Abstract
Presenilin 1 and 2 (PS) are critical components of the γ-secretase complex that cleaves type I transmembrane proteins within their transmembrane domains. This process leads to release of proteolytically processed products from cellular membranes and plays an essential role in signal transduction or vital functions as cell adhesion. Here we studied the function of presenilins in cell-matrix interaction of wild-type and PS knock-out mouse embryonic fibroblasts. We found for PS1-/- cells an altered morphology with significantly reduced sizes of focal adhesion sites compared with wild type. Cell force analyses on micropatterned elastomer films revealed PS1-/- cell forces to be reduced by 50%. Pharmacological inhibition confirmed this function of γ-secretase in adhesion site and cell force formation. On the regulatory level, PS1 deficiency was associated with strongly decreased phosphotyrosine levels of focal adhesion site-specific proteins. The reduced tyrosine phosphorylation was caused by a down-regulation of c-Src kinase activity primarily at the level of c-Src transcription. The direct regulatory connection between PS1 and c-Src could be identified with ephrinB2 as PS1 target protein. Overexpression of ephrinB2 cytoplasmic domain resulted in its nuclear translocation with increased levels of c-Src and a full complementation of the PS1-/- adhesion and phosphorylation phenotype. Cleavage of full-length EB2 and subsequent intracellular domain translocation depended on PS1 as these processes were only found in WT cells. Therefore, we conclude that γ-secretase is vital for controlling cell adhesion and force formation by transcriptional regulation of c-Src via ephrinB2 cleavage.
PS12 and PS2 are aspartyl proteases forming the active components of the γ-secretase complex. This complex cleaves type 1 transmembrane proteins within their transmembrane domains (1-3). A prerequisite for γ-secretase cleavage is shedding of the ectodomains of the respective transmembrane proteins close to their transmembrane domains (4, 5). Subsequently, cleavage of the remaining transmembrane protein stubs by γ-secretase leads to the release of the two cleavage products from the membrane (2). The freed products of some proteins play important roles in different signaling pathways e.g. in cell-cell adhesion or cell differentiation (1, 2, 6). γ-Secretase also cleaves the amyloid precursor protein, thereby producing the neurotoxic amyloid β-peptide that precipitates in amyloid plaques in Alzheimer disease (1, 7). Another example can be found in notch signaling, which is essential for embryonal development. PS-dependent cleavage of notch leads to release of the so called notch intracellular domain. This domain translocates to the nucleus where it acts as a transcriptional coactivator (2). In vivo studies revealed strong phenotypes for PS1-/- and even more for PS1-/-PS2-/- double knock-out mice that closely resemble notch mutant mice, whereas PS2-/- mice are barely affected (8). These observations argue for the ability of PS1 to complement for PS2 deficiency but not the other way around. Because of the wide range of substrates, many additional mechanisms are affected by presenilin deficiency (2, 5, 9-11). One is the ephrinB/Eph receptor mediated cell-cell adhesion. Because ephrinB1 and ephrinB2 are targets of PS1 and additionally bind to cellular sarcoma protein kinase (c-Src), a protein vitally involved in focal adhesion (FA) formation, it is speculated that PS might be an important regulator for cell-cell interaction as well as for switching cell function from a more sessile to a more dynamic, moving phenotype. Such morphological change would subsequently go along with the formation of cell-matrix interactions (5, 9, 12-14).
Cell adhesion to an extracellular matrix defines a very important process in tissue formation, cell survival, embryonal development, and migration processes. Cells adhere to the extracellular matrix by forming complex structures called focal adhesion sites or FAs. At focal adhesion sites the extracellular matrix surrounding the cell is coupled to the actin cytoskeleton inside the cell (15). In detail, FAs consist of a plaque of membrane-spanning integrin heterodimers connected at their cytoplasmic domains to a wide variety of proteins which eventually form a connection to the cytoskeleton. The integrin extracellular domains bind to the extracellular matrix (16). Because of this strong coupling between the inner actin cytoskeleton and the outer extracellular matrix, forces generated by the actin-myosin machinery are transmitted at focal adhesions (17, 18).
FAs as well as the attached actin cytoskeleton are highly dynamic structures able to grow or shrink upon internal or external force application. This regulation makes FAs a center of integration of many signaling transduction pathways, playing roles in adhesion, mechanosensing, and migration or in proliferation, apoptosis, and differentiation (19-22). Moreover, they are themselves places of intense regulatory events. One of the central regulatory proteins controlling FA formation and adhesion is the cellular sarcoma protein kinase (c-Src, also known as pp60Src or just Src). Upon maturation of young focal adhesion complexes to focal adhesion sites, c-Src becomes activated by processes triggering its autophosphorylation at tyrosine 418. These activating processes may be binding events as to activated focal adhesion kinase or may be caused by receptor activation e.g. by the Eph receptor (9, 22, 23). Upon activation, c-Src itself phosphorylates a wide variety of proteins, as paxillin at tyrosines 31 and 118. This c-Src-dependent tyrosine phosphorylation seems to be an important regulatory factor for FA maturation and stability (22, 24).
In this paper we analyzed the function of PS on cell-matrix interaction. We identified a significant reduction in FA size upon PS1 deficiency or pharmacological inhibition of γ-secretase that was associated with a reduction in cell force formation by >50%. Both effects went along with a decreased tyrosine phosphorylation level of FA-associated proteins and were caused by a PS1-dependent down-regulation of c-Src on the level of transcription as well as on the level of diminished autophosphorylation. These data prove an essential function of γ-secretase not only in cell-cell but also in cell-matrix interaction by regulating FA formation in a c-Src-dependent manner.
EXPERIMENTAL PROCEDURES
Cell Culture and Strains—Wild-type mouse embryonic fibroblasts as well as PS1-/-, PS2-/-, and PS1-/-PS2-/- strains of identical genetic background were described earlier and were kindly provided by Dr. B. DeStrooper (University of Leuven, Belgium, (25). Cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and a 1:100 dilution of an antibiotic solution (10,000 units penicillin and 10 mg streptomycin in 0.9% NaCl; Sigma). Typically, cells were grown to about 70% confluency except for immunofluorescence analyses and cell force measurements. Here, cells were seeded at low density of not more than 20% confluency and analyzed as single cells as described below. If necessary, surfaces were covered with 2.5 μg/cm2 fibronectin for 45 min at 25 °C. N-[N-(3,5-Difluorophenacetyl)-l-alanyl]-(S)-phenylglycine t-butyl ester (DAPT) as inhibitor of the γ-secretase complex was dissolved in DMSO and in some experiments added to cell culture medium to a final concentration of 2.5 μm for force measurements and 5 μm for secondary immunofluorescence, Western, and Northern blotting analyses.
Construction of GFP-c-Src was performed by amplification of whole human c-Src reading frame from clone IRAVp968B0149D6 (imaGenes GmbH, Berlin, Germany). Primer-incorporated restriction sites for HindIII and BamHI were used for in-frame ligation into pEGFP-N1 (Clontech). The same vector was used to create a full-length EB2-GFP construct. Here, nucleotides 1-1020 from EB2-cDNA clone (NCBI BC105955, clone IRAMp995N1412Q) were amplified and ligated using XhoI and HindIII as restriction enzymes. The resulting plasmids were confirmed by sequencing. The GFP-vinculin construct was provided by B. Geiger (Weizmann Institute). As a dominant negative version of PS1, a D385A point mutant was used (26, 27).
For expression of EphrinB2 (EB2) intracellular domain fused to GFP, plasmid BH351 was constructed containing the last 83 EB2 C-terminal amino acids. The DNA fragment was ligated into pEGFP-N1 (Clontech) using XhoI and HindIII as restriction sites resulting in plasmid EB2ICD-GFP. pEGFP-N1 itself was used as control for pure GFP localization analysis. Transfection of mouse embryonic fibroblasts (MEF) strains was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Preparation of Elastomeric Substrates—Force measurements were performed on micropatterned, ultrasoft, and elastic substrates which were prepared and characterized as described in Cesa et al. (28) using silicon dioxide molds. Elastic substrates were created using a commercial polydimethylsiloxane-based two-component formulation (Sylgard 184, Dow Corning). Here, a short chain vinyl-terminated polydimethylsiloxane was cross-linked with a long chain methylhydrosiloxane-dimethylsiloxane copolymer as base. This system allowed the preparation of cell substrates with highly reproducible elastic parameters (Poisson number of 0.5, Youngs modulus of 13 kilopascals). The exact elastomer characterization was performed as described in Cesa et al. (28). Holes of 1 cm in diameter were drilled in cell culture dishes. Subsequently, microstructured elastic substrates were glued to their bottoms. Cells were plated on polydimethylsiloxane substrates at a density of 5000 cells/cm2 and examined 2-24 h after seeding.
