prepared in a lysis buffer containing 20 mm Tris-HCl, pH 7.5, 0.25 m sucrose, 2 mm EDTA, 2 mm EGTA, 2 mm Na3VO4, 2 mm NaF, 2 mm Na4P2O7, 1 mm phenylmethylsulfonyl fluoride, and a mixture of protease inhibitors (Complete, Roche Applied Science). PKC ζ/λ was immunoprecipitated from cell extracts (0.5 mg of protein) using 1 μg of a rabbit polyclonal antibody against PKC ζ/λ (sc-216, Santa Cruz Biotechnology). Immunoprecipitates were collected on protein A/G-agarose and incubated for 8 min at 30 °C in 100 μl of buffer containing 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 0.1 mm Na3VO4, 1 mm NaF, 0.1 mm Na4P2O7, 0.1 mm phenylmethylsulfonyl fluoride, 4 μg phosphatidylserine, 50 μm [γ-32P]ATP, and 40 μm PKCε peptide. Reactions were stopped by the addition of 10 μl of 5% acetic acid. Aliquots of reaction mixtures were spotted on P-81 filter papers, washed in 5% acetic acid, and counted for 32P radioactivity.
Statistical Methods—Data were subjected to analysis of variance, and statistical comparisons were made by using Dunnett's or Student's t test. Statistical significance was defined as p < 0.05.
RESULTS
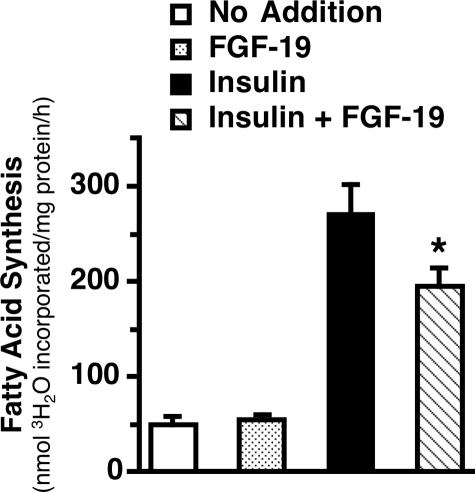
Effect of FGF-19 on the Rate of Fatty Acid Synthesis and the Expression of Enzymes Involved in Fatty Acid Synthesis and Fatty Acid Oxidation—Previous studies have shown that administration of recombinant human FGF-19 to obese/diabetic mice decreases adipose accretion and triacylglycerol levels in the liver and blood (8, 9). As hepatic fatty acid synthesis plays an important role in controlling triacylglycerol accumulation in adipose tissue, liver, and blood, we determined the effects of recombinant human FGF-19 on fatty acid synthesis in primary cultures of rat hepatocytes incubated in the absence and presence of insulin. The rate of fatty acid synthesis was 5.3-fold higher in hepatocytes incubated with insulin relative to hepatocytes incubated without insulin (Fig. 1). The addition of FGF-19 (50 ng/ml) suppressed the insulin-induced increase in fatty acid synthesis by 23% but had no effect on fatty acid synthesis in the absence of insulin. These data suggest that alterations in hepatic fatty acid synthesis play a role in mediating the inhibitory effect of FGF-19 on triacylglycerol levels in intact animals and that FGF-19 inhibits hepatic fatty acid synthesis by suppressing the action of a key hormone that mediates the activation of lipogenesis in response to dietary carbohydrate.
FIGURE 1.
FGF-19 suppresses the stimulatory effect of insulin on fatty acid synthesis. Primary hepatocyte cultures were prepared, and rates of fatty acid synthesis were measured as described under “Experimental Procedures.” Fatty acid synthesis measurements were made during the last 3 h of a 24-h treatment period with insulin (50 nm) in the absence and presence of FGF-19 (50 ng/ml). Values are means ± S.E. of three experiments. The asterisk indicates that the mean is significantly (p ≤ 0.05) lower than that of cells incubated with insulin in the absence of FGF-19.
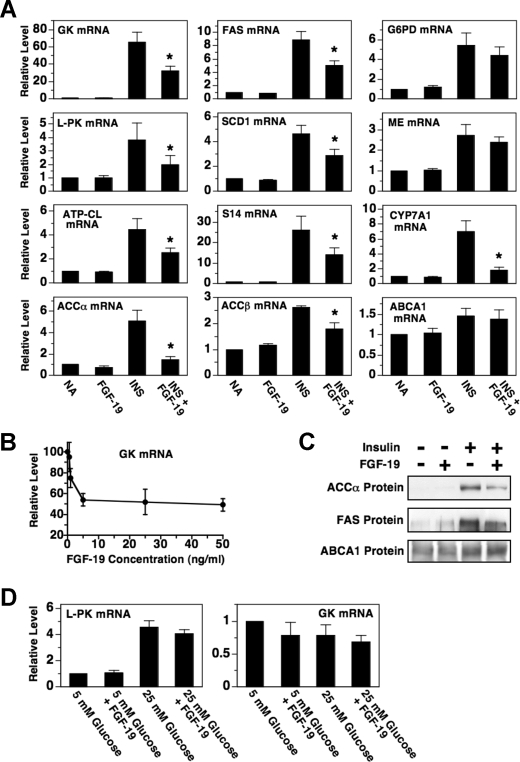
The stimulatory effect of insulin on fatty acid synthesis is mediated in part by an increase in the expression of enzymes comprising the fatty acid synthesis pathway (15). This led us to investigate whether FGF-19 modulates the expression of lipogenic enzymes in hepatocytes incubated with or without insulin. The addition of FGF-19 to the culture medium suppressed the ability of insulin to increase the abundance of mRNA encoding GK, l-PK, ATP-CL, ACCα, FAS, SCD1, and S14 by 38-55% (Fig. 2A). Further analyses demonstrated that FGF-19 inhibited GK mRNA abundance in a dose-dependent manner with a maximal effect observed at 5 ng/ml (Fig. 2B). This concentration of FGF-19 is similar to that required for maximal inhibition CYP7A1 expression in human hepatocytes (7). The inhibitory effect of FGF-19 on ACCα and FAS mRNA abundance was associated with a decrease in ACCα and FAS protein concentration (Fig. 2C). In contrast to the data for GK, l-PK, ATP-CL, ACCα, FAS, SCD1, and S14, FGF-19 had no effect on the ability of insulin to increase the abundance of mRNA encoding G6PD and ME, enzymes that supply reducing equivalents for fatty acid synthesis (Fig. 2A). FGF-19 had no effect on the expression GK, l-PK, ATP-CL, ACCα, FAS, SCD1, S14, G6PD, and ME in the absence of insulin. These data indicate that FGF-19 suppresses insulin-induced fatty acid synthesis, at least in part, by decreasing the expression of enzymes comprising this metabolic pathway.
FIGURE 2.
