Abstract
The Vaccinia virus H1 gene product, VH1, is a dual specificity phosphatase that down-regulates the cellular antiviral response by dephosphorylating STAT1. The crystal structure of VH1, determined at 1.32 Å resolution, reveals a novel dimeric quaternary structure, which exposes two active sites spaced ∼39 Å away from each other. VH1 forms a stable dimer via an extensive domain swap of the N-terminal helix (residues 1-20). In vitro, VH1 can dephosphorylate activated STAT1, in a reaction that is competed by the nuclear transport adapter importin α5. Interestingly, VH1 is inactive with respect to STAT1 bound to DNA, suggesting that the viral phosphatase acts predominantly on the cytoplasmic pool of activated STAT1. We propose that the dimeric quaternary structure of VH1 is essential for specific recognition of activated STAT1, which prevents its nuclear translocation, thus blocking interferon-γ signal transduction and antiviral response.
Dual specificity phosphates (DSPs)2 comprise a growing subclass of protein-tyrosine phosphatases, which dephosphorylate both phosphotyrosine and phosphoserine/threonine residues. The first identified DSP, VH1, is the product of the Vaccinia virus gene H1 (1). To date, the small VH1 (∼20 kDa) is the prototype of a family of VH1-like DSPs found in plants, yeast, insects, and higher eukaryotes (2). The human genome encodes at least 38 VH1-like phosphatases, which regulate many critical aspects of the cell cycle (3). VH1-like DSPs share a common catalytic mechanism, which is mediated by a catalytic triad consisting of a cysteine, an arginine, and an aspartic acid, usually present in the context of an extended consensus motif (4). The structural organization of the minimum catalytic core of VH1-like DSPs is known from the crystal structures of several members of the VH1-like family, such as VHZ (5) and VHR (6). All known DSPs share a common topology with members of the classical protein-tyrosine phosphatases, with the most marked structural difference being in the architecture of the active site. To accommodate both phosphotyrosine and phosphothreonine/serine residues, DSPs present a shallow catalytic cleft only ∼6 Å deep. In contrast, the catalytic cysteine residue of classical protein-tyrosine phosphatases sits at the bottom of a ∼9-Å-deep pocket, which selectively recognizes bulky phosphotyrosines (6). In vitro, VH1 and many other VH1-like DSPs are characterized by resistance to okadaic acid and sensitivity to sodium vanadate (1). Sodium vanadate acts as a potent inhibitor of cysteine-phosphatases by covalently labeling the cysteine group in the active site (4).
The gene encoding VH1 is highly conserved among poxviruses and essential for the viability of Vaccinia virus in tissue cultures (7). VH1 is expressed in the late stage of viral infection, and ∼200 molecules of VH1 are packaged within the virion (7). The conservation of the VH1 gene in poxviruses as well as its essential role for virus viability emphasize VH1 involvement in a critical step of the virus life cycle. Recent evidence indicates that VH1 functions to overcome host defense mechanisms during infection by blocking interferon-γ (IFN-γ) signaling. This function is dependent on the ability of VH1 to specifically dephosphorylate STAT1 (signal transducer and activator of transcription 1) (8, 9).
IFN-γ is a key cytokine involved in protection against viral infection (10). IFN-γ secreted in response to viral infection activates the Jak1 and -2 tyrosine kinase pathway, which leads to phosphorylation and activation of STAT1 (11, 12). In the cytoplasm, phosphorylated STAT1 is recognized by importin α5, which together with importin β mediates its rapid translocation into the cell nucleus (13, 14). Here, activated STAT1 stimulates transcription of target genes, involved in antiviral response. In infected cell lines, Vaccinia virus inhibits IFN-γ-induced phosphorylation and nuclear import of STAT1 in a dose-dependent manner. This inhibition requires viral uncoating and is absent in Vaccinia virus mutants deficient in VH1 (8). Consistent with this observation, in vitro, recombinant VH1 dephosphorylates activated STAT1 and STAT2 but not STAT3 and STAT5 (9).
In contrast to VH1, human cells contain a nuclear VH1-like DSP, VHR, that specifically dephosphorylates activated STAT5 to inactivate its function (15). VHR becomes competent for STAT5 inactivation after phosphorylation at tyrosine 138 by the kinase Tyk2, which also mediates phosphorylation of STAT5 (15). With the exception of VH1 and VHR, the specific cellular substrates for most VH1-related DSPs have not been identified. The molecular basis for the recognition of the substrate and, in turn, the physiological regulation of DSP-mediated dephosphorylation is likely to play a critical role in the progression of cell cycle and in many viral infections. In this paper, we have used a combination of crystallographic, biochemical, and biophysical techniques to characterize the structure and function of the Vaccinia virus dual specificity phosphatase VH1.
EXPERIMENTAL PROCEDURES
Cloning and Protein Expression—The gene encoding VH1 was cloned into the NcoI and BamHI sites of the pET-14b vector (Novagen), which expresses VH1 fused to an N-terminal His6 tag. A PreScission Protease cleavage site was engineered between the His6 tag and the first residue of VH1 (plasmid pET14b-PP-VH1). pET14b-PP-VH1 was expressed in the Escherichia coli BL21(DE3) strain for 16 h at 22 °C. Recombinant His-tagged VH1 was purified by metal chelate affinity chromatography using Qiagen nickel-agarose beads. The His6 tag was cleaved off by incubating with PreScission Protease, followed by gel filtration chromatography on a Superdex 200 column (GE Healthcare) in 150 mm sodium chloride, 20 mm HEPES, pH 7.5, 3 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride. Purified, untagged VH1 was then concentrated to 15 mg ml-1 using a Millipore concentrator (cut-off 10 kDa). The gene encoding full-length STAT1 (FL-STAT1) or STAT1 lacking N-terminal residues 1-132 (Δ132-STAT1) were introduced in an engineered pMal vector containing a PreScission Protease cleavage site (plasmids pMal-PP-STAT1 and pMal-PP-Δ132STAT1). STAT1 constructs were expressed and phosphorylated in E. coli strain TKX-1 (Stratagene, La Jolla, CA), which harbors an inducible plasmid encoding a tyrosine kinase gene (pTK). Expression, in vivo phosphorylation, and purification of FL-STAT1 and Δ132-STAT1 were performed as previously described (16). The plasmid encoding ΔIBB-importin α5 (residues 67-512) was expressed and purified as described (17).