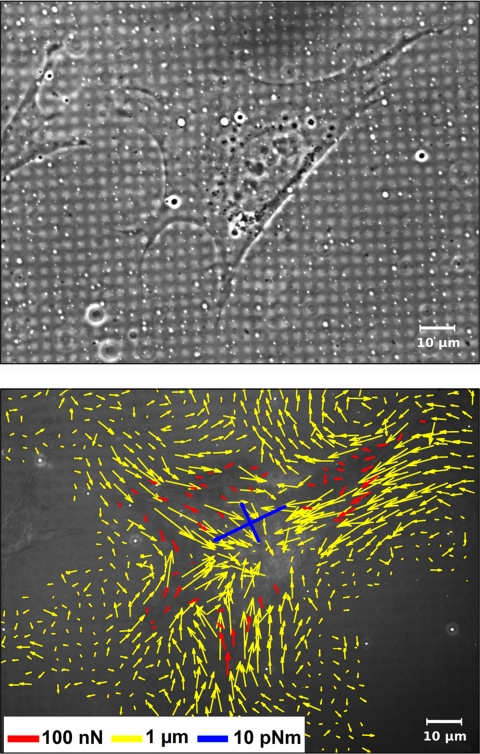
Force Measurements and Image Processing—The algorithm for cell force calculation (28-30) required as input parameters the deformation field of the flat substrate and the locations of the FAs. These structures were assumed to be the only force application sites. Cells were randomly chosen for force analysis. Deformation fields were determined by normalized cross-correlation with a synthetic template resembling the microstructures (28). For force calculations FAs were marked interactively in reflection interference contrast microscopy (RICM) images. A typical force analysis starting with a deformation field determined from an RICM image and ending with calculated cell forces applied at every single focal adhesion point is given in Fig. 1. The additionally shown generalized first moment is a measure for the sum of all contractile forces applied by a cell. For mathematical definition and discussion of the generalized first moment (GFM) tensor, see Cesa et al. (28). In Figs. 1 and 3 we present the GFM tensor by its eigenvectors scaled by the respective eigenvalue. In cases where just one number is stated, the sum of both eigenvalues is given.
FIGURE 1.
From substrate deformation to cell force. MEF WT cells were incubated on micropatterned elastic substrates (Young's modulus 13 kilopascals) for 1 day and analyzed in phase contrast (top) as well as in RICM to detect FAs and the deformed micropattern (not shown). The deformation field (yellow arrows) was determined by comparison to a regular grid using digital image processing. Employing elasticity theory, cell forces were calculated at every single FA (red arrows). Additionally a measure for the sum of all contractile forces is given as a generalized first moment (blue arrows).
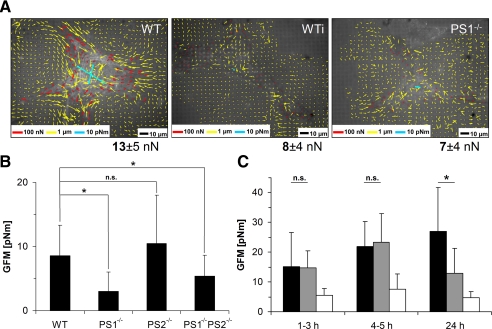
FIGURE 3.
Cell force analysis upon PS deficiency. Wild-type as well as PS mutant strains were incubated on 13-kPa elastic polydimethylsiloxane substrates for 1 day and subsequently analyzed for cell force application. Cell forces were characterized at every FA (A) as well as by the GFM (B and C). A, cell force evaluation for WT (left, wild-type cells treated with DAPT for 24 h (WTi, middle) and PS1-/- mutant cells (right). Color coding is as in Fig. 1. Below each micrograph the average force per focal adhesion is given together with its S.D. B, GFMs for randomly chosen cells. n(WT) = 86, n(PS1-/-) = 36, n(PS2-/-) = 72, n(PS1-/-PS2-/-) = 64. Error bars denote S.D. C, WT (black), DAPT-treated WT (gray) and PS1-/- (white) cells were analyzed for cell force application over time. Cell force analysis started immediately after spreading. Generalized first moments are given at indicated times. For each result given in A and C, at least eight cells were analyzed. Note the differences in average GFMs between B and C. These differences result from different cell picking methods. In B all cells in a given area were analyzed, and in C only cells exhibiting focal adhesions already at early stages were chosen. Therefore, this selection is biased toward strongly contracting cells.
For the analysis of FA size and number, the following method was used. Cells were transfected with enhanced GFP-vinculin, which is an established marker for FAs. Micrographs of enhanced GFP fluorescence were acquired with a laser scanning microscope (LSM 510, Carl Zeiss, Jena, Germany) focused to yield the best contrast of FAs. A mask of the cell area mc was generated by interactively marking the borders of the cell. All image processing operations were only applied to pixels within this mask. In the following, all kernel sizes are given in pixels (0.11 × 0.11 μm). First, images were denoised by smoothing with a gaussian kernel (5 × 5 pixels). Subsequently, illumination gradients were removed by a high pass filter. For this filter the image was further smoothed with a 57 × 57-pixel gaussian kernel and subtracted from the denoised image. For contrast enhancement the local z-score zl was calculated for each pixel (zl = (x - xm)/σ, where x is the gray value at the pixel, and xm and σ denote the mean and S.D. of the gray values in a 45 × 45 pixel-sized square centered on the pixel). Segmentation of FAs was now done by considering areas with zl > 0.9 which were larger than 20 pixels. In the centers of very large FAs zl had minima within some cases lower values than 0.9. To test if holes within FAs were erroneous or not, we identified all pixels directly neighboring such a hole and calculated the median μn and S.D. σn of their gray values. If the median gray value of the pixels within the hole exceeded μn-σn we considered the hole erroneous and closed it. FAs were counted, and their sizes were calculated by multiplying the numbers of related pixels with the pixel size.
Labeling intensity for various general as well as phosphospecific antibodies against focal adhesion proteins was determined using ImageJ as software (Version 1.36). Micrographs were acquired using a laser scanning microscope (LSM 510) with identical microscope settings. Images were opened in ImageJ as 8-bit grayscale images. After surrounding randomly selected FAs with the polygon selection tool, we measured the mean intensity value in the selected area as well as the size of the area.
Microscopy Techniques—Living cells were analyzed using a Zeiss Axiovert 200 inverted microscope equipped with an incubation chamber maintained at 37 °C and 5% CO2. Live cell imaging was performed in phase contrast as well as in RICM using an antiflex EC PlanNeofluar 63×/1.25 oil Ph3 objective with an RICM reflector module and a pre-placed green interference filter selecting the 546-nm line of the mercury arc lamp. Image acquisition was performed using an ORCA ER CCD camera (Hamamatsu Photonics, Hamamatsu, Japan) and Open Box as software (Version 1.77, Informationssysteme Schilling, Munich, Germany). Immunofluorescence analyses were performed using a laser scanning microscope (LSM 510, Zeiss, Jena, Germany) with filter sets appropriate for simultaneous detection of green (Cy2) and red (Cy3) fluorescent light. As objective, a PlanApochromat (63x/1.4, differential interference contrast) was used. The same microscope setup was used to analyze the GFP-c-Src and GFP-vinculin construct. Transfected cells were illuminated using the 488-nm argon-ion laser line. Fluorescence was detected with a 505-530-nm bandpass filter.
Immunofluorescence—To stain adhesion sites and parts of the cytoskeleton, cells were fixed in 4% paraformaldehyde dissolved in cytoskeleton buffer (CB: 150 mm NaCl, 5 mm MgCl2, 5 mm EGTA, 5 mm glucose, 10 mm MES, pH 6.1) for 30 min at 37 °C followed by a rinsing step in 30 mm glycine in CB. Cells were permeabilized in 10% Triton X-100 in CB for 5 min and blocked in 10% normal goat serum for additional 45 min. All antibodies were diluted in blocking solution. Antibody incubations were performed at room temperature for 1 h in a humidified chamber with thorough washing steps between the primary and secondary antibodies. Samples were mounted in Gel-Mount (Biomeda) containing 50 mg/ml 1,4-diazabicyclo[2.2.2]octane (Sigma). Antibodies used were against vinculin (clone hVin-1, Sigma) and phosphotyrosine (p420, BD Biosciences). The secondary antibodies were coupled to Cy3 (Dianova). F-actin was stained with phalloidin coupled to Alexa488 (Invitrogen). All secondary antibodies were F(ab′)2 fragments against mouse or anti-rabbit IgGs produced in goat and conjugated to Cy2 and Cy3, respectively (Dianova).