FGF-19 suppresses the stimulatory effect of insulin on the expression of lipogenic enzymes. A, primary hepatocyte cultures were prepared and incubated in Waymouth's medium. At 20 h of incubation, the medium was replaced with one of the same composition supplemented with or without FGF-19 (50 ng/ml), insulin (50 nm), or insulin plus FGF-19. After 24 h of treatment, total RNA was isolated, and the abundance of the indicated mRNAs was measured by quantitative real-time PCR. The level of mRNA in cells incubated with no additions (NA) was set at 1, and the other values were adjusted proportionately. Values are means ± five experiments. The asterisk indicates that the mean is significantly (p ≤ 0.05) lower than that of cells incubated with insulin in the absence of FGF-19. B, effect of different concentrations of FGF-19 (0 to 50 ng/ml) on the abundance of GK mRNA in hepatocytes incubated in the presence of insulin. The level of GK mRNA in cells incubated with insulin in the absence of FGF-19 was set at 100, and the other values were adjusted proportionately. C, the effect of FGF-19 on the concentration of ACCα, FAS, and ABCA1. Total cell extracts were prepared from hepatocytes incubated with or without FGF-19, insulin, or insulin plus FGF-19 for 48 h. The abundance of ACCα protein, FAS protein, and ABCA1 protein in cell extracts was measured by Western analysis. The data are representative of three independent experiments. D, effect of FGF-19 on the ability of glucose to stimulate l-PK expression. Hepatocytes were incubated in RPMI containing low (5 mm) or high (25 mm) glucose with or without FGF-19. After 24 h of treatment, total RNA was isolated, and the abundance of l-PK mRNA and GK mRNA was measured by quantitative real-time PCR. The level of mRNA in cells incubated with 5 mm glucose in the absence of FGF-19 was set at 1, and the other values were adjusted proportionately. Values are means ± S.E. of four experiments.
ACCβ is a mitochondrial ACC isoform that regulates fatty acid oxidation by modulating the allosteric inhibition of carnitine palmitoylacyltransferase-1 by malonyl-CoA (16). As observed with ACCα, treatment with FGF-19 inhibited the ability of insulin to increase the abundance of ACCβ mRNA in hepatocytes in culture (Fig. 2A). Such an effect would promote an elevation in carnitine palmitoylacyltransferase-1 activity and fatty acid oxidation. Thus, in addition to changes in fatty acid synthesis, alterations in fatty acid oxidation may contribute to the reduction in triacylglycerol levels in animals treated with FGF-19.
Another pathway that signals changes in carbohydrate consumption to lipogenic genes is activated by an elevation in glucose metabolism (17). A metabolite(s) derived from glucose metabolism enhances the activity of carbohydrate response element-binding protein, a transcription factor that increases the transcription of the genes encoding l-PK, ACCα, FAS, and S14. To find out whether FGF-19 modulates the stimulatory effect of glucose on lipogenic enzyme expression, we determined the effect of FGF-19 on the expression of l-PK in hepatocytes incubated in RPMI lacking insulin and containing low glucose (5 mm) or high glucose (25 mm). Increasing the concentration of the medium from 5 to 25 mm glucose stimulated a 4.7-fold increase in the abundance of l-PK mRNA (Fig. 2D). Treatment with FGF-19 did not alter the ability of glucose to stimulate l-PK expression. As a negative control, neither glucose nor FGF-19 modulated the expression of GK mRNA. These results indicate that FGF-19 does not directly regulate the activity of the glucose-signaling pathway. This conclusion is supported by data demonstrating that FGF-19 has no effect on the expression of l-PK, ACCα, FAS, and S14 in hepatocytes incubated in Waymouth's medium lacking insulin and containing a high glucose (27.5 mm) (Fig. 2A). Previous studies have shown that the stimulatory effect of insulin on l-PK gene transcription is mediated by an increase in glucose signaling activity arising from the insulin activation of GK expression (18, 19). Thus, the inhibitory effect of FGF-19 on insulin-induced l-PK expression is likely mediated by an indirect effect on glucose signaling activity.
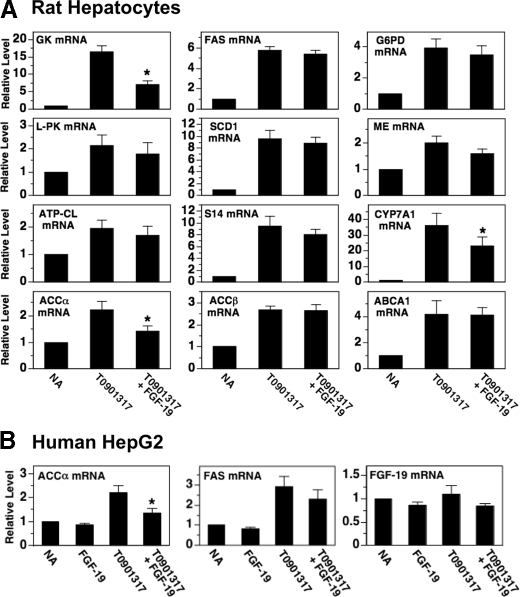
A third signaling pathway that controls lipogenic enzyme expression is activated by agonists of the liver X receptor (20, 21). LXR is bound and activated by naturally occurring oxysterols. Ligand-bound LXR enhances lipogenic gene transcription by interacting with LXR response elements on target promoters (22, 23). Tobin et al. (24) have shown that the insulin-induced increase in the hepatic expression of GK, ACCα, FAS, and SCD1 is dependent on the presence of LXR. The observation that FGF-19 inhibits insulin regulation of GK, ACCα, FAS, and SCD1 (Fig. 2) prompted us to investigate whether FGF-19 modulated LXR signaling activity. Incubating hepatocytes with FGF-19 suppressed the ability of a synthetic LXR agonist (T0-901317) to increase the abundance of mRNA encoding GK and ACCα but had no effect on the ability of T0-901317 to increase the abundance of mRNA encoding l-PK, ATP-CL, FAS, SCD1, S14, G6PD, and ME (Fig. 3A). FGF-19 also had no effect on T0-901317-induced expression of ACCβ mRNA and ABCA1 mRNA. The inability of FGF-19 to modulate T0-901317 regulation of several previously characterized LXR target genes (i.e. FAS, SCD1, ABCA1) suggests that alterations in LXR activity per se are not involved in mediating the inhibitory effects of FGF-19 on insulin action. FGF-19 may inhibit T0-901317-induced expression of GK and ACCα by modulating the activity of a coregulatory protein(s) that interacts with LXR in a gene-specific manner.
FIGURE 3.
FGF-19 suppresses the stimulatory effect of the LXR agonist T0-901317 on the expression of GK, ACCα, and CYP7A1. Primary rat hepatocytes (A) and human HepG2 cells (B) were incubated in serum-free Waymouth's medium supplemented with or without FGF-19 (50 ng/ml), T0-901317 (6 μm), or T0-901317 plus FGF-19. After 24 h of treatment, total RNA was isolated, and the abundance of the indicated mRNAs was measured by quantitative real-time PCR. The level of mRNA in cells incubated with no additions (NA) was set at 1, and the other values were adjusted proportionately. Values are means ± five experiments. The asterisk indicates that the mean is significantly (p ≤ 0.05) lower than that of cells incubated with T0-901317 in the absence of FGF-19.
To investigate whether FGF-19 was effective in modulating lipogenic enzyme expression in human cells, we determined the effect of FGF-19 on the abundance of ACCα mRNA and FAS mRNA in human hepatoma cells (HepG2) incubated in the absence and presence of T0-901317. Treatment of HepG2 cells with FGF-19 suppressed the ability of T0-901317 to increase ACCα mRNA levels but had no effect on the ability of T0-901317 to increase FAS mRNA levels (Fig. 3B). These data indicate that the gene-specific effect of FGF-19 on LXR activity is conserved in humans.
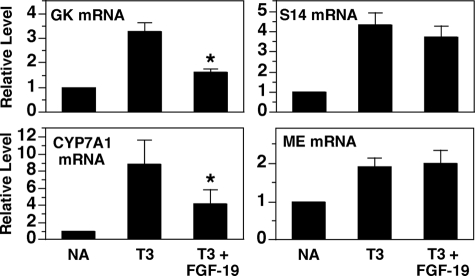
We next determined the ability of FGF-19 to modulate the activity of other nuclear receptor pathways. Previous studies have shown that T3 activates the transcription of GK, S14, and ME by interacting with nuclear T3 receptors bound to these genes (25-27). In Fig. 4, we show that FGF-19 treatment suppresses the ability of T3 to increase the abundance of mRNA encoding GK but has no effect on the ability of T3 to increase the abundance of mRNA encoding S14 and ME. Thus, as with the LXR pathway, FGF-19 inhibits the T3 pathway in a genespecific manner.
