Abstract
The ubiquitous thioredoxin fold proteins catalyze oxidation, reduction, or disulfide exchange reactions depending on their redox properties. They also play vital roles in protein folding, redox control, and disease. Here, we have shown that a single residue strongly modifies both the redox properties of thioredoxin fold proteins and their ability to interact with substrates. This residue is adjacent in three-dimensional space to the characteristic CXXC active site motif of thioredoxin fold proteins but distant in sequence. This residue is just N-terminal to the conservative cis-proline. It is isoleucine 75 in the case of thioredoxin. Our findings support the conclusion that a very small percentage of the amino acid residues of thioredoxin-related proteins are capable of dictating the functions of these proteins.
The thioredoxin fold is the core scaffold of numerous proteins that control disulfide redox activity in the cell (1-3). These redox proteins share very little sequence homology, but all of them incorporate the four-stranded β-sheet, three flanking α-helices, and the redox-active CXXC motif of the TRX5 fold (Fig. 1A). The archetype of the family is thioredoxin (4), a disulfide reductase that maintains a reducing cytosolic environment. Other TRX fold redox proteins include the Dsb proteins (1), which regulate the formation of disulfide bonds in prokaryotes, and protein-disulfide isomerase (5), which catalyzes the oxidation and shuffling of disulfides in the endoplasmic reticulum of eukaryotic cells.
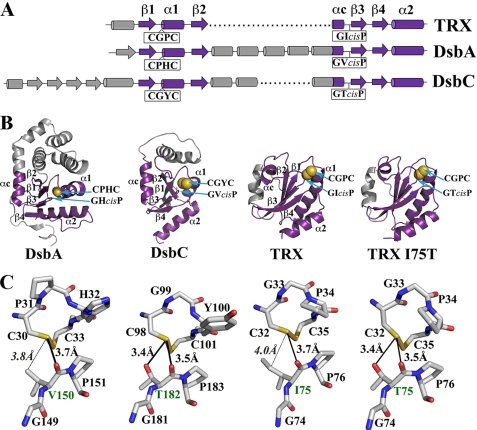
FIGURE 1.
A, schematic representation of the secondary structure elements of E. coli thioredoxin, DsbA, and DsbC (DsbG has secondary structure elements similar to those of DsbC). The characteristic elements of the thioredoxin fold (β1 α1 β2 and β3 β4 α2 motifs linked by a short α helix (αc)) are shown in violet, and insertions in the thioredoxin fold are colored in gray. The positions of the CXXC catalytic motif and the cis-proline loop in each protein are indicated. B, ribbon diagrams of the structures of E. coli DsbA (PDB code 1FVK (54)), the thioredoxin domain of DsbC (oriented using DsbA as a reference; PDB code 1EEJ (39)), thioredoxin (PDB code 2TRX (55)), and thioredoxin I75T variant (PDB code 3DYR (this work)). The thioredoxin domains are shown in violet, the inserted domains in gray, and the active site cysteines in a yellow space-filling representation. The thioredoxin fold elements, CXXC and cis-proline loop, are labeled. C, active sites of E. coli DsbA, DsbC, thioredoxin, and thioredoxin I75T variant showing the catalytic CXXC motif and the cis-proline loop. The distances between the sulfur of the N-terminal nucleophilic cysteine (Cys-30, Cys-98, and Cys-32 for DsbA, DsbC, and thioredoxin, respectively) and the main chain oxygen of the cisPro-1 residue (Val-150, Thr-182, Ile-75, and Thr-75 for DsbA, DsbC, thioredoxin, and thioredoxin I75T, respectively) are indicated. For DsbC and thioredoxin I75T, the distances to the hydroxyl group of Thr-182 and Thr-75 are shown (for comparison, the distances to the CG1 atom of Ile-75 in thioredoxin and the CG2 of Val-150 in DsbA are also indicated). Hydrogen bonds are represented by a thick black line. In the absence of a hydrogen bond, distances are shown with a thin black line. The cisPro-1 residue is labeled in green. This figure was generated using MacPyMOL (15).
This wide range of redox activities of TRX fold proteins is thought to be a consequence of modifications to the common scaffold, which result in different redox properties. Thus, the redox potential of Escherichia coli thioredoxin is very reducing, at -271 mV (6, 7), whereas that of the oxidizing periplasmic protein E. coli DsbA is -120 mV (8). Thioredoxin fold proteins that participate in a wide range of thiol disulfide exchange reactions, such as the eukaryotic protein-disulfide isomerases, have intermediate redox potentials (around -160 mV (9)).