Immunoblotting—Cells were cultured in 6-cm cell culture dishes (Nunc) to 50-70% confluency. After washing with cold phosphate-buffered saline, 250 μl of lysis-buffer (10 mm Tris, 158 mm NaCl, 1 mm EDTA, 0,1% SDS, 1% sodium deoxycholate, 1% Triton X-100, 1% protease inhibitor for mammalian cells, Sigma) per culture dish was added. After 30 min of incubation at 4 °C, lysed cells were harvested and centrifuged at 10,000 × g for 15 min at 4 °C. 50 μg of protein from crude cell extracts were separated on a 10% SDS-polyacrylamide gel and subsequently analyzed by Western blotting. Protein contents were equalized using tubulin as constitutively expressed standard. Subsequently, protein levels for c-Src (184Q20, BIOSOURCE), paxillin (5H11, BIOSOURCE), vinculin (hVin-1, Sigma), and zyxin (polyclonal, Sigma) were determined. The antibody against tubulin (clone YL1/2) was purchased from Chemicon. Phosphotyrosine-specific primary antibodies were against Tyr-418 of c-Src (Chemicon) and Tyr-31 of paxillin (Santa Cruz Biotechnology). Tyr-397-specific antibodies for phosphorylated focal adhesion kinase came from Chemicon. Protein crude extracts for western analyses using phospho-specific antibodies were prepared in the presence of phosphatase inhibitor (1:100 dilution, Sigma). Secondary antibodies (Sigma) were coupled to alkaline phosphatase. Immune complexes were visualized with an alkaline phosphatase immunoblot assay kit (Bio-Rad). Quantification of Western blots was performed with the ImageJ software using the gel analyzing function of the program.
Northern Blots and Quantitative Real-time PCR (qRT-PCR)—Cells were cultured in 6-cm cell culture dishes (Nunc) to 50-70% confluency. For northern analyses, cells were washed with cold phosphate-buffered saline, and 1 ml of Trizol (Invitrogen) was added. Cells were harvested and homogenized in a Dounce homogenizer. Further RNA isolation was done according to the manufacturer's instructions. 5.5 μg of total RNA were separated by gel electrophoresis using 1.25% agarose gels containing formaldehyde. For normalization of total RNA amounts, the 28 S rRNA band was used. RNA was transferred using the Turbo-Blot system (Schleicher and Schuell). After transfer, RNA was cross-linked to the membrane by UV irradiation. Hybridization probes were produced using digoxigenin-labeled nucleotides. Detection was performed using horseradish peroxidase-coupled anti-digoxigenin antibodies. Quantification of northern blots was performed with ImageJ software using the gel analyzing function of the program.
For qRT-PCR analyses, cells were grown for 2 days after transfection with EB2-ICD. Transfection efficiencies were determined before total RNA isolation (RNeasy Plus mini kit, Qiagen, Venlo, The Netherlands) and cDNA synthesis using Random Primers and ThermoScript™ reverse transcriptase (Invitrogen). Relative quantitative gene expression was analyzed using Power SYBR® Green PCR Master Mix (Applied Biosystems) and specific primers for glyceraldehyde-3-phosphate dehydrogenase as the internal control and c-Src (Eurofins MWG) according to the manufacturer's instructions. PCR products were monitored with an ABI STEP ONE sequence detection system (Applied Biosystems) and analyzed with ΔΔCt (threshold values) relative quantitation.
Statistical Analysis—Statistical analyses were performed using Origin (Version 7.5) as software (OriginLab, Northampton, MA). All data were analyzed using one way analysis of variance. It is based on Student's t test assuming equal variances and normal distribution. Data were called significantly different for p < 0.05.
RESULTS
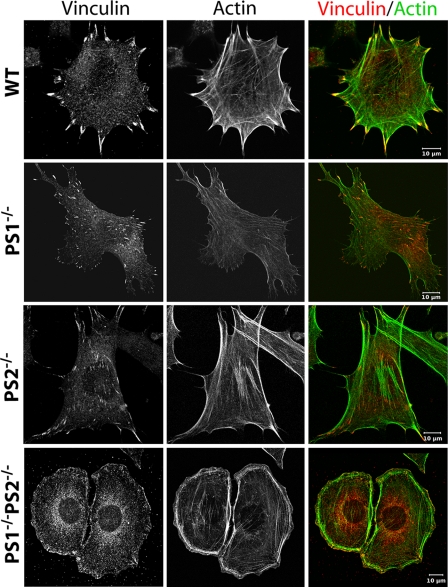
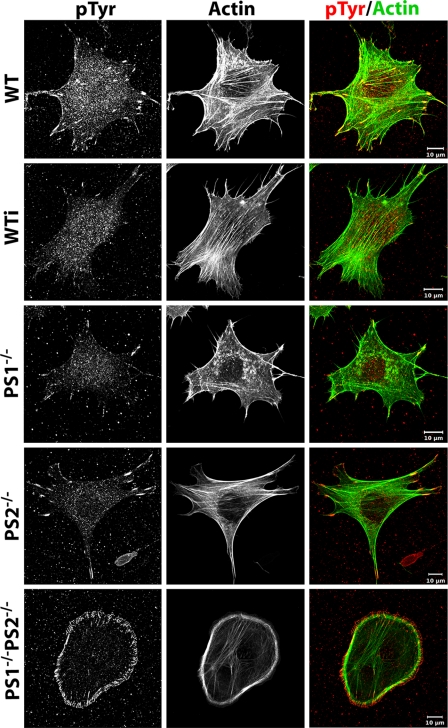
PS1 Affects the Focal Adhesion Site and Actin Cytoskeleton Formation—Former experiments have shown an important function of PS in cell-cell adhesion with an additional putative regulatory function in cell-matrix stability (5, 9, 12). To characterize the exact function of PS in cell-matrix adhesion processes wild-type (WT), presenilin 1 single as well as presenilin 2 single knock-out (PS1-/- and PS2-/-) and presenilin 1 and 2 double knock-out (PS1-/-PS2-/-) MEF were cultured and used for immunofluorescence analyses. As cell-matrix adhesion as well as cell movement strongly depend on focal adhesion sites (FA) and the actin cytoskeleton, both structures were analyzed (Fig. 2). WT cells were characterized by well visible focal adhesion sites when labeled with vinculin as an established marker protein for FAs. These sites with an average size of 0.9 μm2 were mainly located at the cortex of cells and had a classical elongated, sometimes triangular shape. To every FA, thick F-actin bundles called stress fibers were connected. Often these stress fibers spanned the whole cell, connecting two adjacent focal adhesion sites (Fig. 2 and Table 1). A very similar phenotype in respect of adhesion structures and actin cytoskeleton was found for MEF PS2-/- cells, indicating a minor influence of PS2 in cell-matrix adhesion. Very different results were found for PS1-/- cells. Here, the number of FAs was significantly reduced by 40%. The average size of FAs was additionally diminished by 30-40% and reached 0.65 μm2 on average (Table 1). Furthermore, their spatial distribution changed from a prominent cortical to a disperse localization. Prominent actin stress fibers were much less abundant, sometimes absent (Fig. 2). Faint actin bundles failed to span the whole cell (Fig. 2). Instead, they often formed an irregular meshwork with end point connections to the dispersed FAs. Interestingly, the overall cell shape remained unaffected for PS1-/- as well as PS2-/- cells compared with WT. In contrast, double mutation of PS1 and PS2 fully changed the MEF cell shape to a rounded form. These cells were additionally characterized by hundreds of very small adhesion sites located all around the cell cortex. In double knock-out cells, FAs established a longitudinal orientation in the direction of the center of the cell. Stress fibers spanning the cell were almost absent, whereas actin bundles forming the cortex of the cell were prominent.
FIGURE 2.
Focal adhesions and actin cytoskeleton of MEF mutants. WT and PS mutant strains were incubated for 1 day, fixed, and subsequently stained for vinculin (left) and actin (middle). An overlay of both stainings is given on the right. Note the reduced size of FAs and stress fibers in PS1-/- compared with WT and the complete switch in cell morphology in PS1-/-PS2-/- double mutant strains. Throughout the whole experiment all cell types were treated identically and analyzed at a confocal microscope with unchanged microscopic and image processing configurations compared with WT.
TABLE 1.