FIGURE 4.
FGF-19 suppresses the stimulatory effect of T3 on the expression of GK, S14, and CYP7A1. Primary hepatocyte cultures were prepared and incubated in Waymouth's medium. At 20 h of incubation, the medium was replaced with one of the same composition supplemented with or without T3 (100 nm) or T3 plus FGF-19. After 24 h of treatment, total RNA was isolated, and the abundance of the indicated mRNAs was measured by quantitative real-time PCR. The level of mRNA in cells incubated with no additions (NA) was set at 1, and the other values were adjusted proportionately. Values are means ± four experiments. The asterisk indicates that the mean is significantly (p ≤ 0.05) lower than that of cells incubated with T3 in the absence of FGF-19.
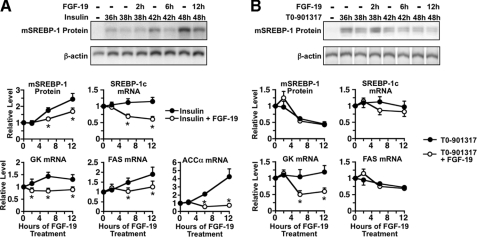
Effect of FGF-19 on the Ability of Insulin and T0-901317 to Increase the Abundance of Mature SREBP-1c—Previous studies have demonstrated that SREBP-1c plays a key role in mediating the effect of insulin on lipogenic enzyme gene transcription (28). SREBP-1c is the most abundant SREBP-1 isoform expressed in liver. It is synthesized as a 125-kDa precursor protein that is anchored to the endoplasmic reticulum. To become transcriptionally active, precursor SREBP-1c is translocated to the Golgi where it is cleaved by two proteases, resulting in the release of the N-terminal segment of SREBP-1c, referred to as mature SREBP-1c (mSREBP-1c). mSREBP-1c is transported into the nucleus where it interacts with the promoter/regulatory region of several lipogenic genes including FAS, ATP-CL, SCD1, and S14. Insulin activates lipogenic enzyme gene transcription in part by increasing the concentration of mSREBP-1c (28). To investigate the mechanism by which FGF-19 inhibits insulin-induced expression of lipogenic enzymes, time course experiments were performed to determine the effect of FGF-19 on the concentration of mSREBP-1. In hepatocytes previously incubated with insulin for 36 h, the addition of FGF-19 caused a 30-38% decrease in the concentration of mSREBP-1c after 6 and 12 h of treatment (Fig. 5A). FGF-19 caused a similar reduction in the concentration of SREBP-1c mRNA after 6 and 12 h of treatment, suggesting that FGF-19 acted at a pretranslational step to decrease mSREBP-1c concentration. The effect of FGF-19 on the abundance of ACCα mRNA and FAS mRNA followed a time course similar to that of mSREBP-1c (Fig. 5A). These results suggest that alterations in mSREBP-1c concentration play a role in mediating the effect of FGF-19 on the insulin-induced expression of ACCα and FAS. In contrast to the effect of FGF-19 on the abundance of ACCα mRNA and FAS mRNA, FGF-19 decreased the abundance of GK mRNA at a time (2 h) that preceded changes in mSREBP-1c concentration. This observation indicates that FGF-19 also inhibits insulin action by a mechanism that is independent of changes in mSREBP-1c concentration. This conclusion is congruent with that of previous work demonstrating that insulin induction of GK gene transcription is not dependent on alterations in mSREBP-1c expression (13, 29, 30).
FIGURE 5.
Time course of the effect of FGF-19 on the expression of SREBP-1c, GK, FAS, and ACCα. Primary hepatocyte cultures were prepared and incubated in Waymouth's medium containing insulin (A) or T0-901317 (B). At 36 h of incubation, FGF-19 (50 ng/ml) was added to the culture medium, and the incubation was continued for 0, 2, 6, and 12 h. Cells were harvested, and total cell extracts were prepared as described under “Experimental Procedures.” mSREBP-1 was measured by Western analysis. Top panels, Western analysis of mSREBP-1 from a representative experiment. Bottom panels, signals for mSREBP-1 were quantitated. In another set of plates treated in a similar manner, total RNA was isolated and the abundance of the mRNAs encoding SREBP-1c, GK, FAS, and ACCα was measured by quantitative real-time PCR. The level of mSREBP-1 protein and mRNA in cells treated with insulin for 36 h and FGF-19 for 0 h was set at 1, and the other values were adjusted proportionately. Values are means ± five experiments. An asterisk indicates that the mean is significantly (p ≤ 0.05) different compared with that of cells treated with insulin for the same time period.
LXR agonists also increase lipogenic enzyme expression by increasing the concentration of mSREBP-1c (31, 32). These observations led us to investigate whether alterations in mSREBP-1c concentration play a role in mediating the inhibitory effect of FGF-19 on T0-901317-induced expression of GK and ACCα. In hepatocytes previously incubated with T0-901317 for 36 h, the addition of FGF-19 had no effect on the concentration of mSREBP-1c protein and SREBP-1c mRNA after 2, 6, and 12 h of treatment (Fig. 5B). In contrast, FGF-19 caused a 50-55% decrease in the abundance of GK mRNA after 6 and 12 h of treatment. In other experiments, FGF-19 also had no effect on the abundance of mSREBP-1c protein and SREBP-1c mRNA when it was added at the beginning of a 24-h incubation with T0-901317 (data not shown). These findings indicate that FGF-19 inhibits T0-901317-induced expression of GK and ACCα via a mechanism that is independent of alterations in mSREBP-1c concentration.
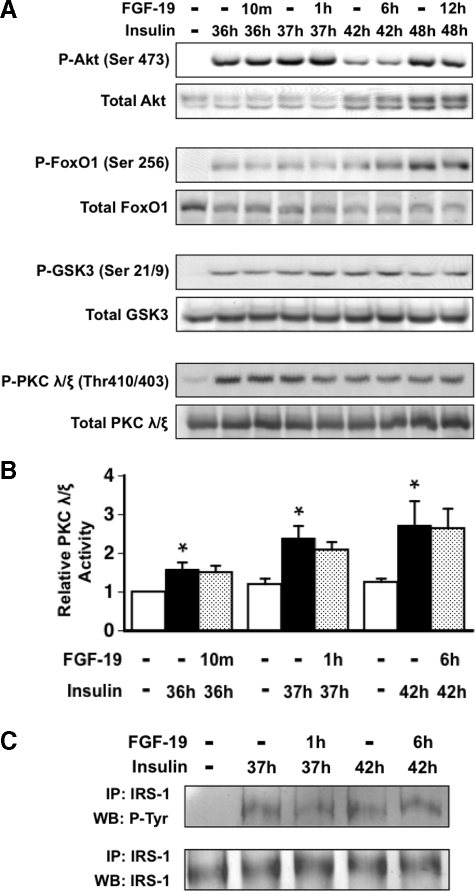
Effect of FGF-19 on the Activity of the Signaling Pathway Mediating the Stimulatory Effect of Insulin on Lipogenic Enzyme Expression—We next investigated the mechanism by which FGF-19 suppressed the stimulatory effect of insulin on the expression of SREBP-1c and lipogenic enzymes. Previous studies have shown that insulin increases SREBP-1c expression by binding to the insulin receptor and triggering a signaling cascade resulting in the recruitment of phosphoinositide 3-kinase to the plasma membrane and the conversion of phosphatidylinositol 4,5-bisphosphate to phosphatidylinositol 3,4,5-trisphosphate (33). Phosphatidylinositol 3,4,5-trisphosphate facilitates additional signaling events leading to the activation of Akt and atypical forms of protein kinase C (PKC λ/ζ). The results from gain-of-function and loss-of-function studies indicate that Akt and PKC λ/ζ play a role in mediating the effects of insulin on expression of GK, FAS, SCD1, and SREBP-1c (34-38). This led us to investigate whether FGF-19 modulates the activity of Akt and PKC λ/ζ in rat hepatocyte cultures. Results from Western analyses demonstrated that FGF-19 had no effect on the ability of insulin to increase the phosphorylation of Akt (Ser473) and the phosphorylation of Akt down-stream targets (FoxO1 (Ser256) and GSK-3 (Ser21/9)) (39, 40) that regulate the expression of SREBP-1c and lipogenic enzymes (Fig. 6A). Treatment with FGF-19 also had no effect on the ability of insulin to stimulate PKC λ/ζ phosphorylation (Thr410/403) (Fig. 6A) and PKC λ/ζ activity (Fig. 6B). These data indicate that alterations in Akt and PKC λ/ζ signaling activity are not involved in mediating the effects of FGF-19 on hepatic insulin action.