Analytical Ultracentrifugation Experiments—VH1 in 20 mm HEPES, pH 7.5, 150 mm sodium chloride was analyzed in a Beckman XL-A analytical ultracentrifuge under velocity sedimentation mode. 450 μl of sample and 400 μl of reference buffers were loaded into separate compartments of a 12-mm path length Epon centerpiece cell. Runs were performed at 50,000 rpm and 10 °C. Absorbance values were collected at a wavelength of 275 nm using three different protein concentrations (5, 50, and 100 μm). The data were fit to a continuous sedimentation coefficient (c(s)) distribution model, and an estimated molecular mass was obtained with the program SEDFIT (Peter Schuck, National Institutes of Health).
Crystallization and Structure Determination—VH1 was crystallized under 62% polyethylene glycol 400, 100 mm Tris at pH 8.0. Crystals of VH1 were screened at beamline F1 at the Cornell High Energy Synchrotron Source on an ADSC Q-270 CCD detector. Diffraction data were reduced to intensities using the programs of the HKL-2000 package (18) and further analyzed using software of the CCP4 package (19). The best diffracting crystals belong to space group C2221 and diffract X-rays past 1.3 Å resolution. The structure was solved by molecular replacement in MolRep (19) using the structure of Variola virus DSP (Protein Data Bank code 2P4D) (20) as a search model. The initial solution was refined in Refmac using rigid body refinement, followed by several rounds of restrained refinement alternated with manual building. A phosphate ion in the active site and 300 ordered water molecules were modeled in Fo - Fc electron density using the program Coot (21). Protein model and solvent were then further subjected to several cycles of positional and anisotropic B-factor refinement in SHELX-97 (22), using all reflections between 15 and 1.32 Å resolution. The final model has an Rwork and Rfree of 17.1 and 18.5%, respectively, and excellent stereochemistry (Table 1). All structural figures were made using the program PyMol (23).
TABLE 1.
X-ray data collection and refinement statistics for the VH1 structure
The numbers in parenthesis refer to the statistics for the outer resolution shell (1.37-1.32 Å).
| Parameter | Value |
|---|---|
| Data collection statistics | |
| Wavelength (Å) | 0.918 |
| Space group | C2221 |
| Unit cell dimensions (Å) | a = 63.82, b = 38.69, c = 134.99 |
| Angles (degrees) | α = β = γ = 90 |
| Resolution range (Å) | 20-1.32 |
| B value from Wilson plot (Å2) | 19.3 |
| Total observations | 1,982,346 |
| Unique observations | 35,905 |
| Completeness (%) | 90.9 (51.4) |
| Rsyma (%) | 5.7 (35.7) |
| <I>/<σ(I)> | 65.6 (2.5) |
| Refinement statistics | |
| No. of reflections (15-1.32Å) | 33,998 (1,250) |
| Rwork/Rfreeb (%) | 17.1/18.5 (36.2/38.4) |
| No. of water molecules | 300 |
| B value of model (Å2) | 26.4 |
| r.m.s. deviation from ideal bond length (Å) | 0.008 |
| r.m.s. deviation from ideal bond angles (degrees) | 1.186 |
| Ramachandran plot (%) | |
| Core region | 90 |
| Allowed region | 10 |
| Generously allowed region | 0 |
| Disallowed region | 0 |
Rsym = ∑i,h|I(i,h) - <I(h)>|/∑i,h|I(i,h)|, where I(i,h) and <I(h)> are the ith and mean measurement of intensity of reflection h.
The Rfree value was calculated using 5% of the data.
Circular Dichroism Analysis—CD spectra were recorded using an AVIV 62A DS spectropolarimeter equipped with a Neslab CFT-33 refrigerated recirculator. A rectangular quartz cuvette with a path length of 1 cm was used to perform the CD measurements of VH1 at a final protein concentration of 6.0 μm in 20 mm sodium phosphate (pH 8.0) and 100 mm NaCl. To measure the temperature-induced unfolding of dimeric VH1, we recorded variations in ellipticity at 218 nm as a function of temperature in 1 °C increments. Samples were equilibrated for 60 s after each 1 °C increment, and CD spectra were recorded with an integration time of 15 s. Reversibility of unfolding was checked by slowly cooling unfolded VH1 to 25 °C followed by a second scan. Under these conditions, VH1 unfolding was completely irreversible.