PS-dependent adhesion and force properties
WT, wild-type inhibited by DAPT for 24 h (WTi), and PS1−/− cells (PS1−/−) as well as the same cells transfected with a c-Src-GFP construct (PS1−/−r) were grown on 13-kilopascal soft polydimethylsiloxane substrates and analyzed for the given features. The number of cells analyzed is indicated for each strain. Where given in the left field, the analyzed cell number was identical for each strain. FA phosphotyrosine intensities are shown relative to average WT intensity and are normalized to FA size. ND, not determined. All values are given with S.D. Note that area/FA was analyzed in immunostaining against phosphotyrosine for the rescue strain, whereas all other strains were characterized using vinculin as GFP-tagged version.
| WT | WTi | PS1−/− | PS1−/−r | PS1−/− EB2ICD | |
|---|---|---|---|---|---|
| FA/cell | 110 ± 80 (n = 6 cells) | 100 ± 40a (n = 6 cells) | 60 ± 30b (n = 7 cells) | ND | ND |
| Cell area (μm2) (n = 10 cells) | 2350 ± 1600 | 1950 ± 400a | 1200 ± 700a | ND | ND |
| FA area/cell (μm2) | 87 ± 61 (n = 6 cells) | 66 ± 13a (n = 6 cells) | 40 ± 19a (n = 7 cells) | ND | ND |
| Area/FAs (μm2) | 0.9 ± 0.2 (n = 645 FA) | 0.7 ± 0.2b (n = 608 FA) | 0.7 ± 0.1b (n = 430 FA) | 0.9 ± 0.9c (n = 80 FA) | 1.0 ± 0.5c (n = 100 FA) |
| Force/FA (nN) | 13 ± 5 (n = 10 cells) | 8 ± 4a (n = 19 cells) | 7 ± 4b (n = 8 cells) | ND | ND |
| GFM (pNm) | 27 ± 15 (n = 10 cells) | 13 ± 8a (n = 19 cells) | 5 ± 2b (n = 10 cells) | ND | ND |
| FA phosphorylation (% intensity of WT) | 100 ± 10 (n = 101 FA) | 64 ± 15b (n = 95 FA) | 64 ± 19b (n = 100 FA) | 102 ± 49c (n = 80 FA) | 88 ± 24c (n = 100 FA) |
Not significantly different from WT (p > 0.05).
Significantly different from WT (p < 0.05).
Significantly different from PS1−/− but not from WT.
Deficiency of PS1 Reduces Cell Force Formation—FAs connect the contractile actin-myosin cytoskeleton to the extracellular matrix. Their reduced size as well as a modified actin cytoskeleton implies the possibility of an affected cell force generation in PS1-deficient cells. For a quantitative measurement of cell forces, MEF WT as well as PS mutant strains were seeded on micropatterned, soft polydimethylsiloxane substrates. Displacements of the regular micropattern were visualized by RICM, and the generating forces were calculated from these data. All cells within randomly chosen areas were analyzed to prevent preselection bias. For single FA sites of WT cells, analyses revealed forces in the range of 13 nN (Fig. 3A) with a generalized first moment of about 8.5 pNm (σ = 4.7 pNm; n = 86; Fig. 3B and Table 1). Comparable forces for single focal adhesions (data not shown) as well as similar generalized first moments were determined for PS2-/- cells. PS1-/- cells instead applied forces in the range of 7 nN per focal adhesion with a generalized first moment of 3.0 pNm (σ = 3.1 pNm; n = 36; Fig. 3, A and B, Table 1). Therefore, PS1-/- cells were significantly reduced in force application by more than 60%. This is in line with their reduced size of focal adhesions. Cell force analyses for PS1-/-PS2-/- cells revealed diminished cell force applications by ∼40% compared with WT with a generalized first moment of 5.4 pNm (σ = 3.3 pNm, n = 64). However, as shown above, PS1-/-PS2-/- cells exhibited an altered shape with a regular ring of adhesion sites around the cortex. Such modified morphology argued for a basically different type of cell adhesion and, therefore, made a direct comparison to WT as well as single mutant strains questionable.
PS1 can serve also cellular functions independent of γ-secretase activity (31, 32). To assess whether the role of PS1 in cell-matrix adhesion involves the activity of the γ-secretase complex, we pharmacologically inhibited the protease activity with DAPT (33, 34). For inhibition analyses, freshly trypsinated wild-type cells were seeded on a micropatterned elastic substrate and immediately incubated with DAPT. The cell shape of wild-type cells remained unaffected over time. In contrast, immunostaining for vinculin identified a significantly reduced average size of focal adhesion sites of 0.7 μm2 after 24 h of DAPT treatment compared with 0.9 μm2 in untreated control cells (Table 1). After spreading, force application increased equally for WT as well as DAPT-treated WT cells within the first 5 h. In contrast, PS1-/- cells were at no time able to apply significant forces and remained on a low level for the whole 24-h period of force analysis (Fig. 3C). Interestingly, forces of WT cells stayed constant from 5 h to 24 h of incubation. In contrast, applied forces at every FA as well as the generalized first moment dropped in DAPT-treated WT cells from identical values as WT after 5 h to values close to PS1-/- cells after 24 h (Fig. 3, A and C; see also Table 1 with additional analyzed cells for WT and PS1-/-). Cell analyses directly after spreading depended on well spread and adhered cells. This resulted in a preselection to stronger cells but was done identically for all cell types or experimental conditions. The resulting forces were, therefore, higher than in Fig. 3B, where randomly chosen cells were evaluated.
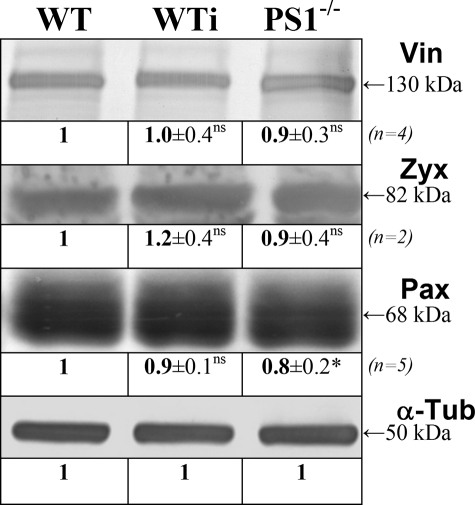
Tyrosine Phosphorylation Is Impaired in PS1-/- cells—The changes in size of focal adhesion sites as well as in force generation could be caused at multiple levels of regulation. In a first step we analyzed the overall concentration of various adhesion site-specific proteins to exclude a more general influence of PS1 on adhesion. Protein levels of vinculin, paxillin, and zyxin were analyzed in protein extracts from WT and PS1-/- as well as WT cells incubated with DAPT for 24 h. As shown in Fig. 4, proteins levels of vinculin, zyxin, and paxillin were unaffected by PS1 inhibition or deficiency, arguing for unaffected focal adhesion protein expression upon PS1 deficiency.
FIGURE 4.
Independency of FA protein expression upon PS deficiency. WT and PS1-/- as well as DAPT treated WT cells (WTi) were harvested after 24 h, and crude protein extracts were analyzed in Western blot analyses. Protein concentrations were equalized using tubulin (α-Tub) as constitutively expressed marker protein. Crude extracts were subsequently analyzed for the indicated proteins. All relative protein concentrations including S.D. given below the bands are normalized with the original band intensity determined in the α-tubulin control. In addition, all values of each line were divided by the corresponding WT value. The apparent molecular weight of the given protein bands is indicated on the right. Please note that Western analyses against zyxin and paxillin showed some unspecific background staining (not shown). Vin, vinculin; Zyx, zyxin; Pax, paxillin; *, significantly different from wild-type (p < 0.05); ns, not significantly different (p > 0.05).
Because earlier experiments indicated an influence of PS on ephrinB1 putatively affecting c-Src activity (5, 9, 24), we analyzed in a next step the phosphotyrosine levels of FAs in WT and PS mutant strains. These analyses were performed in fixed cells using a phosphotyrosine-specific antibody and revealed high levels of phosphorylation in FAs of WT and PS2-/- cells. In contrast, PS1-/- and wild-type cells cultured in the presence of DAPT for 24 h were characterized by a strongly decreased tyrosine phosphorylation (Fig. 5). Such a reduction in phosphotyrosine-specific staining of focal adhesion sites was not only due to the decreased size of focal adhesions. Additionally, labeling intensities normalized to the size of FAs revealed a significant reduction of 40% in PS1-/--as well as DAPT-treated cells compared with WT (Table 1). As control, immunofluorescent staining of identical FAs for vinculin, identified though smaller focal adhesion sites for PS1-deficient cells, compared with WT, but their normalized gray values remained largely unaffected (data not shown).
FIGURE 5.
Tyrosine phosphorylation of focal adhesion sites. WT and PS mutant strains as well as WT cells treated with DAPT for 24 h (WTi) were grown on glass, fixed, and subsequently stained for phosphotyrosine (left) as well as actin (middle). Their overlay is given in color (right). Scale bar, 20 μm. Note the strongly diminished phosphotyrosine level in PS1-/- cells as well as in DAPT-treated cells. Image acquisition is as described in Fig. 2.