FIGURE 6.
Effect of FGF-19 on the activity of the signaling cascade mediating the stimulatory effect of insulin on the expression of lipogenic genes. Primary hepatocyte cultures were prepared and incubated in Waymouth's medium containing insulin. At 36 h of incubation, FGF-19 (50 ng/ml) was added to the culture medium, and the incubation was continued for 10 min, 1 h, 6 h, and 12 h. Cells were harvested, and total cell extracts were prepared as described under “Experimental Procedures.” A, Western analyses were performed using antibodies against phosphorylated Akt (Ser473), phosphorylated FoxO1 (Ser256), GSK-3α/β (Ser21/9), and PKC λ/ζ (Thr410/403) and total AKT, FoxO1, GSK-3α/β, and PKC λ/ζ. The data are representative of four independent experiments. B, PKC λ/ζ activity was measured as described under “Experimental Procedures.” PKC λ/ζ activity in cells incubated in the absence of insulin and FGF-19 for 36 h was set at 1, and the other values were adjusted proportionately. Values are means ± three experiments. An asterisk indicates that the mean is significantly (p ≤ 0.05) different compared with that of cells incubated in the absence of insulin and FGF-19 for the same time period. C, cell extracts were incubated with an antibody against IRS-1, and the resulting immunoprecipitates (IP) were subjected to Western blot analysis (WB) using antibodies against phosphotyrosine and IRS-1. The data are representative of three independent experiments.
The insulin-induced recruitment of phosphoinositide 3-kinase to the plasma membrane is mediated by adaptor proteins that are phosphorylated by the insulin receptor tyrosine kinase. These adaptors are referred to as insulin receptor substrates (33). To investigate whether FGF-19 modulates insulin signaling activity upstream of Akt and PKC λ/ζ, we determined the effect of FGF-19 on the tyrosine phosphorylation of IRS-1, the principal IRS isoform mediating the insulin regulation of lipogenic enzyme expression (41, 42). Treatment with FGF-19 had no effect on the ability of insulin to stimulate the tyrosine phosphorylation of IRS-1 (Fig. 6C). This finding provides further evidence that FGF-19 has no effect on the insulin-signaling pathway controlling the expression of SREBP-1c and lipogenic enzymes.
Effect of FGF-19 on the Activity of Signaling Pathways That Inhibit Lipogenic Enzyme Expression—The observation that FGF-19 had no effect on the activity of the insulin-signaling cascade led us to investigate the role of other signaling pathways in mediating FGF-19 regulation of lipogenic enzyme expression. Previous studies have shown that the inhibitory effect of glucagon and polyunsaturated fatty acids on insulin-induced SREBP-1c expression is dependent on the activation of protein kinase A (PKA) and ERK1/2, respectively (43-45). Other studies have shown that activation of p38 MAPK and AMPK decreases the expression of ACCα, FAS, and SREBP-1c in hepatocytes (46-48). These observations prompted us to determine the effect of FGF-19 on the activity of p38 MAPK, AMPK, and ERK and the concentration of cAMP, the second messenger that activates PKA. Treatment of hepatocytes with FGF-19 in the presence of insulin had no effect on the abundance of the active phosphorylated form of p38 MAPK (Thr180/Tyr182) and AMPK (Thr172) (supplemental Fig. S1A) and the concentration of cAMP (supplemental Table S2). In contrast, FGF-19 treatment caused a rapid (≤10 min) and sustained (≥12 h) increase in the abundance of the active phosphorylated form of ERK1/2 (Thr202/Tyr204) (supplemental Fig. S1A). Thus, FGF-19 enhances the activity of ERK1/2 but has no effect on the activity of p38 MAPK, AMPK, and PKA. We next determined the effect of a cell-permeable inhibitor of MAPK/ERK kinase 1/2 (U0126) on the regulation of GK and SREBP-1c by FGF-19. Treatment of hepatocytes with U0126 inhibited the ability of FGF-19 to increase ERK phosphorylation but had no effect on the ability of FGF-19 to inhibit insulin-induced expression of GK mRNA and SREBP-1c mRNA (supplemental Fig. S1, B and C). Similar results were obtained with the MAPK/ERK kinase 1/2 inhibitor, PD98059. These observations indicate that FGF-19 regulation of SREBP-1c and lipogenic enzyme expression is not mediated by alterations in the activity of p38 MAPK, AMPK, PKA, and ERK.
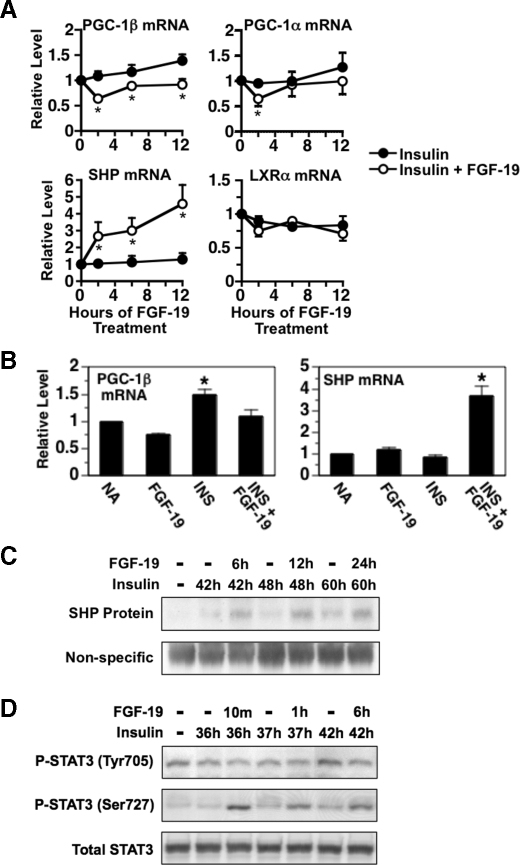
The Effect of FGF-19 on Lipogenic Enzyme Expression Is Associated with Changes in the Expression of PGC-1β, SHP, and Phosphorylated STAT3—Another possible mechanism by which FGF-19 could inhibit the insulin-induced expression of lipogenic enzymes is by modulating the activity of nuclear proteins that regulate the activity/expression of SREBP-1c and/or other factors controlling lipogenic enzyme gene transcription. One such factor is PGC-1β, a coactivator that enhances the ability of SREBP-1c to activate FAS and SCD1 gene transcription (49). Previous studies have shown that the expression of PGC-1β is regulated by hormonal and nutritional status and that alterations in PGC-1β expression play a role in mediating the induction of lipogenic enzyme expression by high-fat feeding (49, 50). This led us to investigate whether FGF-19 modulates PGC-1β expression. In hepatocytes previously incubated with insulin for 36 h, the addition of FGF-19 caused a rapid (<2 h) and sustained (>12 h) decrease in the abundance of PGC-1β mRNA (Fig. 7A). The magnitude of the effect was 37% after 12 h of FGF-19 treatment. FGF-19 also decreased PGC-1β expression when it was added at the beginning of incubation with insulin (Fig. 7B). In contrast to the results for PGC-1β, treatment with FGF-19 had no effect on the abundance of mRNA encoding PGC-1α, a PGC-1 isoform that lacks the ability to coactivate SREBP-1c (49). These findings provide support for a role of PGC-1β in mediating the inhibitory effect of FGF-19 on insulin-induced expression of lipogenic enzymes.