Thioredoxin-related proteins provide an attractive model for the study of how protein function is dictated by sequence and three-dimensional structure; this is because their functions are, in part, determined by their redox properties, which in turn, are easy to quantify. For example, mutations in thioredoxin that make its redox potential more oxidative complement null mutations in the oxidase DsbA (10, 11). A detailed understanding of how thioredoxin fold sequence affects redox properties provides an excellent opportunity to relate sequence and function. Previous work has focused on the role of the CXXC “redox rheostat” active site in determining the properties of thioredoxin-related proteins (3, 12, 13). Experiments that exchange the X-X dipeptide of one thiol-disulfide oxidoreductase with that of another generally result in an oxidoreductase with a redox potential partially shifted in the direction of the oxidoreductase protein that served as the source of the dipeptide (3, 12-14).
Although the X-X dipeptide is important, it is not the only feature that influences thioredoxin fold protein function. Another extremely well conserved but much less studied region of thioredoxin-like proteins is the loop containing a cis-proline, which closely approaches the CXXC motif (5). Biophysical studies of E. coli thioredoxin and DsbA indicate that the conserved cis-proline residue plays a significant role in the stability and structure of these proteins (16, 17). Other studies have identified additional interesting roles for the cis-proline loop. For example, mutation in E. coli DsbA of the cis-proline residue causes accumulation of mixed disulfide intermediates with substrates in vivo (18), suggesting that the cis-proline plays a role in substrate release. Furthermore, studies on human thioredoxin revealed that its cis-proline prevents metal binding by the active site thiolates (19). Structural evidence from both thioredoxin and DsbA indicates that the cis-proline loop is involved in substrate binding (20-22). Finally, mutations in the residue just N-terminal to the cis-proline (the cisPro-minus1 residue) in DsbG give rise to variants that gained DsbC-like isomerase activity (23).
The side chain of the cisPro-minus1 residue is usually within 4 Å of the reactive N-terminal cysteine of the CXXC motif, giving this residue the potential to be involved in hydrogen bonding and hydrophobic interactions with the nucleophilic cysteine and thereby to modulate its activity (see Fig. 1).
To investigate the role of the cisPro-minus1 residue, this study tests the effect of mutations on the activities and redox properties of thioredoxin fold proteins. We examined four divergent members of the thioredoxin family: E. coli DsbA, a strong oxidase; E. coli thioredoxin, which is relatively reducing; E. coli DsbC a protein-disulfide isomerase; and E. coli DsbG, a thioredoxin-related protein with redox properties similar to those of the isomerase DsbC but with no known in vivo substrates. The results show that the cisPro-minus1 residue is universally important in determining the redox properties of thioredoxin fold proteins. It also seems to be crucial in regulating the ability of these proteins to interact with both upstream and downstream substrates.
EXPERIMENTAL PROCEDURES
Bacterial strains and plasmids used in the study are listed in supplemental Table S1. Plasmid construction, site-directed mutagenesis, protein purification, reduction, oxidation, concentration determination, and sequence alignment are described in the supplemental “Experimental Procedures.”
Redox Potential Measurement—The redox potentials of wild type (WT) DsbA, DsbG, DsbC, and their mutants and the thioredoxin mutants CPHC and I75T/CPHC were measured by incubation in degassed redox buffers containing various concentrations of GSH/GSSG, as described by Bessette et al. (24). The redox potentials of thioredoxin and other mutants were determined by redox equilibria with cDsbD, mainly as described by Collet et al. (25). The redox potential of cDsbD (-241 mV) was used as a standard.