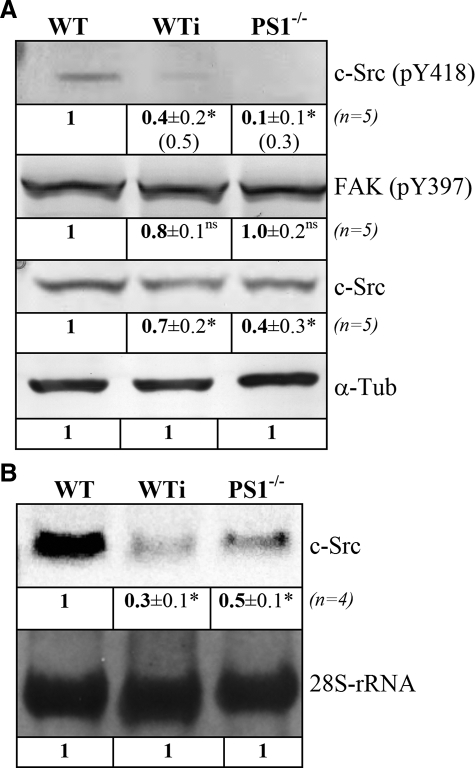
γ-Secretase Regulates Expression of c-Src—Tyrosine phosphorylation in focal adhesion sites is mainly performed by activated c-Src kinase (22). Because tyrosine phosphorylation was significantly affected upon PS1 deficiency, we analyzed the activation status of this kinase as well as its expression level in WT and PS1-/- MEFs. An essential regulatory event in c-Src activation is defined by its autophosphorylation at tyrosine 418. We, therefore, analyzed the phosphorylation status at this site in WT and PS1-/- as well as WT cells treated with DAPT. As given in Fig. 6A, c-Src phosphorylated at Tyr-418 was strongly decreased by 90% in the absence of PS1 compared with WT. Similar results were found for DAPT-treated WT cells with a reduction of 60% in c-Src phosphorylated at Tyr-418.
FIGURE 6.
Levels of c-Src phosphorylation and expression. A, Western analyses. Crude protein extracts from WT and PS1-/- as well as WT cells treated with DAPT (WTi) for 24 h were separated by SDS-page and used for Western blot analyses against c-Src. In addition, phosphotyrosine-specific antibodies against tyrosine 418 of c-Src and tyrosine 397 of focal adhesion kinase were used. Protein concentrations were equalized using α-tubulin (α-Tub) as a constitutively expressed standard. Numbers given in brackets for c-Src (pY418) in WTi and PS1-/- indicate protein amounts normalized to total levels of c-Src. B, Northern analyses. Same cell types as in A were harvested after 24 h, and total RNA was isolated. Total RNA was separated under formaldehyde conditions, and RNA concentrations were equalized for 28 SrRNA. Northern blot analyses were performed with digoxigenin-labeled probes against c-Src mRNA. Relative changes in protein (A) or mRNA (B) levels as well as their S.D. are indicated below each signal and were determined as described in Fig. 4. *, significantly different from wild-type (p < 0.05); ns, not significantly different (p > 0.05).
As such, results might have been caused by either regulation of autophosphorylation or regulation of expression; Northern as well as Western analyses for c-Src were performed. Protein levels of c-Src were reduced by 30% in DAPT-treated WT cells and by 60% in PS1-/- cells (Fig. 6A). Northern analyses identified an almost identical reduction of c-Src transcripts. Here, levels of c-Src mRNA were reduced by 50% in PS1-/- cells and by almost 70% upon DAPT incubation (Fig. 6B). Reduced levels of c-Src protein were, therefore, likely caused by transcriptional down-regulation rather than increased protein degradation. These data elucidate a vital function of PS1 in c-Src transcriptional as well as posttranslational regulation as the 60% reduced protein amount of c-Src in PS1-/- cells remained almost completely in an inactive, non-phosphorylated state.
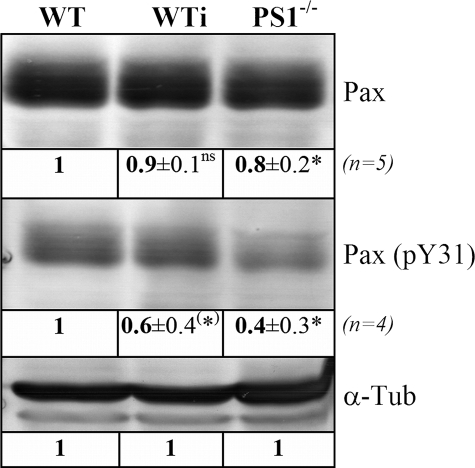
To analyze the lack of c-Src activity on FA phosphorylation in more detail, paxillin phosphorylation was also characterized. Paxillin represents an important bridging factor within FAs and is known to be phosphorylated at tyrosine residues 31 and 118 by activated c-Src (24). Reduced phosphorylation at Tyr-31 in PS1-deficient (60% reduction) as well as in DAPT-treated WT cells (40% reduction) compared with WT confirmed the strong correlation between reduced c-Src protein levels upon PS1 deficiency and diminished tyrosine phosphorylation (Fig. 7). Total paxillin protein levels remained basically unaffected.
FIGURE 7.
Dependence of paxillin phosphorylation on PS1. Crude protein extracts from WT and PS1-/- as well as DAPT-treated WT cells (WTi) were separated by SDS-PAGE and equalized for α-tubulin (α-Tub). Subsequently, Western blot analyses against paxillin (Pax) and phosphorylated tyrosine 31 of paxillin (Pax pY31) were performed. Relative changes in protein levels were determined as described in Fig. 4 and are indicated below each signal. Western analyses were performed four times with each antibody from independent protein isolations. *, significantly different from wild-type (p < 0.05); (*), significantly different from wild-type (p = 0.05); ns, not significantly different (p > 0.05).
Selective Inhibition of c-Src Kinase by γ-Secretase—c-Src is not the only kinase responsible for tyrosine phosphorylation of focal adhesion proteins. To characterize the specificity of PS1 on c-Src expression, we additionally analyzed the autophosphorylation status of focal adhesion kinase. Upon autophosphorylation on Tyr-397, focal adhesion kinase becomes activated, forming a binding site for activated c-Src (35, 36). Using phosphotyrosine 397-specific focal adhesion kinase antibodies, no changes in focal adhesion kinase activity could be found in PS1-deficient cells as compared with WT (Fig. 6A). In addition, rescue experiments using a c-Src-GFP fusion construct expressed in WT and PS1-/- cells revealed a complete reversion of phosphotyrosine levels of focal adhesions in the PS1-/- cells (Table 1). Together, these data indicate a specific effect of PS1 on cell-matrix interaction and force application via c-Src.
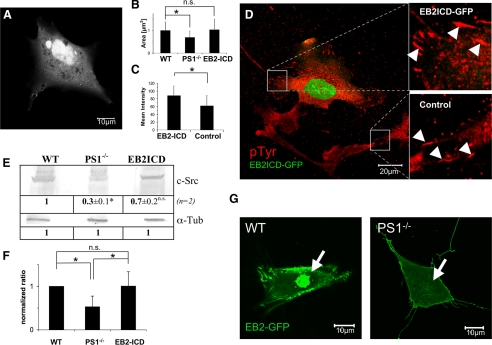
EphrinB2 Cytoplasmic Domain Is Translocated in a PS1-dependent Manner into the Nucleus—To complete the signal transduction pathway from PS1 function to c-Src activity, we checked γ-secretase targets for a putative influence on c-Src. Because two of them, ephrinB1 and ephrinB2, were also known binding partners of c-Src, we analyzed ephrinB2 in more detail. Experiments given above revealed c-Src regulation mainly at the transcriptional level. We, therefore, tested ephrinB2 intracellular domain (EB2-ICD) for transcriptional co-activator function as already known from notch or EB1 upon γ-secretase cleavage (1, 5). A construct of EB2-ICD fused to GFP was constructed and expressed in WT as well as PS1-/- cells, and its localization was compared with GFP only. Life cell imaging revealed intense nuclear localization of EB2ICD-GFP (Fig. 8A) compared with a more cytoplasmic localization of pure GFP. Although toxic at high concentrations or enhanced incubation times, a full-length EB2-GFP construct, expressed in WT and PS1-/- cells, identified the dependence of EB2-ICD translocation on γ-secretase function. Although nuclear localization could be observed for EB2-GFP in WT 20 h after transfection, no such signal was present in PS1-/- cells (Fig. 8G). These data prove EB2 to be cleaved by γ-secretase and that an EB2 cleavage product is transduced into the nucleus.
FIGURE 8.