In Vitro Dephosphorylation Assay—The VH1 phosphatase assay was performed with phosphorylated FL-STAT1 (residues 1-712) and Δ132-STAT1 (residues 133-712) and repeated in the presence of double-stranded DNA. Each reaction contained 50 μg of STAT1 and a 2-fold molar excess of VH1 in a 50-μl reaction. For the DNA-bound STAT1 phosphatase reactions, we incubated STAT1 on ice for 10 min with a 2-fold molar excess of a 38-mer double-stranded DNA oligonucleotide containing two tandem cfosM67 promoter elements spaced by 10 nucleotides (5′-ACGGTTTCCCGTAAATTGACGGATTTCCCGTAAATGGC-3′). VH1 was added to the reaction at 37 °C, and time points were taken at 0, 30, 60, 120, and 240 min. As a negative control, we used catalytically inactive VH1 (C110S). VH1-mediated dephosphorylation was measured by Western blot analysis using the horseradish peroxidase-conjugated anti-phosphotyrosine (PY20) antibody (Exalpha Biologics). The dephosphorylation assay in the presence of importin α5 was performed by incubating 50 μg of STAT1 with increasing quantities of importin α5 in a relative importin α5/monomeric STAT1 molar ratio of 0.125:1, 0.25:1, 0.5:1, 1:1, and 2:1 for 10 min. A 10-fold molar excess of VH1 was added, and the reaction was incubated for 240 min at 37 °C, followed by SDS-PAGE and Western blot analysis as described above. All dephosphorylation reactions were repeated a minimum of three times. The relative amount of phosphorylated versus dephosphorylated STAT1 was quantified using the NIH ImageJ software and the relative intensities of Tyr(P)701 were plotted using the program SigmaPlot.
RESULTS
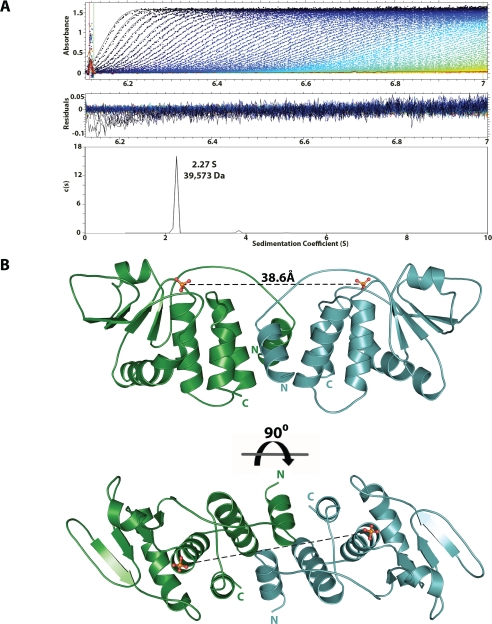
VH1 Forms a Stable Dimer in Solution—VH1 was cloned, expressed, and purified as described under “Experimental Procedures.” After proteolytically removing the His tag, the oligomeric state of the phosphatase was investigated by sedimentation velocity analysis. Fig. 1A shows a typical sedimentation profile of VH1, obtained in 20 mm HEPES, pH 7.5, and 0.15 m sodium chloride at 10 °C. The sedimentation boundary exhibits monophasic behavior, which is indicative of a single major (>99.9%) component in solution, migrating with a sedimentation coefficient of 2.27 S (s20,w = 3.079 S). Conversion of the distribution of the apparent sedimentation coefficient to molecular mass for three independent runs revealed a value of ∼39,810 ± 1,747 Da. This value agrees well with a dimer of VH1, which has an expected molecular mass of ∼40,501 Da. In addition, VH1 dimerization was concentration-independent under the range of concentrations tested (5-100 μm), suggesting a low dimerization constant. Notably, the addition of 10% DMSO prior to sedimentation analysis was found to break the VH1 dimer into a homogeneous monomer, which migrated as a ∼20-kDa species (data not shown).
FIGURE 1.
The Vaccinia virus dual specificity phosphatase VH1 forms a dimer. A, sedimentation velocity profile of VH1 measured in 0.15 m sodium chloride at 10 °C. Top, raw absorbance at 275 nm plotted as a function of the radial position. Data at intervals of 20 min are shown as dots for sedimentation at 50,000 rpm. The monophasic sedimentation boundaries suggest that VH1 exists in a single species of homogeneous oligomeric state. Middle, the residuals between fitted curve and raw data. Bottom, the fitted distribution of the sedimentation coefficient calculated for VH1 (2.27 S and s20,w = 3.079 S) corresponds to an estimated molecular mass of ∼39,573 Da. Given the predicted size of VH1 (∼20 kDa), this indicates that in solution, VH1 exists as a dimer. B, ribbon diagram of the dimeric structure of catalytically inactive VH1 determined at 1.32 Å resolution. The distance between the two active sites in the dimer is ∼39 Å.
Crystal Structure of VH1 at 1.32 Å Resolution—To shed light on the three-dimensional structure of VH1, we crystallized the phosphatase under concentrated solutions of polyethylene glycol 400. Aiming at high resolution diffraction studies, we used a catalytically inactive mutant of VH1, where the cysteine 110 in the active site is replaced by a serine (C110S) (8). This mutant is more stable in solution than the wild-type VH1 and thus more prone to crystallization. Large crystals of VH1(C110S) were obtained using the His6-tagged protein but yielded low resolution diffraction data. After removing the N-terminal His tag with PreScission Protease, untagged VH1(C110S) was subjected to a high throughput crystallization screen at the Hauptman-Woodward Medical Research Institute (24). This screening identified several crystallization conditions, which were subsequently refined in house. The best diffracting crystals were obtained under 62-82% polyethylene glycol 400, at pH 7-8, and typically diffracted past 1.3 Å resolution. VH1 crystals have a centered orthorhombic unit cell (Table 1) containing one monomer of VH1 in the asymmetric unit and ∼40% solvent content. The structure of VH1 was determined by molecular replacement and refined to an Rfactor and Rfree of 17.1 and 18.5%, respectively, including reflections between 15 and 1.32 Å resolution (Fig. 1B). The final model also includes 300 water molecules and has an overall B-factor of ∼26 Å2. In the crystal structure, a dimer of VH1 is built by 2-fold crystallographic symmetry. The structure adopts a slightly elongated shape of ∼85 Å in length, ∼31 Å in height, and ∼28 Å in width. The quaternary structure of VH1 superimposes well (r.m.s. deviation ∼1.46 Å) to the closely related Variola virus DSP, which was recently determined at 1.8 Å resolution (Protein Data Bank code 2P4D) (20). Like VH1, the Variola virus DSP crystallized as a homodimer. Both in Vaccinia and Variola DSP structures, the dimerization interface is rigid, which renders the phosphatase dimer shaped as a thick brick (Fig. 1B). VH1 accounts for ∼55% folded secondary structure elements, with ∼45% α-helices and 10% β-strands. In the 1.32 Å crystal structure of VH1, all residues are clearly visible, with the exception of the initial 4 and the last 3 amino acids, for which the electron density is weak.