FIGURE 7.
Effect of FGF-19 on the expression of PGC-1β, PGC-1α, SHP, LXRα, and phosphorylated STAT3. A, primary hepatocyte cultures were prepared and incubated in Waymouth's medium containing insulin. At 36 h of incubation, FGF-19 (50 ng/ml) was added to the culture medium, and incubation was continued for 0, 2, 6, and 12 h. Cells were harvested, total RNA was isolated, and the abundance of the mRNAs encoding PGC-1β, PGC-1α, SHP, and LXRα was measured by quantitative real-time PCR. The level of mRNA in cells treated with insulin for 36 h and FGF-19 for 0 h was set at 1, and the other values were adjusted proportionately. Values are means ± four experiments. An asterisk indicates that the mean is significantly (p ≤ 0.05) different compared with that of cells treated with insulin for the same time period. B, primary hepatocyte cultures were prepared and incubated in Waymouth's medium. At 20 h of incubation, the medium was replaced with one of the same composition supplemented with or without FGF-19, insulin, or insulin plus FGF-19. After 24 h of treatment, total RNA was isolated, and the abundance of PGC-1β mRNA and SHP mRNA was measured by quantitative real-time PCR. The level of mRNA in cells incubated with no additions (NA) was set at 1, and the other values were adjusted proportionately. Values are means ± four experiments. An asterisk indicates that the mean is significantly (p ≤ 0.05) higher than that of any other treatment. Total cell extracts were prepared from hepatocytes treated as described in A. SHP protein (C) and phosphorylated STAT3 (Tyr705 and Ser727)(D) were measured by Western analysis. The data are representative of four independent experiments.
A second factor that modulates lipogenic enzyme expression is SHP, an atypical nuclear hormone receptor that represses gene transcription by inhibiting the recruitment of coactivators and/or enhancing the recruitment of corepressors to target promoters (51). Previous studies have shown that ablation of the SHP gene increases the expression of GK, l-PK, ACCα, and FAS in hepatocytes (52-54). To assess the role of SHP in FGF-19 regulation of lipogenic genes, we determined the effect of FGF-19 on SHP expression. In hepatocytes previously incubated with insulin for 36 h, the addition of FGF-19 stimulated a rapid (<2 h) and sustained (>12 h) increase in the abundance of SHP mRNA (Fig. 7A). The magnitude of this effect was 4.2-fold after 12 h of FGF-19 treatment. FGF-19 also increased SHP mRNA abundance when it was added at the beginning of incubation with insulin (Fig. 7B). Interestingly, FGF-19 treatment had no effect on SHP mRNA levels in hepatocytes incubated in the absence of insulin (Fig. 7B). Under these conditions, FGF-19 also had no effect on expression of lipogenic enzymes (Fig. 2A). The results from Western analysis indicated that FGF-19 treatment also increased the concentration of SHP protein in hepatocytes (Fig. 7C). The magnitude of this effect was 3.2-fold after 12 h of FGF-19 treatment. Thus, the effect of FGF-19 on lipogenic enzyme expression is correlated with alterations in SHP expression. These finding provides support for a role of SHP in mediating the reduction in lipogenic enzyme expression caused by FGF-19.
Another nuclear factor that controls SREBP-1c activity is STAT3. Previous studies have shown that ablation of the STAT3 gene stimulates the expression of SREBP-1c and FAS, whereas expression of a constitutively active form of STAT3 has the opposite effect (55, 56). Other work has shown that the transcriptional activity of STAT3 is activated by tyrosine phosphorylation (Tyr705) and serine phosphorylation (Ser727) of STAT3 (57). This prompted us to determine whether FGF-19 modulates the level of phosphorylation at these sites. In hepatocytes previously incubated with insulin for 36 h, addition of FGF-19 increased the phosphorylation of Ser727 (Fig. 7D). The phosphorylation of Tyr705 and the concentration of total STAT3 were not affected by FGF-19 treatment. These findings are consistent with a role of STAT3 in mediating the inhibitory action of FGF-19 on lipogenic enzyme expression.
DISCUSSION
The results of the present study describe a mechanism by which FGF-19 administration reverses obesity, hypertriglyceridemia, hepatic steatosis, and glucose intolerance in rodent models of metabolic syndrome. FGF-19 suppresses the ability of insulin to stimulate hepatic fatty acid synthesis. This effect is mediated by a reduction in the insulin-induced expression of enzymes comprising the fatty acid synthesis pathway including GK, l-PK, ATP-CL, ACCα, FAS, and SCD1. FGF-19 also suppresses the stimulatory effect of T0-901317 on expression of GK and ACCα and the stimulatory effect of T3 on expression of GK. Thus, the beneficial effects of FGF-19 on metabolic syndrome may also be mediated by an inhibition of the oxysterol- and T3-signaling pathways controlling lipogenic enzyme expression.
The present study also provides evidence that FGF-19 attenuates the inhibitory effect of insulin on hepatic fatty acid oxidation. FGF-19 suppresses the ability of insulin to stimulate the expression of ACCβ, a key regulator of mitochondrial fatty acid oxidation (16). This observation is consistent with previous studies demonstrating that FGF-19 overexpression in mice causes a reduction in respiratory quotient (8). FGF-19-induced changes in ACCβ expression and hepatic fatty oxidation may also play a role in reversing metabolic syndrome, as ablation of the ACCβ gene decreases triacylglycerol accumulation in the liver and adipose tissue and increases energy expenditure and glucose tolerance (58).
How does FGF-19 suppress the stimulatory effect of insulin on the expression of lipogenic enzymes? The results of the present study indicate that at least two distinct mechanisms mediate this phenomenon. One mechanism involves changes in the concentration of mSREBP-1c, a transcriptional activator of the genes encoding FAS, ATP-CL, SCD1, and S14 (28). FGF-19 inhibits the ability of insulin to increase the concentration of mSREBP-1c, an effect that is closely associated with a reduction in FAS, ATP-CL, SCD1, and S14 expression. A second mechanism is not dependent on changes in mSREBP-1c concentration: FGF-19 inhibits the ability of insulin to stimulate the expression of GK, a gene that is not regulated by mSREBP-1c in hepatocytes (13, 29, 30).
The inhibitory effect of FGF-19 on insulin induction of mSREBP-1c concentration was accompanied by a reduction in SREBP-1c mRNA concentration, indicating that FGF-19 acted at a pretranslational step to control mSREBP-1c concentration (Fig. 5A). Previous studies have shown that insulin increases SREBP-1c gene transcription and that this effect is triggered by an increase in the activity of LXR bound to the SREBP-1c promoter (31, 32). Several lines of evidence indicate that the inhibitory effect of FGF-19 on insulin-induced SREBP-1c mRNA abundance is not mediated by changes in LXR activity. First, FGF-19 is not effective in suppressing the stimulatory effect of T0-901317 on the expression of SREBP-1c and lipogenic genes regulated by SREBP-1c (i.e. ATP-CL, FAS, SCD1, and S14) (Figs. 3 and 5B). This finding indicates that FGF-19 does not modulate the transcriptional activity of ligand-bound LXR on the SREBP-1c promoter. Second, FGF-19 is not effective in suppressing the insulin-induced expression of ABCA1, a well-defined LXR target gene (Fig. 2A). This observation indicates that FGF-19 does not regulate the concentration of an endogenous LXR agonist. Third, FGF-19 has no effect on the expression of mRNA encoding LXRα, the principal LXR isoform expressed in liver (Fig. 7A). This finding indicates that FGF-19 does not modulate the maximal capacity of LXR to stimulate gene transcription.