Determination of pKa Values—To determine the pH-dependent ionization of the nucleophilic cysteine, we recorded the specific absorbance of the thiolate anion at 240 nm (26). We also monitored the pH-dependent absorbance of the oxidized form as a reference. Measurements were carried out at 25 °C in a buffer consisting of 10 mm K2HPO4, 10 mm boric acid, 10 mm sodium succinate, 1 mm EDTA, and 200 mm KCl, pH 7.5, for DsbA, DsbC, DsbG, and mutants, with an average initial protein concentration of 20 μm. The pH of the protein solution was lowered to 2.2 by the stepwise addition of aliquots of 0.2 m HCl. Absorbances at 240 and 280 nm were recorded on a Cary 50BIO UV-visible spectrophotometer and corrected for the volume increase. For thioredoxin and thioredoxin mutants, measurements were carried out at 25 °C in a buffer consisting of 10 mm NaH2PO4, 10 mm sodium citrate, 10 mm sodium borate, pH 8.5. The pH of the protein solution was lowered to 3 by the stepwise addition of aliquots of 0.2 m HCl. Absorbances at 240 and 280 nm were recorded on a Cary 100BIO UV-visible spectrophotometer and corrected for the pH decrease and volume increase. All buffers were degassed, and purged with nitrogen before experiments. The pH dependence of the thiolate-specific absorbance signal (S = (A240/A280)reduced/(A240/A280)oxidized) was fit according to the Henderson-Hasselbach equation.
Reductase Activity Assay—The ability of DsbA, DsbG, thioredoxin, DsbC, and variants to catalyze the reduction of human insulin in the presence of DTT was tested essentially as described by Holmgren (27). A stock solution of 872 μm insulin was freshly prepared in 0.1 m potassium phosphate buffer, pH 7.0, and 2 mm EDTA before each assay. The reaction mixtures were prepared directly in cuvettes using 0.1 m potassium phosphate buffer, pH 7.0, 2 mm EDTA, and 0.33 mm DTT with 5 μm enzyme in a final volume of 0.8 ml. The reactions were started by adding insulin to a final concentration of 131 μm. After thorough mixing, the cuvettes were placed in the spectrophotometer, and measurements were performed at 650 nm for 120 min. In all experiments, the uncatalyzed reduction of insulin by DTT was monitored in a control reaction without the addition of thioredoxin-related oxidoreductases.
The ability of thioredoxin and variants to catalyze the reduction of human insulin in the presence of thioredoxin reductase and NADPH was tested as described by Lundström and Holmgren (28). The reaction mixtures were prepared directly in cuvettes using 0.1 m potassium phosphate buffer, pH 7.0, 2 mm EDTA, 0.2 mm NADPH, 0.1 μm thioredoxin reductase, 5 μm thioredoxin, and variants in a final volume of 0.8 ml. The reactions were started by adding insulin to a final concentration of 100 μm.
Oxidase Activity Assay—The oxidative folding of hirudin was tested essentially as described by Quan et al. (11). In brief, oxidase activity was determined by the rate at which oxidized DsbA, DsbG, thioredoxin, or their variants donate a disulfide bond to reduced hirudin. The fluorescence excitation at 295 nm increases when oxidized DsbA, DsbG, or thioredoxin loses its active site disulfide bond; however, hirudin itself shows no fluorescence change during the reaction. The assay was performed on a KinTek SF-2004 stop flow instrument in single-mixing mode. The traces consisted of a minimum of three to five successive stop flow experiments, and the data for each trace were fit individually to a single exponential equation to obtain the pseudo first-order rate constant Kobs. Averaged Kobs values were plotted against the hirudin concentration. The slope is the observed second-order rate constant of the reaction, which indicates the oxidase activity.
Isomerase Activity Assay with Scrambled Hirudin as a Substrate—Scrambled hirudin refolding was tested essentially as described by Hiniker et al. (23). Hirudin samples were diluted to 24 μm and incubated with or without 24 μm reduced DsbA, DsbG, thioredoxin, or their variants in 100 mm sodium phosphate, 1 mm EDTA, pH 7.0. Folding reactions were quenched by addition of 10% (v/v) formic acid following incubation at room temperature at various time points. The reaction products were separated by reverse-phase HPLC on a Vydac™218TP54 C18 column at 55 °C using an acetonitrile gradient (19-25%, 30 ml) in 0.1% (v/v) trifluoroacetic acid. The eluted proteins were detected by their absorbance at 220 nm.
Refolding of Scrambled RNase A—To determine the in vitro isomerase activity of DsbA V150T and DsbC T182V, we also utilized the scrambled RNase A refolding assay. Reduced denatured RNase A (0.5 mg/ml) was incubated in 50 mm Tris-HCl, pH 8.5, and 6 m GdmCl for at least 3 days in the dark at room temperature to prepare scrambled RNase A (29). The randomly reoxidized RNase A was concentrated, and after acidifying the solution, the oxidation of disulfide bonds was confirmed using Ellman's assay. Reshuffling of scrambled RNase A (40 μm) was carried out by incubation in 100 mm phosphoric acid-NaOH, pH 7, 1 mm EDTA, with 10 μm oxidized DsbC, DsbCT182V, DsbA, or DsbA V150T (DsbA, DsbC, and mutants were oxidized with 1.7 mm copper(II)[1,10-phenanthroline]). The reactions were started by the addition of DTT to a final concentration of 10 μm. As a positive control, we carried out an additional reaction using folded RNase A without any thioredoxin fold protein. The assay was performed at 25 °C, and samples were taken at several time points and assayed for RNase A activity by monitoring cCMP hydrolysis spectrophotometrically at 296 nm for 2.5 min. The fraction of native RNase A (%) was plotted against incubation time.