A DSP-active Site at 1.32 Å Resolution—The high resolution structure of VH1 presented in this paper, the highest ever obtained for a DSP, gives us a unique detailed view of the enzyme active site. At 1.32 Å resolution, anisotropic refinement of the thermal motion allows accurate determination of the atomic position and displacement of individual atoms. The first interesting feature of dimeric VH1 lies in the position of the active sites. In a dimer of VH1, the two active sites are spaced ∼39 Å from each other and slightly offset with respect to an axis running along the dimer and perpendicular to the dimerization interface (Figs. 1B and 2A). The active site is formed by a shallow pocket ∼6 Å in diameter. The catalytic triad consists of Arg116, Asp79, and the Cys110, which, in the catalytically inactive mutant, is replaced by a serine. The catalytic triad in VH1 is superimposable to that seen in the Variola homologue (r.m.s. deviation for Arg116, Asp79, and the Cys110 is 0.88, 1.10, and 0.81 Å, respectively). However, the residue immediately adjacent to the catalytic Cys110, Ala111, is replaced by a valine in the Variola DSP. Ala111 in Vaccinia and Val111 in Variola superimpose poorly, compared with the rest of the two structures (r.m.s. deviation ∼1.90 Å); in Variola Ala111 is translated ∼2 Å away from the active site as compared with VH1.
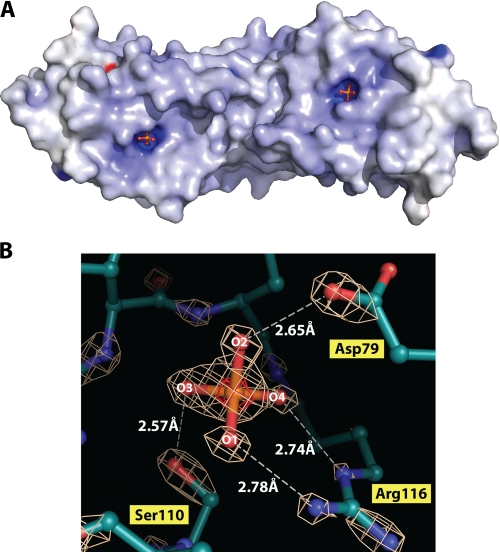
FIGURE 2.
Structural view of the VH1 active site at 1.32 Å resolution. A, electrostatic potential surface diagram of dimeric VH1. The two active sites form ∼6-Å-deep pockets on the surface of the dimer completely exposed to the solvent. B, enlargement of VH1 active site visualized crystallographically at 1.32 Å resolution. The final model, refined to an Rfree of 18.5% is superimposed to the final 2Fo - Fc electron density map contoured at 5.0σ above background (white). The anomalous signal of phosphate ion trapped in the active site is shown in red and is contoured at 6σ above background.
In the crystal structure of VH1, the active site is occupied by a phosphate ion, which is visible as an ∼8σ peak in an electron density difference map calculated with coefficient Fo - Fc (Fig. 2B). The presence of a phosphate ion can be determined unambiguously at this resolution based on the atomic charge of phosphate, which is slightly smaller than sulfate. The atomic charge calculated for a phosphate via the Mulliken population analysis is ∼1.32 versus 2.0 for sulfate (expressed in units of absolute electron charge) (25). When a sulfate ion is placed in the active site and refined against the crystallographic data, an Fo - Fc electron density difference map computed with phases calculated from the refined model (contoured at 3σ above noise) has a clear peak of negative density surrounding the sulfate (data not shown). This negative density is not observed when a phosphate ion is placed in the active site and refined to 1.32 Å resolution, which suggests that phosphate, and not sulfate, is trapped in VH1 active sites. Likewise, the B-factor of the bound phosphate ion is significantly lower than that of average protein atoms in the structure (∼14 Å2 versus ∼26 Å2), which emphasizes the avidity of the active site for this ion. As reported for other DSP, the invariant Arg116 functions in substrate binding and in transition state stabilization, whereas the conserved Asp79 acts as a general acid catalyst protonating the leaving group (4). The guanidinium group of Arg116 orients two phosphate oxygens, indicated as O1 and O4 in Fig. 2B. The O1 is positioned 2.78 Å away from the positively charged guanidium group of Arg116. At higher contour (5σ), the density around O1 is perfectly spherical and distinct from the phosphorous density, which is indicative of an ionized state. At a nearly identical distance of 2.74 Å, the oxygen at position O4 contacts the ε-nitrogen of Arg116. At 1.32 Å resolution, the electron density for O4 is merged with that of the phosphorous atom, suggesting partial orbital overlap between the two atoms. Shorter distances are observed for the other two oxygen atoms. O2 is positioned 2.65 Å away from the general acid catalyst Asp79, whereas the catalytically inactive Ser110 engages in a strong hydrogen bond with O3, positioned only 2.57 Å away. This shorter distance supports the idea that in the catalytically active DSP, a H+ spontaneously transfers under physiological conditions from the catalytic Cys110 to the O3, yielding a thiolate intermediate. In the structure of the catalytic inactive VH1, the hydroxyl group of Ser110 hydrogen-bonds O3, and the electron density between P and O3 is continuous and thick even at 5σ, indicating the presence of a double bond between the two atoms.
