Abstract
The mammalian SWI/SNF chromatin-remodeling complex is essential for the multiple changes in gene expression that occur during differentiation. However, the basis within the complex for specificity in effecting positive versus negative changes in gene expression has only begun to be elucidated. The catalytic core of the complex can be either of two closely related ATPases, BRM or BRG1, with the potential that the choice of alternative subunits is a key determinant of specificity. Short hairpin RNA-mediated depletion of the ATPases was used to explore their respective roles in the well characterized multistage process of osteoblast differentiation. The results reveal an unexpected role for BRM-specific complexes. Instead of impeding differentiation as was seen with BRG1 depletion, depletion of BRM caused accelerated progression to the differentiation phenotype. Multiple tissue-specific differentiation markers, including the tightly regulated late stage marker osteocalcin, become constitutively up-regulated in BRM-depleted cells. Chromatin immunoprecipitation analysis of the osteocalcin promoter as a model for the behavior of the complexes indicates that the promoter is a direct target of both BRM- and BRG1-containing complexes. BRG1 complexes, which are required for activation, are associated with the promoter well before induction, but the concurrent presence of BRM-specific complexes overrides their activation function. BRM-specific complexes are present only on the repressed promoter and are required for association of the co-repressor HDAC1. These findings reveal an unanticipated degree of specialization of function linked with the choice of ATPase and suggest a new paradigm for the roles of the alternative subunits during differentiation.
The mammalian SWI/SNF complex is an evolutionarily well conserved ATPase-powered chromatin-remodeling assembly consisting of approximately 10 subunits (see Fig. 1A). This complex (also known as the BAF complex) coordinates the disruption of nucleosomes to permit the binding of various transcription factors, an activity crucial for proper differentiation and development (reviewed in Refs. 1-5).
FIGURE 1.
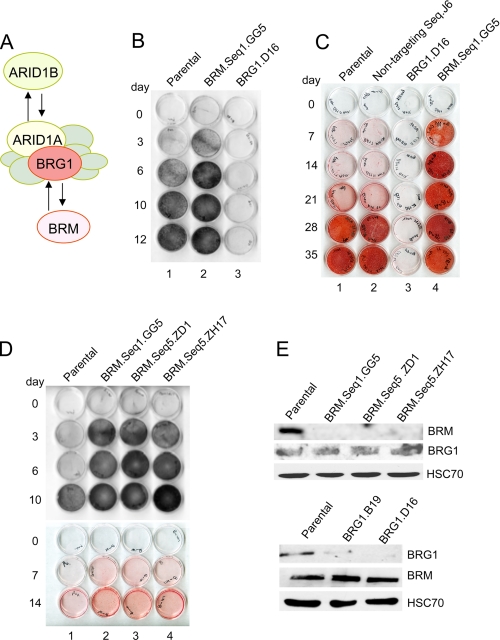
Differentiation phenotypes in BRM- and BRG1-depleted cells. A, the multisubunit SWI/SNF chromatin-remodeling complex contains a core ATPase, either BRG1 or BRM, plus seven or more non-catalytic subunits. Of these, ARID1A and ARID1B are also mutually exclusive alternatives. Either ATPase can associate with either ARID family member (25, 35), such that there are at least four distinct subsets of the SWI/SNF complex. B, parental and knockdown cell cultures were induced for the time intervals indicated, fixed with methanol, and reacted with the substrate BCIP/NBT to reveal alkaline phosphatase activity; positive cells stain purple-black. Seq, sequence. C, induced cell monolayers were stained at later time intervals with Alizarin Red S, which indicates the presence of calcium-containing compounds in the cell matrix. D, the phenotype of three independent BRM knockdown lines, generated with two different shRNA sequences, was analyzed as described in panels B and C. E, total cell lysate from parental and knockdown lines (indicated above the lanes) was probed with antibodies specific to either BRM or BRG1, as indicated to the right. An antibody probe for the constitutively expressed HSC70 protein was used as a loading control.
The entity known as the mammalian SWI/SNF complex actually consists of a small series of compositionally distinct assemblies distinguished by the presence of alternative subunits. The choice of ARID family subunit (ARID1A or ARID1B) is a determinant of complexes with generally opposing roles in cell cycle control (Ref. 6 and reviewed in Ref. 7). The complexes also contain either of two closely related alternative ATPases: Brahma (BRM)3 or Brahma-related gene 1 (BRG1). Although BRM and BRG1 share a high degree of amino acid sequence identity, they are not equally important for development. Brg1-null mice die at a pre- or peri-implantation stage (8), indicating a critical developmental role for BRG1. In contrast, Brm-null mice are viable and fertile, exhibiting only mild abnormalities that include a larger animal size and deregulated cell growth control in derived fibroblasts (9). This study also showed an increased level of BRG1 in the animal tissues in the absence of BRM, and several studies indicate that BRG1- and BRM-containing SWI/SNF complexes play largely compensatory roles in cell cycle control (e.g. Ref. 10 and reviewed in Refs. 11 and 12). Due to these phenotypes, it has been generally thought that BRM plays a similar but mostly auxiliary role to BRG1 in regulation of tissue-specific gene expression (reviewed in Ref. 5). However, few studies have compared the roles of BRM and BRG1 directly in differentiation models, and where considered (e.g. Ref. 13), BRM was generally confirmed as non-essential with relatively little other detail.
Given the ARID family evidence that alternative subunits can be important functional determinants (6, 7), we decided to probe more closely the question of the roles of BRG1 and BRM in differentiation. The ATPases were depleted individually by shRNA approaches in a differentiation model chosen for its well ordered multistep nature, with the expectation that such a model might reveal subtle differences in the requirement for each of the respective complexes. Osteoblast precursors including the mouse calvaria-derived MC3T3-E1 line undergo a tightly regulated differentiation process when induced with appropriate agents such as ascorbic acid and a source of organic phosphate (14-18). An important advantage of this model is that differentiation proceeds through discreet stages with predictable timing, providing a window for observing subtle changes in the rate of differentiation in addition to overall inhibition of differentiation. The expectation was that BRG1 depletion would block differentiation, whereas BRM depletion might cause modest delay. However, the studies described here unexpectedly revealed a programmatic role for BRM-containing complexes in repression of BRG1-dependent differentiation. Deficiency of BRM does not correlate with impaired differentiation; in contrast, it results in an accelerated rate of mineralization with constitutively higher levels of expression of osteogenic markers. These results reveal a new aspect of the alternative ATPases, identifying them as determinants of SWI/SNF complexes with opposing roles across a whole program of tissue-specific gene expression.