Elam and colleagues (59) have recently shown that the induction of SREBP-1c gene transcription is also triggered by an increase in the activity of specificity protein 1 (Sp1) bound on the SREBP-1c promoter. Sp1 enhances SREBP-1c transcription through a functional interaction with LXR bound at an adjacent site. The lack of effect of FGF-19 on LXR activity indicates that FGF-19 is also not effective in suppressing the stimulatory effect of insulin on Sp1 activity. The inability of FGF-19 to modulate insulin activation of LXR and Sp1 on the SREBP-1c promoter is supported by the observation that FGF-19 has no effect on the activity of the proximal portion of the signal transduction pathway mediating the stimulatory effect of insulin on SREBP-1c expression (Fig. 6).
In screening for mechanisms that explain the inhibitory effect of FGF-19 on insulin-induced SREBP-1c expression, we observed that the FGF-19 regulation of SREBP-1c expression was associated with an increase in the expression of the phosphorylated, active form of STAT3 and a decrease in the expression of PGC-1β (Figs. 2, 5, and 7). Previous studies have shown that STAT3 and PGC-1β have opposing actions on SREBP-1c activity. STAT3 is an inhibitor of SREBP-1c gene transcription, whereas PGC-1β is a coactivator that enhances the ability of SREBP-1c to stimulate gene transcription. Other studies have shown that the SREBP-1c binds to its own promoter and participates in mediating the increase in SREBP-1c gene transcription caused by insulin (31, 32). Thus, alterations in the expression of phosphorylated STAT3 and PGC-1β constitute a mechanism explaining the inhibitory effect of FGF-19 on insulin-induced SREBP-1c gene expression.
Other agents that inhibit the stimulatory effect of insulin on SREBP-1c expression include glucagon and polyunsaturated fatty acids (43, 44). The effect of glucagon on SREBP-1c expression is mediated by a decrease in the activity of LXR bound to the SREBP-1c promoter (45). Glucagon inhibits LXR activity by enhancing the PKA-mediated phosphorylation of LXR. The effect of polyunsaturated fatty acids on SREBP-1c expression is mediated by a mechanism that is dependent on the presence of ERK (43). In contrast to the mechanism of action of glucagon and polyunsaturated fatty acids, our findings indicate that FGF-19 inhibits insulin-induced SREBP-1c expression by a mechanism that is not dependent on changes in the activity of LXR, PKA, and ERK (Figs. 2 and 3, supplemental Fig. S1, supplemental Table S2). Thus, mechanism by which FGF-19 inhibits SREBP-1c expression appears to be distinct from that of glucagon and polyunsaturated fatty acids.
We also observed that FGF-19 treatment increased the expression of the transcriptional repressor, SHP, and that this effect preceded or paralleled the reduction in insulin-induced lipogenic enzyme expression (Figs. 2, 5 and 7). Previous studies have shown that SHP represses the expression of GK, l-PK, ACCα, and FAS (52-54) and that the mechanism mediating the inhibitory effect of SHP on FAS gene transcription involves a direct interaction of SHP with liver receptor homologue-1 on the FAS promoter (53). This direct action of SHP on FAS promoter activity constitutes a SREBP-1c-independent mechanism mediating the inhibitory effect of FGF-19 on insulin-induced gene transcription. Such a mechanism may also play a role in mediating the FGF-19 regulation of GK expression, as the GK gene is not regulated by SREBP-1c (13, 29, 30). Previous studies have shown that insulin induction of GK gene transcription is mediated, at least in part, by the activation of hepatocyte nuclear factor-4 (HNF-4) bound on the GK promoter (60, 61). Other studies have shown that SHP interacts with HNF-4 and inhibits its ability to activate transcription (62). The role of SHP-HNF-4 interactions in mediating the inhibitory effect of FGF-19 on GK gene transcription will require further study.
Although FGF-19 has a broad effect on the insulin regulation of enzymes comprising the fatty synthesis pathway, it has no effect on the insulin regulation of ME and G6PD, enzymes supplying reducing equivalents for fatty acid synthesis (Fig. 2). The inability of FGF-19 to modulate the insulin regulation of ME and G6PD may be attributed to the fact that the mechanisms mediating the insulin activation of these enzymes are substantially different than those of other lipogenic enzymes. The stimulatory effect of insulin on malic enzyme gene transcription is mediated by an activator protein 1 (AP-1) motif and an early growth response 1 (Egr-1) binding site in the malic enzyme promoter (63). The stimulatory effect of insulin on G6PD expression is mediated by an increase in the rate of G6PD pre-mRNA processing (64). There is no evidence that AP-1, Egr-1, and pre-mRNA processing play a role in mediating the insulin regulation of other lipogenic enzymes. The lack of effect of FGF-19 on ME and G6PD expression may also be explained by the possibility that ME and G6PD are not targets of regulatory proteins that mediate FGF-19 action (i.e. PGC-1β, STAT3, and SHP).
In summary, we show that FGF-19 suppresses the stimulatory effect of insulin on the rate of fatty acid synthesis and the expression of lipogenic enzymes in hepatocytes. These observations provide a mechanism by which FGF-19 reverses adipose accretion, hepatic steatosis, hyperlipidemia, and type 2 diabetes in obese mice. Another member of the fibroblast growth factor family that is structurally related to FGF-19 is FGF-21. FGF-21 administration also reverses obesity, hepatic steatosis, and type 2 diabetes (65, 66). We postulate that FGF-19 and FGF-21 act via similar mechanisms to reverse diseases associated with metabolic syndrome.
Supplementary Material
Acknowledgments
We thank Dr. Lisa Salati and the members of her laboratory for assistance in preparing hepatocyte cultures.
This work was supported by National Research Initiative Competitive Grant 2007-35206-17845 from the United States Department of Agriculture Cooperative State Research, Education, and Extension Service.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1 and S2.
Footnotes
The abbreviations used are: ACC, acetyl-CoA carboxylase; ABCA1, ATP-binding cassette transporter A1; AMPK, AMP-activated protein kinase; ATP-CL, ATP-citrate lyase; CYP7A1, cholesterol 7α-hydroxylase; ERK, extracellular signal-regulated kinase; FAS, fatty-acid synthase; FGF, fibroblast growth factor; G6PD, glucose-6-phosphate dehydrogenase; GK, glucokinase; GSK, glycogen synthase kinase; IRS, insulin receptor substrate; l-PK, l-pyruvate kinase; LXR, liver X receptor; ME, malic enzyme; MAPK, mitogen-activated protein kinase; PGC, peroxisome proliferator-activated receptor-γ coactivator; PKA, protein kinase A; PKC, protein kinase C; S14, spot 14; SCD1, stearoyl-CoA desaturase-1; SHP, small heterodimer partner; Sp1, specificity protein 1; SREBP-1c, sterol regulatory element-binding protein-1c; mSREBP-1c, mature SREBP-1c; STAT3, signal transducer and activator of transcription 3; T3, 3,5,3′-triiodothyronine.