Effects of EphrinB2 cytoplasmic domain expression in PS1-/- cells. After transfection with EB2ICD-GFP, cells were grown for 2 days. Thereafter, cells were either used for western or RT-PCR analyses. Alternatively, cells were analyzed by confocal live cell microscopy or fixed and immunostained for marker proteins following by confocal imaging. A, nuclear localization of EB2ICD-GFP in living MEF PS1-/- cells. B, quantification of average focal adhesion size. Cells were immunocytochemically labeled for paxillin. Using ImageJ as software, sizes of randomly chosen focal adhesions were determined. WT, n = 100 FAs, 6 cells; PS1-/-, n = 100 FAs, 16 cells; EB2ICD, n = 100 FAs, 11 cells. C, quantification of tyrosine phosphorylation of FAs was performed using a phosphotyrosine specific antibody and ImageJ as software. Values are given in % relative to WT. EB2ICD, n = 150 FAs, 16 cells; PS1-/-, n = 150 FAs, 11 cells. D, exemplary confocal image of phosphotyrosine-immunolabeled cells. EB2ICD-GFP-expressing PS1-/- cells are characterized by a strong nuclear GFP signal and intensively phosphorylated, large FAs (upper square, right). Both are absent in untransfected cells (lower square, right). FAs are indicated by white arrowheads. E, Western analyses of PS1-/- cells and PS1-/- cells additionally expressing EB2ICD-GFP (EB2ICD) were quantified for c-Src protein levels and compared with WT (n = 2). Protein concentrations were equalized using α-tubulin (α-Tub). F, normalized ΔΔCT values with c-Src specific primers in qRT-PCR experiments for the indicated strains (n = 5). Note that the doubled c-Src mRNA level of EB2-ICD-expressing PS1-/- cells (EB2-ICD) was found in the total mRNA pool, although just 10% of all cells were transfected. *, significantly different from WT; ns, not significantly different from WT (p < 0.05). G, WT and PS1-/- cells were transfected with a full-length EB2-GFP construct and analyzed for ephrin nuclear translocation. Note that high level expression of EB2-GFP was toxic to all cells analyzed. Nuclear translocation could, therefore, be analyzed only within the first 20 h after transfection.
Expression of EB2-ICD Fully Rescues Adhesion and Phosphorylation Phenotypes of PS1 Mutants—Reduced transcription of c-Src upon PS1 deficiency resulted in reduced phosphorylation of FA proteins and in FAs of reduced size and diminished force application. We, therefore, analyzed PS1-/- cells for c-Src expression and phenotype restoration after transfection with EB2ICD-GFP. qRT-PCR experiments (n = 5) revealed an increase of c-Src mRNA levels by a factor of 2 in EB2ICD-GFP-transfected cells compared with untransfected PS1-/- cells (Fig. 8F). A similar increase was observed for c-Src in Western analyses (Fig. 8E). Taking into account that these changes in c-Src expression could be observed in mRNA and protein extracts of cultures with EB2ICD-GFP transfection efficiencies of just 10%, the effect of EB2-ICD on c-Src expression is presumably even higher. Additional analyses were performed on the single cell level. Here, transfected cells were fixed and stained for paxillin. Subsequently, only cells with an EB2ICD-GFP signal were randomly chosen, and focal adhesion areas of those cells were determined (Fig. 8B). For transfected WT cells we measured an average FA size of 1.0 μm2 (σ = 0.5 μm2, n = 85 FA, 6 cells). Focal adhesions of untransfected PS1-/- cells were significantly smaller with a mean of 0.7 μm2 (σ = 0.3 μm2, n = 100 FA, 16 cells). For PS1-/- cells expressing EB2ICD-GFP, we observed an average size of 1.0 μm2 (σ = 0.4 μm2, n = 100 FA, 11 cells). As indicated in Fig. 8D, untransfected as well as transfected PS1-/- cells could be analyzed in the same sample and, therefore, displayed ideal conditions for comparison. Replacing paxillin with vinculin as a marker for focal adhesions resulted in identical results for EB2ICD-GFP-transfected PS1-/- cells (FA mean area = 1.1 μm2, σ = 0.6 μm2, n = 75 FA, 5 cells). These results were in good agreement with focal adhesion areas determined before and argue for full restoration of the PS1-/- adhesion phenotype.
This restoration was analyzed in more detail, as c-Src function and, therefore, FA formation depends on phosphorylation of FA proteins. We, therefore, determined tyrosine phosphorylation levels of FAs in EB2ICD-GFP-transfected as well as in untransfected PS1-/- cells. The data showed an ∼30% higher phosphotyrosine level for EB2ICD-GFP-transfected cells than for untransfected PS1-/- cells (Table 1). Phosphorylation levels of EB2ICD-GFP expressing PS1-/- cells were almost identical to those found before in WT cells (88% of WT, σ = 25%, n = 150 FA, 16 cells; PS1-/- = 62% of WT, σ = 27%, n = 150 FA, 11 cells) and, therefore, confirm a fully restored c-Src function upon overexpression of EB2ICD. Furthermore, a dominant negative version of PS1 (D385A) (26, 27) had no visible effect on adhesion formation when expressed in PS1-/- cells (data not shown), further confirming that the PS1-/- phenotype suppression upon EB2 overexpression is mainly caused by a transcriptional regulation of c-Src and not by interaction between c-Src and EB2-ICD.
DISCUSSION
Transmembrane proteins and their manifold functions and specific activations depend on general pathways regulating their protein turnover. Besides vesicular internalization and subsequent degradation by endosomal/lysosomal proteases (37-40), transmembrane proteins can also undergo a two-step process including sequential shedding of their ectodomains and intramembranous cleavage of the resulting stubs by γ-secretase (1-3). The released intracellular domains can also serve multiple functions in signal transductions as evident for that of notch, CD44, or probably amyloid precursor protein where these domains translocate to the nucleus and regulate gene transcription (41-43).
In this work we identified γ-secretase as a regulator of transcription and activation of c-Src kinase. PS deficiency or pharmacological inhibition led to decreased levels of c-Src transcripts, indicating that γ-secretase activity positively regulates c-Src transcription. Interestingly, reduced c-Src transcription was observed specifically in PS1-deficient cells but not in cells lacking PS2. Our data are, therefore, in good agreement with former results demonstrating that PS1 has a much higher contribution to total cellular γ-secretase activity than PS2 (2, 8, 44).
Beside transcriptional regulation, γ-secretase also impaired autophosphorylation of c-Src, arguing for a dual role of PS1 in the regulation of c-Src activity. The autophosphorylation of c-Src characterizes a well described mechanism. Under non-activating conditions, Src kinases are maintained in an autoinhibited state by intramolecular interactions of their SH2 and SH3 domains (45-47). In the inhibited configuration, SH3 binding to a polyproline sequence in the linker region between the SH2 and kinase domains regulates the inactive conformation of the catalytic lobes. Disruption of this interaction by other SH3 ligands can increase Src activity (48). Many of these c-Src-activating binding partners are described ranging from β3-integrins to catenins or ephrinB1 (9, 23, 49-51).
c-Src belongs to a family of tyrosine kinases (52) and is described to be involved in cell spreading and especially focal contact formation (53-55). Knock-out of c-Src was reported to suppress tyrosine phosphorylation in focal contacts and to reduce the size of focal contacts. In addition, these cells showed delayed spreading on fibronectin-coated substrates (53, 56). Our data are fully consistent with these findings and additionally identified a reduced force application at each focal adhesion site, whereas the overall concentration of all studied focal adhesion proteins in the cell remained unaffected upon γ-secretase inhibition. On the other hand, increased c-Src expression strongly increased tyrosine phosphorylation in adhesion structures (57). Data on the viral form of Src (v-Src) as well as expression analyses on cancer cells additionally argue for increased cell motility upon Src up-regulation (58, 59). A recent quantitative microscopic analysis of c-Src highlights the role of c-Src in cell adhesion. In that study c-Src-dependent tyrosine phosphorylation were reported to colocalize with focal complexes (young adhesion sites) and focal adhesions (adhesion sites of intermediate age). In highly stable and old fibrillar adhesions tyrosine phosphorylation was found to be absent, but upon c-Src deficiency no fibrillar adhesions could be formed at all. Additionally, protein composition of cellular adhesions changed with tyrosine phosphorylation levels (60, 61). Those findings strongly argue for a central role of c-Src in assembly of cell-matrix adhesions and especially in their subsequent molecular maturation.
Most important for our model of a direct regulation of c-Src by PS1 was our finding of EB2 domain translocation into the nucleus only in WT and not PS1-/- cells and of c-Src expression regulated by EB2-ICD. Although additional indirect effects on adhesion, caused by PS1 lack of function or a crowding effect in the plasma membrane, cannot be fully excluded, strong nuclear translocation of EB2-ICD identified the signal transducer from PS1 to c-Src. qRT-PCR as well as Western analyses confirmed the direct influence of EB2-ICD on c-Src expression. Furthermore, analyses performed on single cell levels showed that only EB2-ICD-expressing PS1-/- cells were restored in tyrosine phosphorylation of FA proteins and FA size. This proved c-Src to be as active as in WT. Our data now also offer a mechanism that can be used to explain former findings of increased Src-dependent phosphorylation upon ephrinB2 overexpression or injection (14, 62, 63). Furthermore, as for notch or EB1 (2, 5, 64), with EB2 we could identify an additional protein to be functional as a transcriptional coactivator after γ-secretase cleavage.