EXPERIMENTAL PROCEDURES
Materials and Cell Culture—Penicillin and streptomycin were purchased from Mediatech (Herndon, VA). Ascorbic acid, β-glycerol phosphate, sodium phosphate mono and dibasic, Alizarin red S, and protease inhibitors were obtained from Sigma. Fetal bovine serum was purchased from Atlanta Biologicals, and α-MEM was from Irvine Scientific (Santa Ana, CA). G418 was from Invitrogen. Radiochemicals were obtained from PerkinElmer Life Sciences. Culture and differentiation of low passage MC3T3-E1 cells by exposure to ascorbic acid and β-glycerol phosphate has been described previously (18).
shRNA and Isolation of Stable BRM and BRG1 Knockdown Lines—The shRNA sequences were tested in a pSUPER vector (19). For BRM, two targeted constructs were generated using the RNAi Designer at the Clontech site. The 64-bp forward sequence for the first (BRM:seq1rnai) sequence is: 5′-GATCCCCGATCCAGAAGCTCTCCAAATTCAAGAGATTTGGAGAGCTTCTGGATCTTTTTGGAAA-3′ (the 19-bp target sequence is underlined). The respective 19-bp sequence for the alternative BRM sequence (BRM:seq5rnai) is 5′-GTCATAAGCCTGAGGCAAA-3′. The respective 19-bp BRG1 target sequence is 5′-GCCTATGGAGTCCATGCAC-3′ (adapted from Ref. 20). The pSUPER-derived vectors containing the respective knockdown sequences were introduced into MC3T3-E1 cells by lipofection together with a selectable neo marker. G418-resistant clones were amplified and screened by Western blot for BRM or BRG1 expression. Aliquots of low passage depleted lines were frozen as stocks. A control line transfected with a scrambled non-targeting sequence has been described previously (21).
Alkaline Phosphatase Staining—Cell monolayers were rinsed in PBS, fixed in 100% methanol, rinsed with PBS, and then overlaid with 1.5 ml of 0.15 mg/ml BCIP plus 0.3 mg/ml NBT (Promega, Madison, WI) for 30 min and rinsed again with PBS three times.
Mineralization Assay—Cells were induced and plated as described above. The monolayers were washed with PBS, covered with 0.1% alizarin red S for 10 min, and then rinsed with PBS three times and dried.
Northern Blots—Total cell RNA was prepared using TRIzol reagent (Invitrogen) or Tri Reagent (Sigma) according to manufacturer's recommendations. 20 μg of RNA were loaded per lane and separated by electrophoresis through a 1% formaldehyde-agarose gel. The RNA was transferred to a Hybond-N nylon membrane (Amersham Biosciences) and cross-linked by UV irradiation. 32P-labeled probes were prepared using a random primer labeling kit (Roche Applied Science). 500 μCi of [α-32P]ATP. was used per labeling reaction. Between successive probes, blots were stripped by treatment with boiling 0.1% SDS. The osteocalcin probe and plasmid pGB.GAPDH were described previously (22, 23).
Immunoblotting—Cells were washed and harvested in PBS and lysed in p300 lysis buffer (24). Proteins were separated by polyacrylamide gel electrophoresis, transferred to Immobilon-P membrane (Millipore), and visualized using either Western Lighting chemiluminescence reagent Plus (PerkinElmer Life Sciences) or BCIP/NBT (Promega).
Real-time PCR Assay—Real-time assays were performed with the RT2 Profiler™ PCR array: mouse osteogenesis (Super-Array, Frederick, MD, catalogue number PAMM-026), according to the manufacturer's directions. The array contains primer sets for 84 osteogenesis-related genes and five housekeeping genes. The starting amount of RNA used was 1 μg. PCR was carried out on an ABI7500 cycler using the following parameters: 1 cycle for 10 min at 95 °C and 40 cycles for 15 s at 95 °C, 1 min at 60 °C. Data were analyzed using the PCR Array Data Analysis Web Portal.
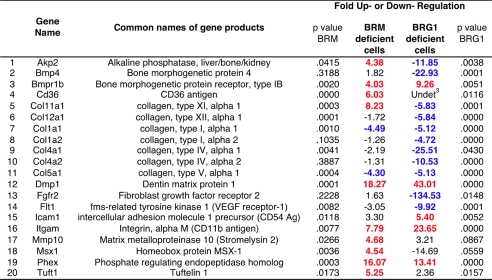
All quality control parameters (genomic DNA control, reverse transcription control, and positive PCR control) were within manufacturer's recommended limits in each assay. Results for each assay were normalized to the average of all five housekeeping genes. The parental cell population and the scrambled sequence line were each analyzed in duplicate, and the averages were compared. The 10 genes that differed more than 4-fold between these two controls were excluded from further consideration. Each of the three independent BRM knockdown lines (GG5, ZD1, and ZD17) was analyzed in duplicate, and the average of the six runs was compared with the average of the parental and scrambled cells to obtain the -fold change. Likewise, the two BRG1 knockdown lines (D16 and B19) were analyzed in duplicate and again compared with the parental and scrambled cells. In accordance with recommended thresholds, a gene expression change is reported in Table 1 if it is greater than 4-fold and the t test p value is less than 0.05.
TABLE 1.
Genes affected by knockdown of BRM or BRG11,2
1 Fold change in gene expression levels was determined as described under “Experimental Procedures.” Positive changes are highlighted in red; negative changes are in blue. A gene expression change is highlighted if it is greater than 4-fold and the t test p value was <0.05.