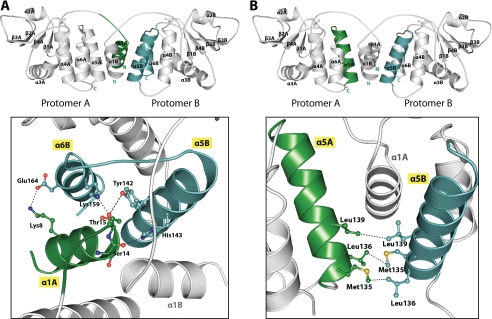
The Dimerization Interface—The dimeric quaternary structure of VH1 is kept in place by an extended domain swap of the N-terminal helix α1 (residues 1-20). In a dimer of VH1, the two N-terminal swapped helices α1A and α1B cross each other at an angle of ∼95°. The molecular arrangement of VH1 swapped helices is very similar to that seen in the structure of the Variola DSP (20), which supports the idea of an evolutionary conservation of the dimerization interface. In VH1, two networks of interactions, for a total of at least 15 specific side chain contacts, stabilize the domain-swapped dimerization interface (Fig. 3). The first set of contacts is generated between the N-terminal swapped α1 of one protomer and the C-terminal helices α5-α6 of the other protomer (Fig. 3A). Here, two sets of residues protruding on the surface of helices α1A and α1B, Lys8/Ser14/Thr15 and Tyr9/Leu13/Leu14, engage in extensive electrostatic and hydrophobic contacts, respectively, with the other VH1 protomer. Notably, the hydroxyl groups of Ser14 and Thr15 are positioned only 2.79 and 2.77 Å away from the ε-nitrogen atoms of His143 and Lys159, respectively, in a position ideal to engage in highly specific and energetic hydrogen bonds. A close salt bridge (3.15 Å) is also seen between Lys8 and Glu164 (Fig. 3A). Given the 2-fold symmetric structure of dimeric VH1, the contacts made by the swapped helices α1A and α1B are identical and mirror images. The second set of contacts stabilizing the VH1 dimerization interface is observed between the C-terminal helix α5A and α5B of two adjacent VH1 protomers (Fig. 3B). This includes three reciprocal hydrophobic contacts between residues Met135, Leu136, and Leu139. Overall, the presence of several hydrophobic contacts at the VH1 dimerization interface explains the observation that 10% DMSO selectively disrupts dimeric VH1 (data not shown).
FIGURE 3.
Structural view of the two binding determinants stabilizing the VH1 dimerization interface. A, six residues on the surface of helix α1, Lys8/Ser14/Thr15 and Tyr9/Leu13/Leu14 engage in extensive electrostatic and hydrophobic contacts, respectively, with the other VH1 protomer. Only the electrostatic contacts made by Lys8/Ser14/Thr15 are shown in a as dashed black lines. B, the interface involving α5 helices of both protomers consists of three hydrophobic contacts that spans beneath the two N-terminal α1 helices of the VH1 dimer.
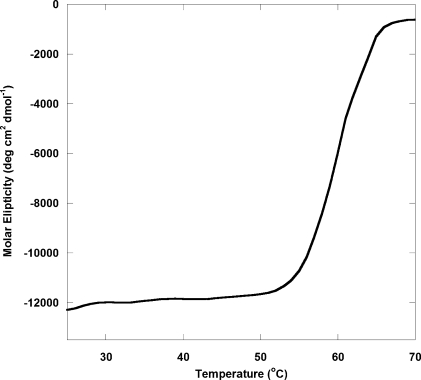
The structural role of the VH1 dimerization interface is also emphasized by the way the protein unfolds in solution. Temperature-induced equilibrium unfolding, recorded by monitoring variation in ellipticity at 218 nm, revealed a steep unfolding transition with an apparent temperature of melting (Tm) of ∼60 °C (Fig. 4). The unfolding transition observed for VH1 from a fully folded dimer to an unfolded monomer indicates high cooperativity. The concentration independence of this unfolding transition (at least in the range of concentrations tested) supports the idea that dimerization of VH1 is a physiological property of this viral phosphatase.
FIGURE 4.
Cooperative thermal denaturation of VH1. Stability of VH1 against thermal denaturation monitored by measuring changes in the ellipticity intensity at 218 nm as a function of temperature. The VH1 concentration used in this experiment was 6 μm. The apparent Tm was ∼60 °C.