The given data, therefore, suggest a model in which γ-secretase cleavage of ephrinB2 leads to increased c-Src expression and activity (Fig. 9). This has an effect on FA phosphorylation vital for mature FA formation. Upon cell motility-inducing signals, transmembrane proteins as ephrin or cadherin can be efficiently excluded from the plasma membrane by internalization (37-40). According to our model, these diminished levels of the responsible PS substrates would subsequently not only result in weakened cell-cell contacts but also in reduced c-Src transcription as well as protein activity. Such a reduction will go along with incomplete focal adhesion maturation and increased adhesion dynamics and would, therefore, also affect cells on the level of cell-matrix interactions to switch cell morphology from a sessile to a motile phenotype.
FIGURE 9.
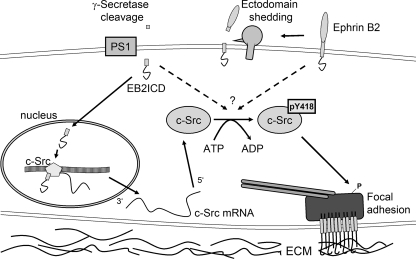
Model of c-Src regulation. Transmembrane proteins as e.g. ephrins or cadherin/catenin are subject of ADAM (a disintegrin and metalloprotease)-dependent shedding and subsequent PS-dependent cleavage processes. This mechanism frees the remaining transmembrane domains from the membrane. Hence, the freed EphrinB2 intracellular domain functions as coactivator for c-Src transcription. In addition, interaction of expressed c-Src with a stub fragment or directly with a transmembrane protein positively regulates c-Src autophosphorylation and, therefore, its function to phosphorylate focal adhesion-specific proteins. These phosphorylation events are essential for maturation of focal adhesion sites and, therefore, for stable cell-matrix interactions. ECM, extracellular matrix.
Acknowledgments
MEF WT and PS mutant strains were kindly provided by Drs. B. DeStrooper and P. Saftig. We gratefully thank C. M. Cesa and N. Hersch for intense help in lithography and cell culture. We additionally thank C. Schaefer for helpful discussions and critical reading of the manuscript. The GFP-vinculin construct was kindly provided by B. Geiger (Weizmann Institute).
This work was supported by Deutsche Forschungsgemeinschaft Sonderforschungsbereich 645. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PS, presenilin; GFP, green fluorescent protein; MEF, mouse embryonic fibroblast; WT, wild type; ICD, intracellular domain; FA, focal adhesion; DAPT, N-[N-(3,5-difluorophenacetyl)-l-alanyl]-(S)-phenylglycine t-butyl ester; EB2, ephrinB2; RICM, reflection interference contrast microscopy; MES, 2-(N-morpholino)ethanesulfonic acid; qRT-PCR, quantitative real-time PCR; GFM, generalized first moment; pNm, piconewtonmeter; nN, nanonewton.
References
- 1.De Strooper, B., Saftig, P., Craessaerts, K., Vanderstichele, H., Guhde, G., Annaert, W., Von Figura, K., and Van Leuven, F. (1998) Nature 391 387-390 [DOI] [PubMed] [Google Scholar]
- 2.De Strooper, B., Annaert, W., Cupers, P., Saftig, P., Craessaerts, K., Mumm, J. S., Schroeter, E. H., Schrijvers, V., Wolfe, M. S., Ray, W. J., Goate, A., and Kopan, R. (1999) Nature 398 518-522 [DOI] [PubMed] [Google Scholar]
- 3.Xia, W., and Wolfe, M. S. (2003) J. Cell Sci. 116 2839-2844 [DOI] [PubMed] [Google Scholar]
- 4.Maretzky, T., Reiss, K., Ludwig, A., Buchholz, J., Scholz, F., Proksch, E., de Strooper, B., Hartmann, D., and Saftig, P. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 9182-9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomita, T., Tanaka, S., Morohashi, Y., and Iwatsubo, T. (2006) Mol. Neurodegener. 1 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgakopoulos, A., Marambaud, P., Efthimiopoulos, S., Shioi, J., Cui, W., Li, H. C., Schutte, M., Gordon, R., Holstein, G. R., Martinelli, G., Mehta, P., Friedrich, V. L., Jr., and Robakis, N. K. (1999) Mol. Cell 4 893-902 [DOI] [PubMed] [Google Scholar]
- 7.Newman, M., Musgrave, I. F., and Lardelli, M. (2007) Biochim. Biophys. Acta 1772 285-297 [DOI] [PubMed] [Google Scholar]
- 8.Herreman, A., Hartmann, D., Annaert, W., Saftig, P., Craessaerts, K., Serneels, L., Umans, L., Schrijvers, V., Checler, F., Vanderstichele, H., Baekelandt, V., Dressel, R., Cupers, P., Huylebroeck, D., Zwijsen, A., Van Leuven, F., and De Strooper, B. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11872-11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgakopoulos, A., Litterst, C., Ghersi, E., Baki, L., Xu, C., Serban, G., and Robakis, N. K. (2006) EMBO J. 25 1242-1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soriano, S., Kang, D. E., Fu, M., Pestell, R., Chevallier, N., Zheng, H., and Koo, E. H. (2001) J. Cell Biol. 152 785-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunkan, A. L., and Goate, A. M. (2005) J. Neurochem. 93 769-792 [DOI] [PubMed] [Google Scholar]
- 12.Zou, K., Hosono, T., Nakamura, T., Shiraishi, H., Maeda, T., Komano, H., Yanagisawa, K., and Michikawa, M. (2008) Biochemistry 47 3370-3378 [DOI] [PubMed] [Google Scholar]
- 13.Davy, A., and Soriano, P. (2005) Dev. Dyn. 232 1-10 [DOI] [PubMed] [Google Scholar]
- 14.Slack, S., Battaglia, A., Cibert-Goton, V., and Gavazzi, I. (2008) Neuroscience 156 175-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badley, R. A., Woods, A., Carruthers, L., and Rees, D. A. (1980) J. Cell Sci. 43 379-390 [DOI] [PubMed] [Google Scholar]
- 16.Zamir, E., Katz, B. Z., Aota, S., Yamada, K. M., Geiger, B., and Kam, Z. (1999) J. Cell Sci. 112 1655-1669 [DOI] [PubMed] [Google Scholar]
- 17.Balaban, N. Q., Schwarz, U. S., Riveline, D., Goichberg, P., Tzur, G., Sabanay, I., Mahalu, D., Safran, S., Bershadsky, A., Addadi, L., and Geiger, B. (2001) Nat. Cell Biol. 3 466-472 [DOI] [PubMed] [Google Scholar]
- 18.Beningo, K. A., Dembo, M., Kaverina, I., Small, J. V., and Wang, Y. L. (2001) J. Cell Biol. 153 881-888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiger, B., Bershadsky, A., Pankov, R., and Yamada, K. M. (2001) Nat. Rev. Mol. Cell Biol. 2 793-805 [DOI] [PubMed] [Google Scholar]
- 20.Singh, N., Talalayeva, Y., Tsiper, M., Romanov, V., Dranovsky, A., Colflesh, D., Rudamen, G., Vitek, M. P., Shen, J., Yang, X., Goldgaber, D., and Schwarzman, A. L. (2001) Exp. Cell Res. 263 1-13 [DOI] [PubMed] [Google Scholar]
- 21.Galbraith, C. G., Yamada, K. M., and Sheetz, M. P. (2002) J. Cell Biol. 159 695-705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wozniak, M. A., Modzelewska, K., Kwong, L., and Keely, P. J. (2004) Biochim. Biophys. Acta 1692 103-119 [DOI] [PubMed] [Google Scholar]
- 23.Arias-Salgado, E. G., Lizano, S., Sarkar, S., Brugge, J. S., Ginsberg, M. H., and Shattil, S. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13298-13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vindis, C., Teli, T., Cerretti, D. P., Turner, C. E., and Huynh-Do, U. (2004) J. Biol. Chem. 279 27965-27970 [DOI] [PubMed] [Google Scholar]