2 Of the 84 genes on the array, 10 (Col10a1, Csf2, Fn1, Igf1, Mmp9, Serpinh1, Smad2, Smad3, Sox9, and Tnf) differed more than 4-fold between the scrambled cell line and the parental population and were therefore regarded as too variable for analysis in the knockdown lines. A further 54 were unaffected by either knockdown. They are; Ahsg, Ambn, Anxa5, Bgn, Bmp1, Bmp2, Bmp3, Bmp5, Bmp6, Bmpr1a, Cdh11, Col12a1, Col14a1, Col3a1, Col6a1, Col6a2, Col7a1, Comp, Csf3, Ctsk, Egf, Enam, Fgf1, Fgf2, Fgf3, Fgfr1, Gdf10, Igf1r, Itga2, Itga2b, Itga3, Itgav, Itgb1, Mmp2, Mmp8, Nfkb, Pdgfa, Runx2, Scarb1, Smad1, Smad4, Sost, Tfip11, Tgfb1, Tgfb2, Tgfb3, Tgfbr1, Tgfbr2, Tgfbr3, Twist1, Vcam, Vdr, Vegfa, and egfb.
3 Undet product was not detected.
ChIP Assays—Chromatin immunoprecipitation (ChIP) assays were performed with the EZ ChIP™ system (Upstate Cell Signaling Solutions, Lake Placid, NY), according to the manufacturer's directions, modified to include preclearing of lysates with 60 μl of a 50% slurry of protein G/salmon sperm DNA for 1 h at 4 °C, and again performed overnight. Negative controls consisted of either IgG or the viral-specific monoclonal antibody 419. Primer sequences are listed in Fig. 3B. PCR conditions were 40 cycles at for 30 s at 95 °C, 30 s at 72 °C, and 30 s at 60 °C.
FIGURE 3.
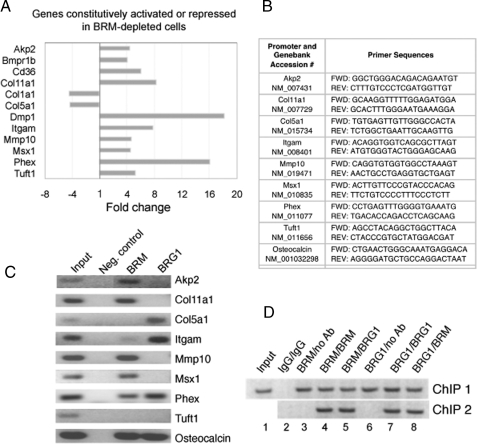
Real-time PCR analysis of gene expression in BRM-depleted cells. A, expression of 84 osteogenesis-associated genes was analyzed by QPCR in wild type and BRM-depleted MC3T3-E1 cells. Genes whose expression changed by more than 4-fold in BRM-depleted cells are shown in the graph above. Most changes were activating, consistent with the phenotypic evidence that BRM complexes act predominantly to repress differentiation. B, the table shows the primers used for the ChIP analysis in panel C. C, the promoters of a selection of the genes identified in panel A were subjected to ChIP analysis with the indicated primers to determine whether the genes are direct targets of BRM complexes. Neg. control, negative control. D, serial ChIP analysis indicates that BRG1 and BRM are present simultaneously on the osteocalcin promoter. The antibodies (Ab) used in ChIP 1/ChIP 2 are indicated above the lanes.
Re-ChIP Assays—Re-ChIP assays were performed with the Active Motif Re-ChIP-IT™ system (Active Motif, Carlsbad, CA), according to the manufacturer's directions. The assay was modified to include overnight incubations of the antibodies at 4 °C, for both the first and the second chromatin IP. Primer sequences for osteocalcin are listed in Fig. 3B. PCR conditions are same as above.
Antibodies—Antibodies of the following specificities were obtained from commercial sources: PEB2αA/RUNX2 (s-19 sc12488), BRM (N-19, sc-6450), BRG1 (H-88 sc-10768), and HSC70 (B-6, sc-7298) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); SNF5/INI1 (612110, BD Biosciences); anti-trimethyl-histone H3 (Lys 4) (catalog number 04-745, Millipore); and HDAC1 (catalog number 2062, Cell Signaling). Monoclonal antibodies specific for p270/ARID1A (PSG3), ARID1B (KMN1), and SV40 T antigen (419) have been described previously (6, 25).
RESULTS
Phenotypic Analysis of BRG1 and BRM Knockdown Lines—DNA sequences encoding shRNA molecules complementary to either Brg1 or Brm were introduced from a plasmid vector by stable integration into low passage MC3T3-E1 cells. In each transfection, colonies appeared at similar frequencies and showed essentially the same doubling time in normal growth medium as a vector-only control. The resultant lines were screened for the ability to respond to an ascorbic acid signal by induction of two key indicators of osteoblast differentiation: increased alkaline phosphatase activity and formation of a mineralized matrix. Alkaline phosphatase is among the earliest markers of osteoblast differentiation. The enzyme is exported to the osteoblast cell surface, where its activity can be visualized in a sensitive in situ assay scored by color development (Fig. 1B). As expected, the BRG1-depleted line (designated D16) showed severely impaired induction of alkaline phosphatase activity (row 3 as compared with row 1). Unexpectedly, the BRM-depleted line (GG5) showed an enhanced level of alkaline phosphatase activity even in non-induced cells (day 0) and an enhanced induction in response to the differentiation signal (row 2 as compared with row 1).
The same patterns were seen when the cells were tested for mineralization activity. Formation of calcium-containing mineralization products in the cell matrix can be detected by staining with Alizarin Red S (Fig. 1C). In this assay, the BRG1-depleted line again behaved as expected, showing virtually no mineralization (row 3 as compared with row 1). In addition, unexpectedly again, but consistent with the alkaline phosphatase induction pattern, the BRM-depleted cells showed accelerated progression to the mineralization phenotype (row 4 as compared with row 1). A control cell line (J6) derived from transfection with a non-targeting sequence (row 2) behaved like the parental cells.