References
- 1.Muoio, D. M., and Newgard, C. B. (2008) Nat. Rev. Mol. Cell Biol. 9 193-205 [DOI] [PubMed] [Google Scholar]
- 2.Postic, C., and Girard, J. (2008) J. Clin. Investig. 118 829-838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savage, D. B., Petersen, K. F., and Shulman, G. I. (2007) Physiol. Rev. 87 507-520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou, G., Myers, R., Li, Y., Chen, Y., Shen, X., Fenyk-Melody, J., Wu, M., Ventre, J., Doebber, T., Fujii, N., Musi, N., Hirshman, M. F., Goodyear, L. J., and Moller, D. E. (2001) J. Clin. Investig. 108 1167-1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ladher, R. K., Anakwe, K. U., Gurney, A. L., Schoenwolf, G. C., and Francis-West, P. H. (2000) Science 290 1965-1967 [DOI] [PubMed] [Google Scholar]
- 6.Inagaki, T., Choi, M., Moschetta, A., Peng, L., Cummins, C. L., McDonald, J. G., Luo, G., Jones, S. A., Goodwin, B., Richardson, J. A., Gerard, R. D., Repa, J. J., Mangelsdorf, D. J., and Kliewer, S. A. (2005) Cell Metab. 2 217-225 [DOI] [PubMed] [Google Scholar]
- 7.Holt, J. A., Luo, G., Billin, A. N., Bisi, J., McNeill, Y. Y., Kozarsky, K. F., Donahee, M., Wang, D. Y., Mansfield, T. A., Kliewer, S. A., Goodwin, B., and Jones, S. A. (2003) Genes Dev. 17 1581-1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu, L., John, L. M., Adams, S. H., Yu, X. X., Tomlinson, E., Renz, M., Williams, P. M., Soriano, R., Corpuz, R., Moffat, B., Vandlen, R., Simmons, L., Foster, J., Stephan, J. P., Tsai, S. P., and Stewart, T. A. (2004) Endocrinology 145 2594-2603 [DOI] [PubMed] [Google Scholar]
- 9.Tomlinson, E., Fu, L., John, L., Hultgren, B., Huang, X., Renz, M., Stephan, J. P., Tsai, S. P., Powell-Braxton, L., French, D., and Stewart, T. A. (2002) Endocrinology 143 1741-1747 [DOI] [PubMed] [Google Scholar]
- 10.Stabile, L. P., Klautky, S. A., Minor, S. M., and Salati, L. M. (1998) J. Lipid Res. 39 1951-1963 [PubMed] [Google Scholar]
- 11.Lowenstein, J. M., Brunengraber, H., and Wadke, M. (1975) Methods Enzymol. 35 279-287 [DOI] [PubMed] [Google Scholar]
- 12.Chomczynski, P., and Sacchi, N. (1987) Anal. Biochem. 162 156-159 [DOI] [PubMed] [Google Scholar]
- 13.Hansmannel, F., Mordier, S., and Iynedjian, P. B. (2006) Biochem. J. 399 275-283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Standaert, M. L., Galloway, L., Karnam, P., Bandyopadhyay, G., Moscat, J., and Farese, R. V. (1997) J. Biol. Chem. 272 30075-30082 [DOI] [PubMed] [Google Scholar]
- 15.Hillgartner, F. B., Salati, L. M., and Goodridge, A. G. (1995) Physiol. Rev. 75 47-76 [DOI] [PubMed] [Google Scholar]
- 16.Saggerson, D. (2008) Annu. Rev. Nutr. 28 253-272 [DOI] [PubMed] [Google Scholar]
- 17.Postic, C., Dentin, R., Denechaud, P. D., and Girard, J. (2007) Annu. Rev. Nutr. 27 179-192 [DOI] [PubMed] [Google Scholar]
- 18.Dentin, R., Pegorier, J. P., Benhamed, F., Foufelle, F., Ferre, P., Fauveau, V., Magnuson, M. A., Girard, J., and Postic, C. (2004) J. Biol. Chem. 279 20314-20326 [DOI] [PubMed] [Google Scholar]
- 19.Koo, S. H., Dutcher, A. K., and Towle, H. C. (2001) J. Biol. Chem. 276 9437-9445 [DOI] [PubMed] [Google Scholar]
- 20.Edwards, P. A., Kennedy, M. A., and Mak, P. A. (2002) Vascul. Pharmacol. 38 249-256 [DOI] [PubMed] [Google Scholar]
- 21.Repa, J. J., and Mangelsdorf, D. J. (2002) Nat. Med. 8 1243-1248 [DOI] [PubMed] [Google Scholar]
- 22.Joseph, S. B., Laffitte, B. A., Patel, P. H., Watson, M. A., Matsukuma, K. E., Walczak, R., Collins, J. L., Osborne, T. F., and Tontonoz, P. (2002) J. Biol. Chem. 277 11019-11025 [DOI] [PubMed] [Google Scholar]
- 23.Talukdar, S., and Hillgartner, F. B. (2006) J. Lipid Res. 47 2451-2461 [DOI] [PubMed] [Google Scholar]
- 24.Tobin, K. A., Ulven, S. M., Schuster, G. U., Steineger, H. H., Andresen, S. M., Gustafsson, J. A., and Nebb, H. I. (2002) J. Biol. Chem. 277 10691-10697 [DOI] [PubMed] [Google Scholar]
- 25.Narkewicz, M. R., Iynedjian, P. B., Ferre, P., and Girard, J. (1990) Biochem. J. 271 585-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petty, K. J., Morioka, H., Mitsuhashi, T., and Nikodem, V. M. (1989) J. Biol. Chem. 264 11483-11490 [PubMed] [Google Scholar]
- 27.Zilz, N. D., Murray, M. B., and Towle, H. C. (1990) J. Biol. Chem. 265 8136-8143 [PubMed] [Google Scholar]
- 28.Foufelle, F., and Ferre, P. (2002) Biochem. J. 366 377-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregori, C., Guillet-Deniau, I., Girard, J., Decaux, J. F., and Pichard, A. L. (2006) FEBS Lett. 580 410-414 [DOI] [PubMed] [Google Scholar]
- 30.Stoeckman, A. K., and Towle, H. C. (2002) J. Biol. Chem. 277 27029-27035 [DOI] [PubMed] [Google Scholar]
- 31.Cagen, L. M., Deng, X., Wilcox, H. G., Park, E. A., Raghow, R., and Elam, M. B. (2005) Biochem. J. 385 207-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen, G., Liang, G., Ou, J., Goldstein, J. L., and Brown, M. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11245-11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi, C. M., Emanuelli, B., and Kahn, C. R. (2006) Nat. Rev. Mol. Cell Biol. 7 85-96 [DOI] [PubMed] [Google Scholar]
- 34.Fleischmann, M., and Iynedjian, P. B. (2000) Biochem. J. 349 13-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iynedjian, P. B., Roth, R. A., Fleischmann, M., and Gjinovci, A. (2000) Biochem. J. 351 621-627 [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto, M., Ogawa, W., Akimoto, K., Inoue, H., Miyake, K., Furukawa, K., Hayashi, Y., Iguchi, H., Matsuki, Y., Hiramatsu, R., Shimano, H., Yamada, N., Ohno, S., Kasuga, M., and Noda, T. (2003) J. Clin. Investig. 112 935-944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono, H., Shimano, H., Katagiri, H., Yahagi, N., Sakoda, H., Onishi, Y., Anai, M., Ogihara, T., Fujishiro, M., Viana, A. Y., Fukushima, Y., Abe, M., Shojima, N., Kikuchi, M., Yamada, N., Oka, Y., and Asano, T. (2003) Diabetes 52 2905-2913 [DOI] [PubMed] [Google Scholar]