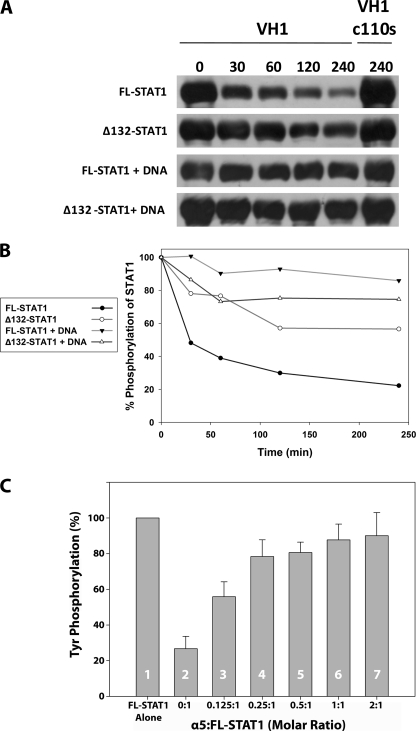
VH1 Dephosphorylates Activated STAT1 in the Absence of DNA—Thus far, only two virion membrane-associated factors, A17 (26) and A14 (27), of Vaccinia virus have been shown to be bona fide substrates of VH1, in vivo. In addition, mounting evidence has shown that activated STAT1 immunoprecipitated from NIH 3T3 cells stimulated with IFN-γ is dephosphorylated by VH1 in vitro (8). To further characterize the activity of VH1, we sought to study the dephosphorylation of STAT1 using purified phosphorylated STAT1. To achieve this, we expressed and purified activated STAT1 using the methodology previously reported by Becker et al. (28). In this bacterial expression and phosphorylation system, STAT1 was coexpressed with the catalytic domain of the Elk receptor tyrosine kinase, which selectively phosphorylates Tyr(P)701, thus activating STAT1. In a time course of dephosphorylation, ∼80% of activated full-length STAT1 (residues 1-712) was dephosphorylated by VH1 over 240 min (Fig. 5, A and B). Tyrosine dephosphorylation at position 701 was determined by anti-Tyr(P) Western blotting. The phosphatase activity was specifically dependent on VH1 and not due to background phosphatases, since the catalytic inactive VH1(C110S) showed no appreciable STAT1 dephosphorylation, even after prolonged incubation (Fig. 5, A and B). We then investigated if VH1 was active with respect to activated Δ132-STAT1, which lacks the N-terminal dimerization domain (residues 1-132) (29). This construct of STAT1 is locked in a dimeric conformation that remains persistently Tyr-phosphorylated in cells after IFN-γ treatment (30, 31). Purified dimeric VH1 displayed decreased efficiency for phosphorylated Δ132-STAT1, yielding ∼50% of dephosphorylation after 240 min (Fig. 5, A and B).
FIGURE 5.
Activated STAT1 is dephosphorylated by VH1 in vitro. A, time course of STAT1 dephosphorylation in the presence of purified VH1. The efficiency of dephosphorylation was monitored by anti-Tyr(P) (anti-pTyr) Western blotting. Full-length STAT1 and Δ132-STAT1 were incubated with a 2-fold molar excess of either active VH1 or catalytically inactive VH1(C110S) for 0-240 min. B, quantification of band intensities from A. C, VH1-mediated dephosphorylation of full-length STAT1 in the presence of increasing amounts of importin α5. Error bars indicate S.D. for three independent dephosphorylation reactions.
To determine if VH1 was active with respect of full-length activated STAT1 bound to DNA, we incubated phosphorylated STAT1 with a 38-mer oligonucleotide containing two tandem cfosM67 promoter elements spaced by 10 nucleotides (16, 32). This mutated version of the cfos promoter is known to cooperatively bind two STAT1 dimers, interacting with high affinity via the N-terminal domains (33). Interestingly, VH1 failed to dephosphorylate DNA-bound STAT1 even after 240 min of incubation (Fig. 5, A and B). Likewise, no dephosphorylation was seen using Δ132-STAT1 bound to the same DNA (Fig. 5, A and B). In a control experiment where VH1 was incubated with phosphorylated STAT1 in the presence of a nonspecific double-stranded DNA oligonucleotide, no significant inhibition of VH1 activity was observed (data not shown), ruling out the possibility that VH1 is directly inhibited by DNA. Therefore, dimeric VH1 can dephosphorylate activated STAT1 but is inactive with respect to the DNA-bound state of the transcription factor.
Importin α5 Protects STAT1 from VH1-mediated Dephosphorylation—Activated STAT1 is imported into the nucleus by a heterodimer of importin α5 bound to importin β (13, 14). The binding of STAT1 to importin α5 requires tyrosine phosphorylation at position 701, which is a specific substrate for VH1. Thus, we asked whether importin α5 can protect activated STAT1 from VH1-mediated dephosphorylation. To answer this question, we repeated the dephosphorylation assay in the presence of increasing concentration of importin α5. Under conditions that yield ∼75% dephosphorylation of activated STAT1 (Fig. 5C, 2), the addition of substoichiometric quantities of importin α5 gave ∼45% dephosphorylation (Fig. 5C, 3). ∼80% protection from VH1-mediated dephosphorylation was observed in the presence of 0.25-0.5 eq of importin α5/monomer of STAT1 (Fig. 5C, 4 and 5). The addition of equal or 2-fold excess of importin α5 over STAT1 increased the protection from VH1 up to 90% (Fig. 5C, 6 and 7). Thus, importin α5 protects STAT1 from VH1-mediated dephosphorylation in a dose-dependent manner. Interestingly, importin α5 does not bind a peptide of STAT1 spanning residues 695-708 and containing Tyr(P)701 (data not shown), suggesting that the transport adaptor does not recognize the Tyr(P)701 directly. Therefore, the decreased susceptibility to VH1 in the presence of importin α5 must be caused by the structural constraints imposed by importin α5 binding to STAT1 more than a direct competition for the Tyr(P)701.
DISCUSSION
It has been 18 years since the identification of the first DSP, VH1 (1). Although this protein is the prototype of the VH1-like family of DSPs, its structural characterization has remained elusive. In this paper, we have determined the high resolution crystal structure of VH1 to 1.32 Å resolution. We demonstrate that, both in solution and in crystal, VH1 adopts a dimeric quaternary structure mediated by an N-terminal domain swap. A dimer of VH1 exposes two active sites spaced ∼39 Å away from each other on the surface of the phosphatase. Consistent with this dimeric quaternary structure, VH1 unfolds following a highly cooperative transition, characterized by an apparent Tm of ∼60 °C.