Because this pattern was so contrary to expectations, additional BRM-depleted lines were constructed using a second, independent, shRNA sequence. Two separate clones (ZD1 and ZH17) were isolated. The pattern of all three BRM-depleted lines is analyzed in Fig. 1D, and all show the same phenotype. In each line, there is a constitutively enhanced level of alkaline phosphatase activity (day 0), accelerated induction of alkaline phosphatase activity, and accelerated progression to the mineralization phenotype. Rather than augmenting BRG1-dependent progression to terminal differentiation, the BRM-containing subset of SWI/SNF complexes participates in an opposing pathway, restraining differentiation.
Western blot analysis with antibodies specific for BRM and BRG1, respectively, indicate that depletion of one ATPase subunit does not have a major effect on expression of the other (Fig. 1E). It is also known from established tumor cell lines that deficiency of BRG1 or BRM, or both, does not otherwise disrupt assembly of the SWI/SNF complex (e.g. Ref. 26).
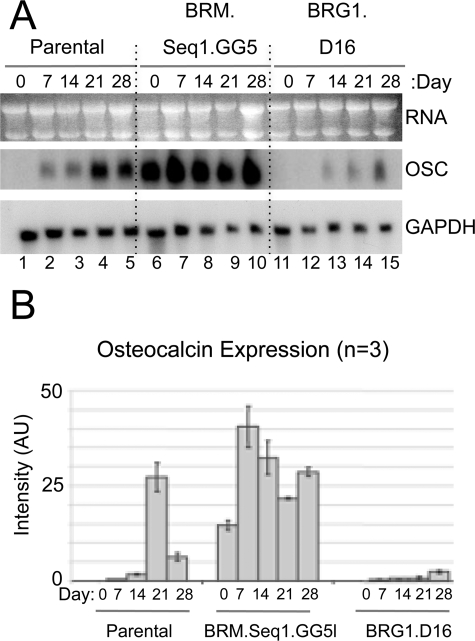
Osteocalcin Is Constitutively Expressed in BRM-depleted Cells—The best studied marker of late stage differentiation in osteoblasts is the mineralized matrix component osteocalcin. The osteocalcin gene (Bglap2) is a well established model for induction of tissue-specific gene expression whose activation has been shown to be dependent on SWI/SNF complex activity in a differentiating rat osteosarcoma cell line (27, 28). To probe the molecular events underlying the phenotypes of the knockdown lines, osteocalcin expression was assessed quantitatively by Northern blot analysis. Normally, osteocalcin expression is barely detectable in non-induced cells. After induction of differentiation, expression increases dramatically in parallel with mineral deposition. The Northern blot in panel A of Fig. 2 shows the typical pattern of osteocalcin (OSC) induction in parental cells as compared with expression in BRM and BRG1 knockdown lines. The BRG1-depleted cells show greatly impaired induction of osteocalcin (lanes 11-15 as compared with lanes 1-5), correlating with the severe defect in mineralization phenotype. In contrast, BRM-depleted cells show strikingly high constitutive expression of osteocalcin (lane 6 as compared with lane 1) and rapid induction to higher levels (lanes 6-10 as compared with lanes 1-5), concordant with the accelerated mineralization phenotype. Results averaged from three independent experiments are shown quantitatively in panel B.
FIGURE 2.
Regulation of osteocalcin expression in BRM- and BRG1-depleted lines. A, parental and knockdown lines were cultured in differentiation medium; total RNA was isolated at days 0, 7, 14, 21, and 28, as indicated, and analyzed by Northern blotting with sequentially applied probes for osteocalcin (OSC) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Seq, sequence. B, Northern blot analysis from three independent experiments was quantified by phosphoimaging, normalized to glyceraldehyde-3-phosphate dehydrogenase signals, averaged, and plotted as arbitrary units (AU) of phosphoimaging values. Error bars indicate the average deviation from the mean.
Real-time PCR Array Analysis of Osteogenesis-associated Gene Expression—The deregulation of the osteocalcin gene indicates that BRM-depleted cells have lost a major promoter repression function. To gauge the extent of genes affected by BRM depletion, an array of 84 osteogenesis-associated genes was analyzed in the osteoblast precursors by quantitative real-time reverse transcription-PCR (QPCR). Each of the three independent BRM knockdown lines (GG5, ZD1, ZD17) was analyzed in duplicate, and the average of the six runs was compared with the average of duplicate runs performed on both parental cells and the J6 control line to obtain the -fold change. Taking the recommended 4-fold difference as the cut-off point, 12 genes scored as affected by BRM depletion in this assay; 10 were up-regulated, and two were down-regulated (Fig. 3A). (Osteocalcin was not present on the array.)
Among the genes constitutively up-regulated in BRM-depleted cells is Akp2, encoding alkaline phosphatase, a major osteoblast differentiation marker whose increase was also apparent at the level of enzyme activity (Fig. 1). The remainder of the list encompasses genes from multiple classes, including those encoding hematopoietic cell-associated antigen CD11b (Itgam) and the widely expressed CD36 antigen, both of which have been observed in differentiating osteoblasts (29, 30). Genes encoding various extracellular matrix components (DMP1, collagen 11, and tuftelin) are activated. In addition, expression of the enzyme-encoding genes Phex and Mmp10 is increased. These enzymes, like alkaline phosphatase, participate in phosphate and matrix metabolism. The list also includes genes encoding one of the receptors for the bone morphogenic proteins (Bmpr1b), as well as the osteogenic transcription factor MSX1. (A list of common names of the genes is shown in Table 1.)
Not every osteoblast marker on the array is constitutively activated in BRM-depleted cells, nor would they be predicted to be as the BRM-depleted cells do not mineralize spontaneously, and only a minority of promoters are thought to be targets of regulation mediated by the SWI/SNF complexes. The two down-regulated genes in the BRM-deficient cells both encode additional members of the large collagen gene family. Down-regulation of these two genes diverges from the general pattern, but the overall profile is clearly consistent with a central role for BRM complexes in restraining precocious osteoblast differentiation.
The effect of BRG1 depletion was analyzed as well. As expected, multiple genes whose expression is characteristic of osteoblast commitment were down-regulated in BRG1-depleted cells. A comparison of the effects of BRG1 versus BRM depletion (Table 1) highlights the largely antagonistic nature of their effects, consistent with the concept that the two ATPases are specificity determinants of complexes with generally opposing roles in osteogenesis.