Kinetics of DsbA V150T Interaction with DsbB—Oxidation of reduced DsbA and variants by DsbB and Q1 was tested according to Tapley et al. (30). Briefly, a typical reaction contained 100 μm freshly reduced DsbA or V150T, 10 μm DsbB, and 200 μm ubiquinone-1 (coenzyme Q1, Sigma). DsbA and a DsbB/coenzyme Q1 mixture were incubated in 50 mm sodium phosphate, 300 mm NaCl, and 0.04% dodecyl maltoside, pH 8.0, at 25 °C before mixing. The absorbance after mixing was recorded at 510 nm. One data set contained three to five successive injections, and data for each trace were analyzed separately. The resulting data were further fit using SigmaPlot and the Michaelis-Menten equation to give Km and kcat values.
Spot Titers for Cadmium Resistance and Copper Resistance—Spot titers for cadmium resistance were performed to quantify the relative oxidase activity of DsbA and variants in vivo. Spot titers for copper resistance were performed to quantify the relative reductase activity caused by mutations in thioredoxin in vivo. Briefly, strains were grown overnight in LB-ampicillin (200 mg/ml) and diluted 1:100 into fresh LB-ampicillin (200 mg/ml). Strains were grown to mid-logarithmic phase at 37 °C and serially diluted into 150 mm NaCl. 2 μl of each dilution was plated onto LB-ampicillin plates (200 mg/ml) with 40 μm cadmium (for oxidase assay) or TB plates with 17 mm CuCl2 (for reductase assay) as described previously (23, 31); cells were grown at 37 °C overnight. All spot titers were performed in duplicate or triplicate.
4-Acetoamido-4′-maleimidylstilbene2,2′-disulfonicacid(AMS) Trapping—Strains were grown overnight in LB-ampicillin (200 mg/ml) and diluted 1:100 into fresh LB-ampicillin (200 mg/ml). Strains were grown to mid-logarithmic phase at 37 °C, and DsbA or mutant protein were expressed by induction using 1 mm isopropyl 1-thio-β-d-galactopyranoside for 10 min. Cells were acid-precipitated overnight equivalent to 0.5 OD units. Acid-precipitated proteins were solubilized in buffered SDS solution containing 10 mg/ml AMS. The samples were incubated in the dark at 37 °C for 1 h. Alkylation was stopped by the addition of reducing SDS loading buffer and analyzed by electrophoresis and Western blotting.
Protein Structure Determination—Purified thioredoxin I75T (15 mg/ml) was crystallized using sitting-drop vapor diffusion. Crystals were grown in 30% polyethylene glycol 8000, 150 mm sodium acetate, and 100 mm sodium cacodylate, pH 6.5. The crystals belong to the C2 space group with unit cell dimensions of a = 58.8 Å, b = 39.4 Å, c = 90.6 Å, and β = 96.6°. There are two molecules in the asymmetric unit. A 2.0 Å resolution data set was collected using X-rays generated by a Rigaku rotating anode x-ray generator. The structure was solved by molecular replacement as implemented in the CNS program (32) using the WT E. coli thioredoxin structure as a search model (2TRX). The model was initially refined using the CNS program (32) with rebuilding in O (33) followed by refinement with Refmac5 (34) from CCP4 suite (35) and finally in Phenix (36). TLS parameters were incorporated during the refinement process. Each thioredoxin molecule in the asymmetric unit was defined as one TLS group. The final model has a working R-factor of 25.1% and a free R-factor of 29.0%. The modest R-factors are largely because the crystal used in data collection was slightly twinned. Data collection and structure refinement statistics are shown in supplemental Table S3.