- 25.Herreman, A., Serneels, L., Annaert, W., Collen, D., Schoonjans, L., and De Strooper, B. (2000) Nat. Cell Biol. 2 461-462 [DOI] [PubMed] [Google Scholar]
- 26.Capell, A., Steiner, H., Romig, H., Keck, S., Baader, M., Grim, M. G., Baumeister, R., and Haass, C. (2000) Nat. Cell Biol. 2 205-211 [DOI] [PubMed] [Google Scholar]
- 27.Tamboli, I. Y., Prager, K., Thal, D. R., Thelen, K. M., Dewachter, I., Pietrzik, C. U., St George-Hyslop, P., Sisodia, S. S., De Strooper, B., Heneka, M. T., Filippov, M. A., Muller, U., van Leuven, F., Lutjohann, D., and Walter, J. (2008) J. Neurosci. 28 12097-12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cesa, C. M., Kirchgessner, N., Mayer, D., Schwarz, U. S., Hoffmann, B., and Merkel, R. (2007) Rev. Sci. Instrum. 78 034301-1-034301-10 [DOI] [PubMed] [Google Scholar]
- 29.Dembo, M., Oliver, T., Ishihara, A., and Jacobson, K. (1996) Biophys. J. 70 2008-2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarz, U. S., Balaban, N. Q., Riveline, D., Bershadsky, A., Geiger, B., and Safran, S. A. (2002) Biophys. J. 83 1380-1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang, D. E., Soriano, S., Xia, X., Eberhart, C. G., De Strooper, B., Zheng, H., and Koo, E. H. (2002) Cell 110 751-762 [DOI] [PubMed] [Google Scholar]
- 32.Prager, K., Wang-Eckhardt, L., Fluhrer, R., Killick, R., Barth, E., Hampel, H., Haass, C., and Walter, J. (2007) J. Biol. Chem. 282 14083-14093 [DOI] [PubMed] [Google Scholar]
- 33.Dovey, H. F., John, V., Anderson, J. P., Chen, L. Z., de Saint Andrieu, P., Fang, L. Y., Freedman, S. B., Folmer, B., Goldbach, E., Holsztynska, E. J., Hu, K. L., Johnson-Wood, K. L., Kennedy, S. L., Kholodenko, D., Knops, J. E., Latimer, L. H., Lee, M., Liao, Z., Lieberburg, I. M., Motter, R. N., Mutter, L. C., Nietz, J., Quinn, K. P., Sacchi, K. L., Seubert, P. A., Shopp, G. M., Thorsett, E. D., Tung, J. S., Wu, J., Yang, S., Yin, C. T., Schenk, D. B., May, P. C., Altstiel, L. D., Bender, M. H., Boggs, L. N., Britton, T. C., Clemens, J. C., Czilli, D. L., Dieckman-McGinty, D. K., Droste, J. J., Fuson, K. S., Gitter, B. D., Hyslop, P. A., Johnstone, E. M., Li, W. Y., Little, S. P., Mabry, T. E., Miller, F. D., and Audia, J. E. (2001) J. Neurochem. 76 173-181 [DOI] [PubMed] [Google Scholar]
- 34.Micchelli, C. A., Esler, W. P., Kimberly, W. T., Jack, C., Berezovska, O., Kornilova, A., Hyman, B. T., Perrimon, N., and Wolfe, M. S. (2003) FASEB J. 17 79-81 [DOI] [PubMed] [Google Scholar]
- 35.Schaller, M. D., Hildebrand, J. D., Shannon, J. D., Fox, J. W., Vines, R. R., and Parsons, J. T. (1994) Mol. Cell. Biol. 14 1680-1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calalb, M. B., Polte, T. R., and Hanks, S. K. (1995) Mol. Cell. Biol. 15 954-963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker, M., Roberts, R., Enriquez, M., Zhao, X., Takahashi, T., Pat Cerretti, D., Daniel, T., and Chen, J. (2004) Biochem. Biophys. Res. Commun. 323 17-23 [DOI] [PubMed] [Google Scholar]
- 38.Marston, D. J., Dickinson, S., and Nobes, C. D. (2003) Nat. Cell Biol. 5 879-888 [DOI] [PubMed] [Google Scholar]
- 39.Tai, C. Y., Mysore, S. P., Chiu, C., and Schuman, E. M. (2007) Neuron 54 771-785 [DOI] [PubMed] [Google Scholar]
- 40.Balzac, F., Avolio, M., Degani, S., Kaverina, I., Torti, M., Silengo, L., Small, J. V., and Retta, S. F. (2005) J. Cell Sci. 118 4765-4783 [DOI] [PubMed] [Google Scholar]
- 41.Selkoe, D., and Kopan, R. (2003) Annu. Rev. Neurosci. 26 565-597 [DOI] [PubMed] [Google Scholar]
- 42.Ray, W. J., Yao, M., Mumm, J., Schroeter, E. H., Saftig, P., Wolfe, M., Selkoe, D. J., Kopan, R., and Goate, A. M. (1999) J. Biol. Chem. 274 36801-36807 [DOI] [PubMed] [Google Scholar]
- 43.Fortini, M. E. (2001) Curr. Opin. Cell Biol. 13 627-634 [DOI] [PubMed] [Google Scholar]
- 44.Shen, J., Bronson, R. T., Chen, D. F., Xia, W., Selkoe, D. J., and Tonegawa, S. (1997) Cell 89 629-639 [DOI] [PubMed] [Google Scholar]
- 45.Shvartsman, D. E., Donaldson, J. C., Diaz, B., Gutman, O., Martin, G. S., and Henis, Y. I. (2007) J. Cell Biol. 178 675-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin, X., Wang, Y., Ahmadibeni, Y., Parang, K., and Sun, G. (2006) J. Mol. Biol. 357 1263-1273 [DOI] [PubMed] [Google Scholar]
- 47.Cowan-Jacob, S. W., Fendrich, G., Manley, P. W., Jahnke, W., Fabbro, D., Liebetanz, J., and Meyer, T. (2005) Structure 13 861-871 [DOI] [PubMed] [Google Scholar]
- 48.Roskoski, R., Jr. (2004) Biochem. Biophys. Res. Commun. 324 1155-1164 [DOI] [PubMed] [Google Scholar]
- 49.Shattil, S. J. (2005) Trends Cell Biol. 15 399-403 [DOI] [PubMed] [Google Scholar]
- 50.Castano, J., Solanas, G., Casagolda, D., Raurell, I., Villagrasa, P., Bustelo, X. R., Garcia de Herreros, A., and Dunach, M. (2007) Mol. Cell. Biol. 27 1745-1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piedra, J., Miravet, S., Castano, J., Palmer, H. G., Heisterkamp, N., Garcia de Herreros, A., and Dunach, M. (2003) Mol. Cell. Biol. 23 2287-2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingley, E. (2007) Biochim. Biophys. Acta 1784 56-65 [DOI] [PubMed] [Google Scholar]
- 53.Volberg, T., Romer, L., Zamir, E., and Geiger, B. (2001) J. Cell Sci. 114 2279-2289 [DOI] [PubMed] [Google Scholar]
- 54.Brown, M. C., Cary, L. A., Jamieson, J. S., Cooper, J. A., and Turner, C. E. (2005) Mol. Biol. Cell 16 4316-4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katz, B. Z., Romer, L., Miyamoto, S., Volberg, T., Matsumoto, K., Cukierman, E., Geiger, B., and Yamada, K. M. (2003) J. Biol. Chem. 278 29115-29120 [DOI] [PubMed] [Google Scholar]
- 56.Kaplan, K. B., Swedlow, J. R., Morgan, D. O., and Varmus, H. E. (1995) Genes Dev. 9 1505-1517 [DOI] [PubMed] [Google Scholar]
- 57.Takayama, Y., Tanaka, S., Nagai, K., and Okada, M. (1999) J. Biol. Chem. 274 2291-2297 [DOI] [PubMed] [Google Scholar]
- 58.Mitra, S. K., and Schlaepfer, D. D. (2006) Curr. Opin. Cell Biol. 18 516-523 [DOI] [PubMed] [Google Scholar]
- 59.Dehm, S. M., and Bonham, K. (2004) Biochem. Cell Biol. 82 263-274 [DOI] [PubMed] [Google Scholar]
- 60.Zaidel-Bar, R., Milo, R., Kam, Z., and Geiger, B. (2007) J. Cell Sci. 120 137-148 [DOI] [PubMed] [Google Scholar]
- 61.Zaidel-Bar, R., Cohen, M., Addadi, L., and Geiger, B. (2004) Biochem. Soc. Trans. 32 416-420 [DOI] [PubMed] [Google Scholar]
- 62.Takasu, M. A., Dalva, M. B., Zigmond, R. E., and Greenberg, M. E. (2002) Science 295 491-495 [DOI] [PubMed] [Google Scholar]
- 63.Battaglia, A. A., Sehayek, K., Grist, J., McMahon, S. B., and Gavazzi, I. (2003) Nat. Neurosci. 6 339-340 [DOI] [PubMed] [Google Scholar]
- 64.Fischer, A., and Gessler, M. (2007) Nucleic Acids Res. 35 4583-4596 [DOI] [PMC free article] [PubMed] [Google Scholar]