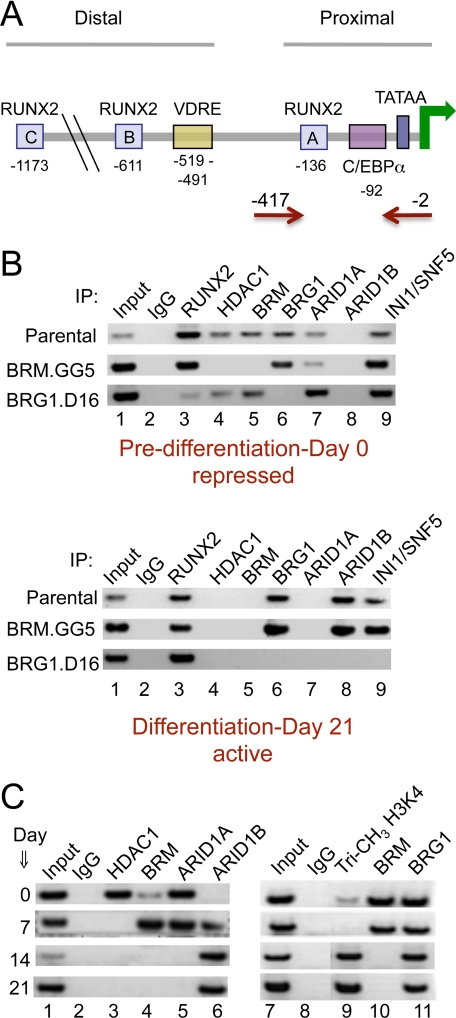
Identification of Direct Targets of BRM-specific Complexes—The gene array results identify a minimum of 10 osteogenesis markers in addition to osteocalcin that are coordinately derepressed as a consequence of BRM deficiency. This does not, however, indicate whether the promoters of these genes are direct targets of BRM complexes. To address this question, a panel of BRM-affected genes whose promoter sequences were readily identifiable was probed by ChIP analysis in non-induced cells (Fig. 3, B and C). At least six genes were revealed in this manner to be direct targets of BRM complexes: Akp2, Col11a1, Mmp10, Msx1, Phex, and osteocalcin. In addition, BRM was weakly detectable on the Itgam promoter. In each of these cases, the promoter occupation pattern is consistent with a role for BRM complexes in promoter repression. BRM was not detected on the Col5a1 or Tuft1 promoters, implying that BRM affects these genes only indirectly. This is of particular note for the divergently regulated Col5a1, but negative results remain inconclusive. In most cases where one of the ATPases was identified in association with the promoter, the occupation was either/or with respect to BRM versus BRG1. However, BRG1 was readily detected along with BRM on the Phex and osteocalcin promoters. Phex is one of the minority cases seen in Table 1 in which BRG1 also appears to contribute a repressor role, so this is consistent with the ChIP results. In contrast, BRG1 is a required activator of osteocalcin, and the apparent presence of both ATPases simultaneously on this promoter implies a more complex mechanism of regulation of this key gene product. The osteocalcin promoter is by far the best characterized of the identified BRM-targeted promoters and was subjected to further detailed analysis. First, a serial ChIP assay was performed to determine whether the two ATPases actually do associate with this promoter simultaneously (Fig. 3D). The antibodies used in ChIP 1/ChIP 2 are indicated above the lanes. The re-ChIP (ChIP 2) confirms that BRG1 is present on the BRM-precipitated promoter DNA (lane 5), and conversely, that BRM is present on the BRG1-precipitated promoter DNA (lane 7).
BRM Complexes Override BRG1-dependent Activation of the Osteocalcin Promoter—The simplest mechanisms by which BRM-specific complexes might repress expression from a particular promoter would be by preventing association of a required activator or co-activator or by facilitating association of a required repressor or co-repressor. Interestingly, the results in Fig. 3B demonstrate that BRM complexes do not necessarily simply compete with BRG1 complexes for promoter association. Prior analysis of the osteocalcin promoter has identified certain other key factors that were considered here. RUNX2/CBFA1 is a major tissue-specific transcriptional activator controlling lineage commitment in osteoblasts (reviewed in Ref. 31) and is known to be associated with the osteocalcin promoter prior to activation (e.g. Refs. 27, 32, 33). In contrast to activation, transcriptional repression typically involves associated histone deacetylase (HDAC) activity. HDAC1 appears to be a key regulator for osteoblast differentiation and has been identified in association with the osteocalcin promoter specifically in the predifferentiation (i.e. repressed) state in primary bone marrow cells (34).
Runx2 was included in the QPCR array, and notably, its expression in the non-induced cells is unaffected by depletion of either ATPase (Table 1, Footnote 2). Prior to differentiation, BRM, BRG1, RUNX2, and HDAC1 can all be seen in association with the promoter in parental cells (Fig. 4B, upper panel, lanes 3-6). Association of RUNX2 is unaffected by BRM depletion in the BRM.GG5 cell line, whereas association of HDAC1 is lost in BRM.GG5 cells. Analysis of the BRG1.D16 line shows that BRG1 complexes, although present on the repressed promoter, are not linked with association of HDAC1. (The association patterns are represented schematically in Fig. 5.) These results combined with the biological phenotype indicate that the promoter is poised for expression in non-induced cells but that expression functions are overridden by the presence of BRM-containing complexes and their HDAC1 affiliate. Depletion of BRM essentially converts the association profile of the key indicators (Fig. 4B, lanes 3-6) from the pattern characteristic of the repressed promoter in parental cells (upper panel) to the pattern characteristic of the active promoter (lower panel). The promoter in the BRM.GG5 line at day 0 is almost as active as the parental promoter at day 21 of differentiation (Fig. 2).
FIGURE 4.