RESULTS
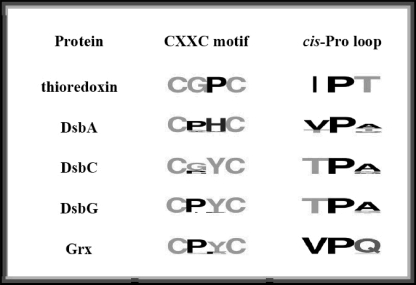
The cisPro-minus1 Residue Regulates the Redox Potential of Thioredoxin Fold Proteins—Sequence alignment of the various subfamilies of thioredoxin fold proteins revealed that the cis-Pro-minus1 residue tends to be conserved within each subfamily but differs between different subfamilies (Table 1). Isoleucine is common in thioredoxins, threonine is common in the prokaryotic disulfide isomerases DsbC and DsbG, and valine is common in DsbA and glutaredoxins. Given this pattern of conservation, the close proximity in three-dimensional space of this residue to the active site (see Fig. 1) and the existence of gain and loss of function mutations that affected this cisProminus1 residue in DsbG (23), we were intrigued by the possibility that the character of the cisPro-minus1 residue helps to determine the divergent properties of the various subfamilies of thioredoxin fold proteins. Thus, DsbA, DsbC, DsbG, and thioredoxin cisPro-minus1 mutants (Table 2) were constructed on the basis of the simple principle of exchanging the side chain common to one subfamily with that present in another subfamily. For example, the cisPro-minus1 residue in the oxidase DsbA is nearly always valine; this was exchanged with threonine, the residue most commonly found in the isomerases DsbC and DsbG, to generate the variant DsbA V150T.
TABLE 1.
Sequence of the CXXC motif and cisPro loop
The Sequences of each protein family surrounding CXXC and cis-proline were compiled and used to generate sequence logos. The size of the amino acid single-letter code is proportional to the occurrence of that amino acid at that position. To determine the conservation of the residues in the CXXC and cis-proline loop in these proteins, we analyzed the proteins from genomes that are as divergent as possible; however, we wanted to avoid comparing proteins that have been evolving over different evolutionary time frames. Thioredoxin and glutaredoxin, for instance, are present in eukaryotes, bacteria, and archaea, and therefore have been evolving for at least 3.8 billion years (53), whereas DsbC and DsbG are restricted to proteobacteria and have probably been evolving for at least ~0.5 billion years (23, 53). Thus we restricted our comparison to genomes that contain an orthologue to DsbC. We used our previous alignment of all species-specific DsbC sequences available in GenBank™ (23) to obtain the list of organisms that contain DsbC. We then obtained the sequence of the individual thioredoxin, DsbA, DsbG, and glutaredoxin orthologues present in these individual genomes using blast by searching with the E. coli homologue.
TABLE 2.
Parameters for DsbA, DsbG, thioredoxin, DsbC, and their variants
N.D., not determined. The lack of redox-dependent fluorescence in DsbC and its mutants means that these cannot be assayed for oxidase activity in this manner. The `+', `-' designations refer to `+++++' the best isomerase activity among all the proteins listed; `-' no detectable isomerase activity.
| Protein | Redox potential | pKa | Reductase | Oxidase | Isomerase |
|---|---|---|---|---|---|
| mV | A−1 min−2 μm−1 | μm−1S−1 | |||
| DsbA | −120 ± 2 | 3.3 ± 0.1 | 1.74 ± 0.06 × 10−4 | 1.76 ± 0.12 | ++++ |
| V150T | −92 ± 1 | 3.5 ± 0.1 | 0.13 ± 0.01 × 10−4 | 2.20 ± 0.014 | ++ |
| DsbG | −127 ± 2 | 3.5 ± 0.1 | <0.01 × 10−4 | 0.0043 ± 0.0004 | − |
| T200L | −178 ± 4 | N.D. | 0.22 ± 0.01 × 10−4 | 0.0043 ± 0.0004 | + |
| T200Ma | −181 ± 3 | N.D. | 0.23 ± 0.1 × 10−4 | N.D. | + |
| Thioredoxin | −271 ± 3 | 7.0 ± 0.2 | 14.9 ± 0.2 × 10−4 | 0.090 ± 0.003 | ++++ |
| 175T | −226 ± 2 | 5.5 ± 0.2 | 33.1 ± 0.3 × 10−4 | 0.123 ± 0.004 | ++++++ |
| DsbC | −143 ± 2 | 4.6 ± 0.1 | 27.6 × 10−4 | N.D. | ++++ |
| T182V | −195 ± 2 | 5.8 ± 0.1 | 33.9 × 10−4 | N.D. | ++++++ |
Parameters for DsbG T200M are from Hiniker et al. (23).