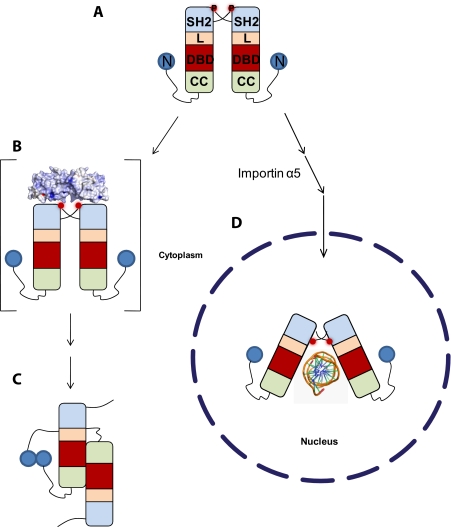
Role of Vaccinia Virus VH1 in STAT1 Inactivation—One of the most striking features of the Vaccinia virus VH1 is its ability to specifically reduce the expression of interferon-γ genes by selectively dephosphorylating STAT1 (8, 9). In this work, we demonstrate that dephosphorylation of activated STAT1 occurs only when STAT1 is not bound to its DNA recognition site, cfosM67. This suggests that VH1 acts on the pool of cytoplasmic STAT1, concomitantly or probably immediately after activation. What is the conformation of STAT1 recognized by VH1? As pointed out by Wenta et al. (33), upon phosphorylation, antiparallel STAT1 adopts a parallel dimeric conformation in equilibrium with tetramers (33). Dimerization of STAT1 is solely mediated by reciprocal interactions of the Tyr(P)701 residues with the SH2 domains (Fig. 6A). Our data indicate that this conformation of activated STAT1 is the likely substrate for dimeric VH1. The observation that importin α5 protects STAT1 from VH1-mediated dephosphorylation supports the hypothesis that VH1 recognizes the cytoplasmic pool of activated STAT1, prior to its nuclear import (Fig. 6B). This points to a possible competition between the host nuclear transport machinery and VH1, which is associated with the Vaccinia virion and released into the host cytoplasm upon infection. Although the structure of activated STAT1 in the absence of DNA is not known, we hypothesize that this conformation is more accessible, exposing the Tyr(P)s to VH1 (Fig. 6B). After VH1-mediated dephosphorylation, inactivated STAT1 reorganizes its structure to adopt an antiparallel conformation mediated by its N-terminal domains (Fig. 6C) (33).
FIGURE 6.
Model for VH1-mediated dephosphorylation of activated STAT1. A, activated STAT1 adopts a parallel conformation stabilized by interactions of the Tyr(P)701 with the SH2 domains (33). L, linker; DBD, DNA-binding domain; CC, coiled-coil domain; N, N-terminal domain; P, phosphate on Tyr701. B, dimeric VH1 is active with respect to this conformation of activated STAT1, which we hypothesize exists in the cytoplasm. C, upon dephosphorylation, STAT1 adopts a dimeric antiparallel conformation stabilized by reciprocal interactions of the N-terminal domains (33). D, as an alternative route, activated STAT1 can be imported into the cell nucleus by a heterodimer of importin α5 and importin β, where it binds to specific promoter sequences. As demonstrated in this paper, the structure of STAT1 bound to DNA is not accessible by VH1, probably due to the poor accessibility of the Tyr(P)s buried against the SH2 domains (29).
As an alternative scenario, activated STAT1 that is recognized by a heterodimer of importin α5/β enters the cell nucleus, where it readily localizes to specific DNA sequences (Fig. 6D). The crystal structure of Δ132-STAT1 bound to DNA reveals that the Tyr(P)701 residues are swapped between two STAT1 protomers and lie ∼40 Å away from each other, intimately buried against the SH2 domains (29). Although the distance between the two Tyr(P)s in this conformation of STAT1 is consistent with the distance between active sites in dimeric VH1, this conformation of STAT1 is not accessible to the phosphatase in vitro (Fig. 6D). In contrast, in the cell nucleus, TC45, the nuclear isoform of the ubiquitously expressed T-cell protein-tyrosine phosphatase, specifically dephosphorylates activated STAT1 to promote its recycling to the cytoplasm (34). Although Vaccinia virus VH1 and TC45 inactivate the STAT1 pathway by dephosphorylating STAT1, there are dramatic differences between these two phosphatases. First, TC45 is an endogenous phosphatase, whereas VH1 is brought into the cell upon Vaccinia virus infection. Second, TC45 dephosphorylates nuclear STAT1 bound to DNA (30), whereas, as demonstrated in this paper, VH1 is completely inactive with respect to DNA-bound STAT1. Third, the tyrosine phosphatase TC45 requires an intact N-terminal domain of STAT1 (residues 1-132) for proper catalytic activity. Deletion of this domain leads to persistent phosphorylation of STAT1 (30). In contrast, VH1 is active, albeit at a reduced rate, with respect to STAT1 lacking the N-terminal domain (Fig. 5, A and B). In conclusion, dimeric VH1 emerges as a distinct virally encoded phosphatase unique in structure, activity, and specificity, which Vaccinia virus and other members of the Poxviridae family (9) have developed to selectively block interferon-γ production and thus prevent host antiviral response.
Acknowledgments
We thank Dr. John Lewis (SUNY Downstate Medical Center), Dr. Uwe Vinkemeier (University of Nottingham, United Kingdom), and Dr. Darren Hart (EMBL-Grenoble, France) for donating the clones for VH1, STAT1, and importin α5, respectively. We are also grateful to Dr. Anamika Patel and Dr. Michael Cosgrove (Syracuse University) for technical assistance with the analytical ultracentrifugation and to Dr. Uwe Vinkemeier (University of Nottingham) for the gift of the 38-mer oligonucleotide containing two tandem cfosM67 elements. We thank Vivian Stojanoff and the staff at National Synchrotron Light Source beamline X6A as well as the macCHESS staff for assistance in data collection.
The atomic coordinates and structure factors (code 3CM3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant GM074846.
Footnotes
The abbreviations used are: DSP, dual specificity phosphatase; IFN-γ, interferon-γ; r.m.s., root mean square; SH2, Src homology 2.