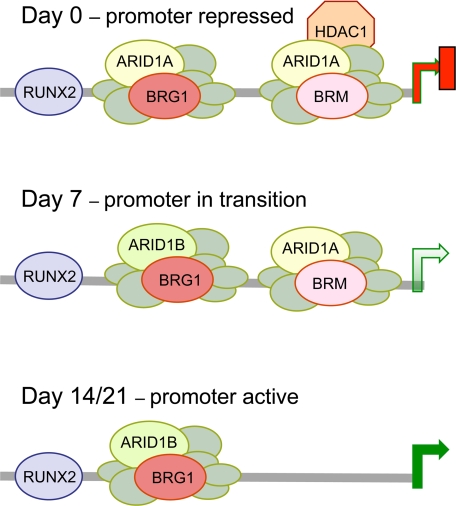
ChIP analysis of the osteocalcin promoter. A, schematic representation of the osteocalcin promoter indicating the locations of the three RUNX2 binding sites (A, B, and C), the vitamin D-responsive element (VDRE), the TATA box, and the CAAT/enhancer-binding protein α (C/EBPα) element identified as a target of SWI/SNF complexes (27). Primer sequences were designed to target the proximal promoter; the red arrows indicate the position of the primers used in the ChIP assays. B, parental or knockdown lines were harvested at day 0 (predifferentiation) or day 21 after induction and analyzed by ChIP assay for the presence of specific SWI/SNF subunits and other factors of interest on the osteocalcin promoter. IP, immunoprecipitation. C, ChIP analysis was performed on parental cells at intermediate time points during differentiation. The dynamics indicate that BRM dissociates between day 7 and day 14, concordant with the sharp rise in osteocalcin expression between days 14 and 21. Dissociation of HDAC1 precedes BRM dissociation, and binding of an ARID1B-containing complex precedes complete dissociation of ARID1A-containing complexes. This suggests the existence of a transition configuration on a partially activated promoter around day 7, shown schematically in Fig. 5. After the transition point, the promoter region becomes strongly trimethylated at histone H3K4, a chromatin mark indicative of active transcription.
FIGURE 5.
Schematic representation of the dynamics of complex association on the proximal osteocalcin promoter. The association of key factors at the osteocalcin promoter at major points during differentiation in normal cells as determined by ChIP analysis in Fig. 4, B and C, is represented schematically. The unlabeled circles represent the invariable subunits of the SWI/SNF complex. The relative positions of the complexes are indicated arbitrarily. HDAC1 is drawn in association with the BRM complex prior to induction to indicate its specific dependence on BRM association. Dissociation of HDAC1 precedes BRM complex dissociation, and binding of an ARID1B-containing complex precedes complete dissociation of ARID1A-containing complexes, indicating the existence of a transition configuration on a partially activated promoter at around day 7.
ARID Family Subunits Associate Differentially with the Osteocalcin Promoter—We recently reported that the SWI/SNF subunits, ARID1A and ARID1B, are specificity determinants of complexes that play repressing versus activating roles, respectively, on pro-proliferative genes (6). These are the only subunits other than the ATPases known to exist as a mutually exclusive pair in most cells. Examination of the association pattern of the ARID family subunits with the osteocalcin promoter (Fig. 4B, lanes 7 and 8) shows ARID1A present only on the repressed promoter and ARID1B present only when the promoter is active. ARID1A association is not dependent on either ATPase individually, so this subunit is likely associated with both complexes on the repressed promoter. Because only ARID1B is on the active promoter, it appears that the BRG1 complex changes from an ARID1A-containing configuration on the repressed promoter to an ARID1B-containing configuration on the active promoter (Fig. 5, schematic). As a further control, the presence of the INI1/SNF5 subunit was also probed. INI1/SNF5 is present in all known subsets of the complex. Its association profile (lane 9) is consistent with this and with a general finding that the presence of an ATPase subunit is required for promoter association of the complex as a whole.
Although the ARID family subunits help to distinguish activator versus repressor complexes, their role is apparently not essential on the osteocalcin promoter as the ARID1A and ARID1B knockdown lines do not show major differences in mineralization phenotypes (data not shown). This is consistent with the ChIP results indicating that the BRG1 complex does not need to switch to an ARID1B subunit to effect constitutive activation of osteocalcin in BRM-depleted cells. However, a complex specifically containing BRG1 itself is clearly required for activation (Fig. 2B). The four different combinations of ARID subunit and ATPase (25, 35) help explain how each of the subunits can be identified on both active and repressed promoters, although their respective roles are not random. The significance of the alternative subunits is only beginning to be addressed, but evidence so far suggests a general pattern in which BRM+ARID1A complexes are linked most closely with repression and BRG1+ARID1B complexes are linked most closely with activation, whereas the other possible combinations, BRM+ARID1B and BRG1+ARID1A, are more variable in their activities.
Disassociation of BRM from the Osteocalcin Promoter Correlates Temporally with Up-regulation of Osteocalcin Expression—To obtain a more dynamic picture of SWI/SNF-mediated regulation of the osteocalcin promoter, the association of key factors was probed at major intermediate time points (Fig. 4C). The results show that BRM is still present at day 7 but dissociates by day 14. This correlates well with the pattern of osteocalcin induction, which rises rapidly between days 14 and 21. The results reveal further that dissociation of HDAC1 (lane 3) precedes dissociation of BRM and that association of ARID1B (lane 6) precedes complete dissociation of ARID1A. The promoter association dynamics imply the existence of a transition point at about day seven when the promoter is undergoing initial stages of activation (Fig. 5, schematic), which correlates with the initial level of activation seen at day 7 in Northern blots. Presumably, following this transition period, other chromatin events occur that lead to full activation. One important event characteristic of activated promoters and associated with increased histone acetylation is trimethylation at histone H3 lysine 4 (H3K4) (36). This modification increases on the osteocalcin promoter at later times (days 14 and 21) (Fig. 4C, lane 9) coordinately with dissociation of the BRM complexes.
DISCUSSION
The presence of alternative subunits in the SWI/SNF complex is a feature unique to higher order eukaryotes that apparently evolved to permit finer tuning in transcriptional regulation. The present results reveal an unanticipated degree of specialization of function linked with the choice of ATPase and suggest a new paradigm for the roles of the alternative subunits during differentiation. The outcome highlights the importance of comparing subunit function within a single biological system and across a whole program of gene expression.