- 38.Taniguchi, C. M., Kondo, T., Sajan, M., Luo, J., Bronson, R., Asano, T., Farese, R., Cantley, L. C., and Kahn, C. R. (2006) Cell Metab. 3 343-353 [DOI] [PubMed] [Google Scholar]
- 39.Sundqvist, A., Bengoechea-Alonso, M. T., Ye, X., Lukiyanchuk, V., Jin, J., Harper, J. W., and Ericsson, J. (2005) Cell Metab. 1 379-391 [DOI] [PubMed] [Google Scholar]
- 40.Zhang, W., Patil, S., Chauhan, B., Guo, S., Powell, D. R., Le, J., Klotsas, A., Matika, R., Xiao, X., Franks, R., Heidenreich, K. A., Sajan, M. P., Farese, R. V., Stolz, D. B., Tso, P., Koo, S. H., Montminy, M., and Unterman, T. G. (2006) J. Biol. Chem. 281 10105-10117 [DOI] [PubMed] [Google Scholar]
- 41.Kubota, N., Kubota, T., Itoh, S., Kumagai, H., Kozono, H., Takamoto, I., Mineyama, T., Ogata, H., Tokuyama, K., Ohsugi, M., Sasako, T., Moroi, M., Sugi, K., Kakuta, S., Iwakura, Y., Noda, T., Ohnishi, S., Nagai, R., Tobe, K., Terauchi, Y., Ueki, K., and Kadowaki, T. (2008) Cell Metab. 8 49-64 [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto, M., Ogawa, W., Teshigawara, K., Inoue, H., Miyake, K., Sakaue, H., and Kasuga, M. (2002) Diabetes 51 1672-1680 [DOI] [PubMed] [Google Scholar]
- 43.Botolin, D., Wang, Y., Christian, B., and Jump, D. B. (2006) J. Lipid Res. 47 181-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foretz, M., Pacot, C., Dugail, I., Lemarchand, P., Guichard, C., Le Liepvre, X., Berthelier-Lubrano, C., Spiegelman, B., Kim, J. B., Ferre, P., and Foufelle, F. (1999) Mol. Cell. Biol. 19 3760-3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto, T., Shimano, H., Inoue, N., Nakagawa, Y., Matsuzaka, T., Takahashi, A., Yahagi, N., Sone, H., Suzuki, H., Toyoshima, H., and Yamada, N. (2007) J. Biol. Chem. 282 11687-11695 [DOI] [PubMed] [Google Scholar]
- 46.Foretz, M., Ancellin, N., Andreelli, F., Saintillan, Y., Grondin, P., Kahn, A., Thorens, B., Vaulont, S., and Viollet, B. (2005) Diabetes 54 1331-1339 [DOI] [PubMed] [Google Scholar]
- 47.Woods, A., Azzout-Marniche, D., Foretz, M., Stein, S. C., Lemarchand, P., Ferre, P., Foufelle, F., and Carling, D. (2000) Mol. Cell. Biol. 20 6704-6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong, Y., Collins, Q. F., An, J., Lupo, E., Jr., Liu, H. Y., Liu, D., Robidoux, J., Liu, Z., and Cao, W. (2007) J. Biol. Chem. 282 4975-4982 [DOI] [PubMed] [Google Scholar]
- 49.Lin, J., Yang, R., Tarr, P. T., Wu, P. H., Handschin, C., Li, S., Yang, W., Pei, L., Uldry, M., Tontonoz, P., Newgard, C. B., and Spiegelman, B. M. (2005) Cell 120 261-273 [DOI] [PubMed] [Google Scholar]
- 50.Handschin, C., and Spiegelman, B. M. (2006) Endocr. Rev. 27 728-735 [DOI] [PubMed] [Google Scholar]
- 51.Bavner, A., Sanyal, S., Gustafsson, J. A., and Treuter, E. (2005) Trends Endocrinol. Metab. 16 478-488 [DOI] [PubMed] [Google Scholar]
- 52.Kerr, T. A., Saeki, S., Schneider, M., Schaefer, K., Berdy, S., Redder, T., Shan, B., Russell, D. W., and Schwarz, M. (2002) Dev. Cell 2 713-720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsukuma, K. E., Wang, L., Bennett, M. K., and Osborne, T. F. (2007) J. Biol. Chem. 282 20164-20171 [DOI] [PubMed] [Google Scholar]
- 54.Wang, L., Huang, J., Saha, P., Kulkarni, R. N., Hu, M., Kim, Y., Park, K., Chan, L., Rajan, A. S., Lee, I., and Moore, D. D. (2006) Mol. Endocrinol. 20 2671-2681 [DOI] [PubMed] [Google Scholar]
- 55.Inoue, H., Ogawa, W., Ozaki, M., Haga, S., Matsumoto, M., Furukawa, K., Hashimoto, N., Kido, Y., Mori, T., Sakaue, H., Teshigawara, K., Jin, S., Iguchi, H., Hiramatsu, R., LeRoith, D., Takeda, K., Akira, S., and Kasuga, M. (2004) Nat. Med. 10 168-174 [DOI] [PubMed] [Google Scholar]
- 56.Ueki, K., Kondo, T., Tseng, Y. H., and Kahn, C. R. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10422-10427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen, Z., Zhong, Z., and Darnell, J. E., Jr. (1995) Cell 82 241-250 [DOI] [PubMed] [Google Scholar]
- 58.Choi, C. S., Savage, D. B., Abu-Elheiga, L., Liu, Z. X., Kim, S., Kulkarni, A., Distefano, A., Hwang, Y. J., Reznick, R. M., Codella, R., Zhang, D., Cline, G. W., Wakil, S. J., and Shulman, G. I. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 16480-16485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng, X., Yellaturu, C., Cagen, L., Wilcox, H. G., Park, E. A., Raghow, R., and Elam, M. B. (2007) J. Biol. Chem. 282 17517-17529 [DOI] [PubMed] [Google Scholar]
- 60.Hirota, K., Sakamaki, J. I., Ishida, J., Shimamoto, Y., Nishihara, S., Kodama, N., Ohta, K., Yamamoto, M., Tanimoto, K., and Fukamizu, A. (2008) J. Biol. Chem. 283 32432-32441 [DOI] [PubMed] [Google Scholar]
- 61.Roth, U., Curth, K., Unterman, T. G., and Kietzmann, T. (2004) J. Biol. Chem. 279 2623-2631 [DOI] [PubMed] [Google Scholar]
- 62.Lee, Y. K., Dell, H., Dowhan, D. H., Hadzopoulou-Cladaras, M., and Moore, D. D. (2000) Mol. Cell. Biol. 20 187-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Streeper, R. S., Chapman, S. C., Ayala, J. E., Svitek, C. A., Goldman, J. K., Cave, A., and O'Brien, R. M. (1998) Mol. Endocrinol. 12 1778-1791 [DOI] [PubMed] [Google Scholar]
- 64.Salati, L. M., Szeszel-Fedorowicz, W., Tao, H., Gibson, M. A., Amir-Ahmady, B., Stabile, L. P., and Hodge, D. L. (2004) J. Nutr. 134 2437S-2443S [DOI] [PubMed] [Google Scholar]
- 65.Coskun, T., Bina, H. A., Schneider, M. A., Dunbar, J. D., Hu, C. C., Chen, Y., Moller, D. E., and Kharitonenkov, A. (2008) Endocrinology 149 6018-6027 [DOI] [PubMed] [Google Scholar]
- 66.Xu, J., Lloyd, D. J., Hale, C., Stanislaus, S., Chen, M., Sivits, G., Vonderfecht, S. N., Hecht, R., Li, L., Lindberg, R. A., Chen, J. L., Jung, D. Y., Zhang, Z., Ko, H. J., Kim, J. K., and Veniant, M. M. (2009) Diabetes 58 250-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.