References
- 1.Guan, K. L., Broyles, S. S., and Dixon, J. E. (1991) Nature 350 359-362 [DOI] [PubMed] [Google Scholar]
- 2.Fauman, E. B., and Saper, M. A. (1996) Trends Biochem. Sci. 21 413-417 [DOI] [PubMed] [Google Scholar]
- 3.Ducruet, A. P., Vogt, A., Wipf, P., and Lazo, J. S. (2005) Annu. Rev. Pharmacol. Toxicol. 45 725-750 [DOI] [PubMed] [Google Scholar]
- 4.Jackson, M. D., and Denu, J. M. (2001) Chem. Rev. 101 2313-2340 [DOI] [PubMed] [Google Scholar]
- 5.Agarwal, R., Burley, S. K., and Swaminathan, S. (2008) J. Biol. Chem. 283 8946-8953 [DOI] [PubMed] [Google Scholar]
- 6.Yuvaniyama, J., Denu, J. M., Dixon, J. E., and Saper, M. A. (1996) Science 272 1328-1331 [DOI] [PubMed] [Google Scholar]
- 7.Liu, K., Lemon, B., and Traktman, P. (1995) J. Virol. 69 7823-7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Najarro, P., Traktman, P., and Lewis, J. A. (2001) J. Virol. 75 3185-3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mann, B. A., Huang, J. H., Li, P., Chang, H. C., Slee, R. B., O'Sullivan, A., Anita, M., Yeh, N., Klemsz, M. J., Brutkiewicz, R. R., Blum, J. S., and Kaplan, M. H. (2008) J. Interferon Cytokine Res. 28 367-380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krause, C. D., and Pestka, S. (2007) Cytokine Growth Factor Rev. 18 473-482 [DOI] [PubMed] [Google Scholar]
- 11.Levy, D. E., and Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3 651-662 [DOI] [PubMed] [Google Scholar]
- 12.Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H., and Schreiber, R. D. (1998) Annu. Rev. Biochem. 67 227-264 [DOI] [PubMed] [Google Scholar]
- 13.Meyer, T., and Vinkemeier, U. (2004) Eur. J. Biochem. 271 4606-4612 [DOI] [PubMed] [Google Scholar]
- 14.Vinkemeier, U. (2004) J. Cell Biol. 167 197-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyt, R., Zhu, W., Cerignoli, F., Alonso, A., Mustelin, T., and David, M. (2007) J. Immunol. 179 3402-3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinkemeier, U., Cohen, S. L., Moarefi, I., Chait, B. T., Kuriyan, J., and Darnell, J. E., Jr. (1996) EMBO J. 15 5616-5626 [PMC free article] [PubMed] [Google Scholar]
- 17.Tarendeau, F., Boudet, J., Guilligay, D., Mas, P. J., Bougault, C. M., Boulo, S., Baudin, F., Ruigrok, R. W., Daigle, N., Ellenberg, J., Cusack, S., Simorre, J. P., and Hart, D. J. (2007) Nat. Struct. Mol. Biol. 14 229-233 [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 19.(1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-763 [DOI] [PubMed] [Google Scholar]
- 20.Phan, J., Tropea, J. E., and Waugh, D. S. (2007) Acta Crystallogr. Sect. D Biol. Crystallogr. 63 698-704 [DOI] [PubMed] [Google Scholar]
- 21.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 22.Sheldrick, G. M., and Schneider, T. R. (1997) Methods Enzymol. 277 319-343 [PubMed] [Google Scholar]
- 23.DeLano, W. L. (2002) PyMol, DeLano Scientific LLC, Palo Alto, CA
- 24.Luft, J. R., Collins, R. J., Fehrman, N. A., Lauricella, A. M., Veatch, C. K., and DeTitta, G. T. (2003) J. Struct. Biol. 142 170-179 [DOI] [PubMed] [Google Scholar]
- 25.Verlinde, C. L., Noble, M. E., Kalk, K. H., Groendijk, H., Wierenga, R. K., and Hol, W. G. (1991) Eur. J. Biochem. 198 53-57 [DOI] [PubMed] [Google Scholar]
- 26.Derrien, M., Punjabi, A., Khanna, M., Grubisha, O., and Traktman, P. (1999) J. Virol. 73 7287-7296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer, J., and Traktman, P. (2003) J. Virol. 77 8857-8871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker, S., Corthals, G. L., Aebersold, R., Groner, B., and Muller, C. W. (1998) FEBS Lett. 441 141-147 [DOI] [PubMed] [Google Scholar]
- 29.Chen, X., Vinkemeier, U., Zhao, Y., Jeruzalmi, D., Darnell, J. E., Jr., and Kuriyan, J. (1998) Cell 93 827-839 [DOI] [PubMed] [Google Scholar]
- 30.Mertens, C., Zhong, M., Krishnaraj, R., Zou, W., Chen, X., and Darnell, J. E., Jr. (2006) Genes Dev. 20 3372-3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong, M., Henriksen, M. A., Takeuchi, K., Schaefer, O., Liu, B., ten Hoeve, J., Ren, Z., Mao, X., Chen, X., Shuai, K., and Darnell, J. E., Jr. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3966-3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner, B. J., Hayes, T. E., Hoban, C. J., and Cochran, B. H. (1990) EMBO J. 9 4477-4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenta, N., Strauss, H., Meyer, S., and Vinkemeier, U. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 9238-9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ten Hoeve, J., de Jesus Ibarra-Sanchez, M., Fu, Y., Zhu, W., Tremblay, M., David, M., and Shuai, K. (2002) Mol. Cell. Biol. 22 5662-5668 [DOI] [PMC free article] [PubMed] [Google Scholar]