These results do not imply that BRM-containing complexes are linked exclusively with repression; indeed tissue-specific gene activation functions have been identified in other contexts (e.g. Refs. 20, 37-39). In particular, a positive role for BRM in retinal cell differentiation was identified recently (40). Conversely, the results also do not imply that BRG1 is linked only with activation of differentiation. Conditional loss of BRG1 in neuronal stem cells results in precocious neuronal differentiation, implying a role for BRG1 complexes in maintaining the neuronal stem cell phenotype (41); this effect may be analogous to what we see with depletion of BRM in osteoblast precursors. Interestingly, ARID1A also appears to play a role in maintaining a stem cell phenotype (42). What is essential in the current context is that the roles of the two ATPases were not compared directly in these other systems, so the distinct question of balanced opposing functions was not addressed.
The results described here suggest that at least one mechanism by which BRM-containing complexes effect repression is by mediating promoter association of the histone deacetylase, HDAC1. This interpretation is supported by analysis elsewhere of the effects of HDAC1 depletion in a rat osteosarcoma cell model. Depletion of HDAC1 via small interfering RNA-mediated knockdown was accompanied by increased alkaline phosphatase activity and heightened expression of osteoblast differentiation markers (34), a phenotype very similar to the phenotype reported here for BRM-depleted cells. These factors are not the only ones active at the promoter, of course, but the present results establish that BRM-containing complexes are essential for repression and that the default condition of the osteocalcin promoter in the absence of a BRM complex is active expression, even without a signal for differentiation.
The present results raise the question of whether BRM acts at the organism level to restrain differentiation of osteoblasts or of other mesenchymally derived tissues such as muscle and cartilage. BRG1 is vital for differentiation and development, but the observation that mice develop relatively normally in the absence of BRM has led to a general view that BRM complexes are largely auxiliary to BRG1 complexes, performing similar but non-essential roles. However, the BRM-null mice do suggest a unique and non-overlapping role for BRM in cell and tissue function. Their most prominent phenotype is a larger body size (∼14% above normal). Both BRG1 and BRM are linked with cell cycle repression, and increased cell proliferation may contribute to the larger body size of BRM-null mice. It is intriguing, however, that the authors of this study also noted a body weight distribution suggesting a disproportionately increased bone and/or muscle mass (9). This accords well with a specific role for BRM in the repression of differentiation in mesenchymally derived tissues. During mammalian development, deficiency of BRM could lead to increased bone mass if osteoblast precursors are primed to progress more quickly to mineralization once stimulated by anabolic factors.
Supplementary Material
Acknowledgments
We thank Kimberly Hsu and Lindsay Kelly for excellent technical assistance and Robert Donnelly, Ph.D., for guidance in the use of real-time PCR. We gratefully acknowledge the support and critical feedback provided by all our colleagues at Temple University School of Medicine and the University of Medicine and Dentistry of New Jersey.
This work was supported, in whole or in part, by National Institutes of Health Grant GM073257 from the USPSH (to E. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a Paper of the Week.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: BRM, Brahma; BRG1, Brahma-related gene 1; shRNA, short hairpin RNA; ChIP, chromatin immunoprecipitation; HDAC, histone deacetylase; PBS, phosphate-buffered saline; NBT, nitro blue tetrazolium; BCIP, 5-bromo-4-chloro-3-indolyl phosphate; QPCR, quantitative real-time reverse transcription-PCR.
References
- 1.Kingston, R. E., and Narlikar, G. J. (1999) Genes Dev. 13 2339-2352 [DOI] [PubMed] [Google Scholar]
- 2.Vignali, M., Hassan, A. H. G., Neely, K. E., and Workman, J. L. (2000) Mol. Cell. Biol. 20 1899-1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohrmann, L., and Verrizer, C. P. (2005) Biochim. Biophys. Acta. 1681 59-73 [DOI] [PubMed] [Google Scholar]
- 4.Smith, C. L., and Peterson, C. L. (2005) Curr. Top. Dev. Biol. 65 115-148 [DOI] [PubMed] [Google Scholar]
- 5.de la Serna, I. L., Ohkawa, Y., and Imbalzano, A. N. (2006) Nat Rev Genet. 6 461-473 [DOI] [PubMed] [Google Scholar]
- 6.Nagl, N. G., Jr., Wang, X., Patsialou, A., Van Scoy, M., and Moran, E. (2007) EMBO J. 26 752-763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blais, A., and Dynlacht, B. D. (2007). Curr. Opin. Cell Biol. 19 658-662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bultman, S., Gebuhr, T., Yee, D., LaMantia, C., Nicholson, J., Gilliam, A., Randazzo, F., Metzger, D., Chambon, P., Crabtree, G., and Magnuson, T. (2000) Mol. Cell. 6 1287-1295 [DOI] [PubMed] [Google Scholar]
- 9.Reyes, J. C., Barra, J., Muchardt, C., Camus, A., Babinet, C., and Yaniv, M. (1998) EMBO J. 17 6979-6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strobeck, M. W., Reisman, D. N., Gunawardena, R. W., Betz, B. L., Angus, S. P., Knudsen, K. E., Kowalik, T. F., Weissman, B. E., and Knudsen, E. S. (2002) J. Biol. Chem. 277 4782-4789 [DOI] [PubMed] [Google Scholar]
- 11.Klochendler-Yeivin, A., Muchardt, C., and Yaniv, M. (2002) Curr. Opin. Genet. Dev. 12 73-79 [DOI] [PubMed] [Google Scholar]
- 12.Roberts, C. W., and Orkin, S. H. (2004). Nat. Rev. Cancer. (2), 133-142 [DOI] [PubMed]
- 13.Griffin, C. T., Brennan, J., and Magnuson, T. (2008) Development (Camb.) 135 493-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quarles, L. D., Yohay, D. A., Lever, L. W., Caton, R., and Wenstrup, R. J. (1992) J. Bone Miner. Res. 7 683-692 [DOI] [PubMed] [Google Scholar]
- 15.Franceschi, R. T., Iyer, B. S., and Cui, Y. (1994) J. Bone Miner Res. 9 843-854 [DOI] [PubMed] [Google Scholar]
- 16.Choi, J. Y., Lee, B. H., Song, K. B., Park, L. S., Sohn, K. Y., Jo, J. S., and and Ryoo, H. M. (1996) J. Cell. Biochem. 61 609-618 [DOI] [PubMed] [Google Scholar]
- 17.Stein, G. S., Lian, J. B., Stein, J. L., Van Wijnen, J., and Montecino, M. (1996) Physiol. Rev. 76 593-629 [DOI] [PubMed] [Google Scholar]
- 18.Beck, G. R., Jr., Zerler, B., and Moran, E. (2001) Cell Growth & Differ. 12 61-83 [PubMed] [Google Scholar]
- 19.Brummelkamp, T. R., Bernards, R., and Agami, R. (2002) Science 296 550-553 [DOI] [PubMed] [Google Scholar]
- 20.Wang, F., Zhang, R., Beischlag, T. V., Muchardt, C., Yaniv, M., and Hankinson, O. (2004) J. Biol. Chem. 279 46733-46741 [DOI] [PubMed] [Google Scholar]
- 21.Nagl, N. G., Jr., Zweitzig, D. R., Thimmapaya, B., Beck, G. R., Jr., and Moran, E. (2006) Cancer Res. 66 1289-1293 [DOI] [PubMed] [Google Scholar]
- 22.Beck, G. R., Jr., Sullivan, E. C., Moran, E., and Zerler, B. (1998) J. Cell. Biochem. 68 269-280 [DOI] [PubMed] [Google Scholar]
- 23.Beck, G. R., Jr., Moran, E., and Knecht, N. (2003) Exp. Cell Res. 288 288-300 [DOI] [PubMed] [Google Scholar]
- 24.Yaciuk, P., and Moran, E. (1991) Mol. Cell. Biol. 11 5389-5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, X., Nagl, N. G., Wilsker, D., Van Scoy, M., Pacchione, S., Yaciuk, P., Dallas, P. B., and Moran, E. (2004) Biochem. J. 383 319-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, W., Cote, J., Xue, Y., Zhou, S., Khavari, P. A., Biggar, S. R., Muchardt, C., Kalpana, G. V., Goff, S. P., Yaniv, M., Workman, J. L., and Crabtree, G. R. (1996) EMBO J. 15 5370-5382 [PMC free article] [PubMed] [Google Scholar]
- 27.Villagra, A., Cruzat, F., Carvallo, L., Paredes, R., Olate, J., van Wijnen, A. J., Stein, G. S., Lian, J. B., Stein, J. L., Imbalzano, A. N., and Montecino, M. (2006) J. Biol. Chem. 281 22695-22706 [DOI] [PubMed] [Google Scholar]
- 28.Montecino, M., Stein, J. L., Stein, G. S., Lian, J. B., van Wijnen, A. J., Cruzat, F., Gutierrez, S., Olate, J., Marcellini, S., and Gutierrez, J. L. (2007) Biochem. Cell Biol. 85 419-425 [DOI] [PubMed] [Google Scholar]
- 29.Reyes-Botella, C., Montes, M. J., Vallecillo-Capilla, M. F., Olivares, E. G., and Ruiz, C. (2002) Cell Physiol. Biochem. 12 359-364 [DOI] [PubMed] [Google Scholar]
- 30.Brodeur, M. R., Brissette, L., Falstrault, L., Luangrath, V., and Moreau, R. (2008) J. Bone Miner Res. 23 326-337 [DOI] [PubMed] [Google Scholar]
- 31.Yang, X., and Karsenty, G. (2002) Trends Mol. Med. 8 340-345 [DOI] [PubMed] [Google Scholar]
- 32.Schroeder, T. M., Kahler, R. A., Li, X., and Westendorf, J. J. (2004) J. Biol. Chem. 279 41998-42007 [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. S., Thomas, D. M., Gutierrez, G., Carty, S. A., Yanagawa, S., and Hinds, P. W. (2006) J. Bone Miner Res. 21 921-933 [DOI] [PubMed] [Google Scholar]
- 34.Lee, H. W., Suh, J. H., Kim, A. Y., Lee, Y. S., Park, S. Y., and Kim, J. B. (2006) Mol. Endocrinol. 20 2432-2443 [DOI] [PubMed] [Google Scholar]
- 35.Inoue, H., Furukawa, T., Giannakopoulos, S., Zhou, S., King, D. S., and Tanese, N. (2002) J. Biol. Chem. 277 41674-41685 [DOI] [PubMed] [Google Scholar]
- 36.Nightingale, K. P., Gendreizig, S., White, D. A., Bradbury, C., Hollfelder, F., and Turner, B. M. (2007) J. Biol. Chem. 282 4408-4416 [DOI] [PubMed] [Google Scholar]
- 37.Glaros, S., Cirrincione, G. M., Muchardt, C., Kleer, C. G., Michael, C. W., and Reisman, D. (2007) Oncogene 26 7058-7066 [DOI] [PubMed] [Google Scholar]
- 38.Kadam, S., and Emerson, B. (2003) Mol. Cell 11 377-389 [DOI] [PubMed] [Google Scholar]
- 39.Marshall, T. W., Link, K. A., Petre-Draviam, C. E., and Knudsen, K. E. (2003) Curr. Opin. Genet. Dev. 13 136-14212672490 [Google Scholar]
- 40.Das, A. V., James, J., Bhattacharya, S., Imbalzano, A. N., Antony, M. L., Hegde, G., Zhao, X., Mallya, K., Ahmad, F., Knudsen, E., and Ahmad, I. (2007) J. Biol. Chem. 282 35187-35201. [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto, S., Banine, F., Struve, J., Xing, R., Adams, C., Liu, Y., Metzger, D., Chambon, P., Rao, M. S., and Sherman, L. S. (2006) Dev. Biol. 289 372-383 [DOI] [PubMed] [Google Scholar]
- 42.Gao, X., Tate, P., Hu, P., Tjian, R., Skarnes, W. C., and Wang, Z. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 6656-6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.