The redox potentials of the mutant proteins were measured (Table 2, supplemental Fig. S1). When threonine replaces the hydrophobic valine or isoleucine residue at the cisPro-minus1 position, the redox potential is substantially increased. Thus threonine exerts a more oxidizing influence than valine. For the DsbA V150T mutant, for instance, the equilibrium constant with glutathione is 1.0 × 10-5 m. This translates to an intrinsic redox potential (Eo′) of -92 mV. DsbA V150T is thus substantially more oxidizing than WT DsbA, Keq = 8.67 × 10-5 m, Eo′=-120 mV, making it as oxidizing as DsbL (37), the most oxidizing disulfide catalyst known. In contrast, when a hydrophobic side chain replaces the more hydrophilic threonine, as in the DsbG T200L, DsbG T200M, or DsbC T182V mutants, the redox potential of proteins is substantially decreased. Thus a hydrophobic residue at the cisPro-minus1 position exerts a reducing influence on the protein (Table 2).
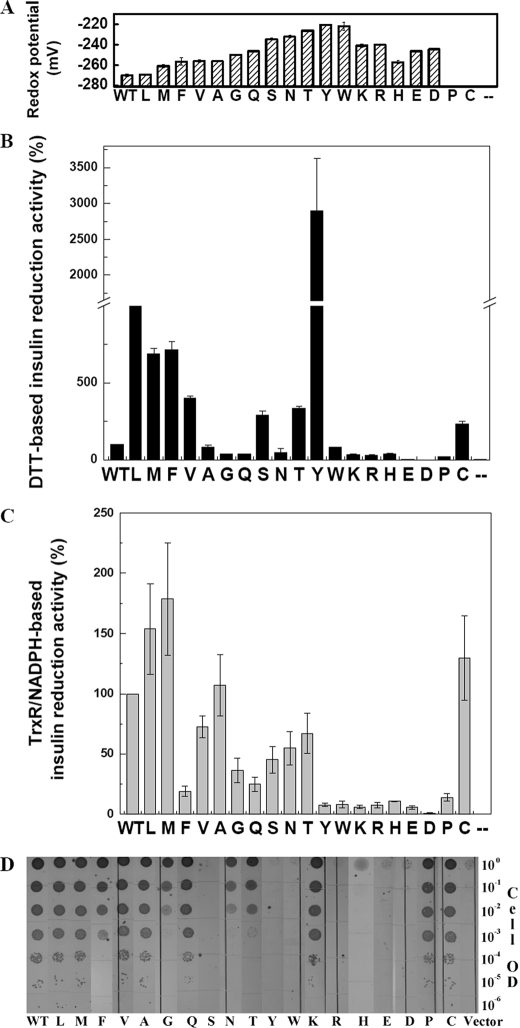
To further explore the effects of side chain substitutions at the cisPro-minus1 position, we examined the effect of altering this residue in thioredoxin (Ile-75) to all other possible amino acid side chains. We characterized the mutants in vivo and in vitro, as shown in supplemental Table S2 and Fig. 2. All substitutions at the Ile-75 position alter the redox potential of the thioredoxin (Fig. 2A). Every substitution made thioredoxin more oxidizing, consistent with the idea that evolution has maximized the reducing power of thioredoxin. These results show that the cisPro-minus1 residue has a very strong influence on the redox properties of thioredoxin proteins. Surprisingly, these changes in redox potential are very similar in magnitude to those within the active site CXXC motif of thioredoxin, suggesting that the cisPro-minus1 residue in thioredoxin fold proteins has just as important a role in determining the redox potential of these proteins as the comprehensively studied CXXC residues (3, 11-14, 38). In contrast, substitutions in the residue just C-terminal to the cis-proline (the cisPro-plus1 position) had very little effect on the redox properties of various thioredoxin fold proteins (data not shown), leading to the conclusion that this residue is not important in determining the redox properties of these proteins.
FIGURE 2.
In vitro and in vivo characterization of thioredoxin Ile-75 variants. The variants are listed as shown in the bottom of each panel. A, redox potentials of thioredoxin and mutants except for the I75C and I75P mutants. Reduced I75C and I75P are too rapidly oxidized by air to allow for an accurate measurement of redox potential. B, in vitro reductase activity of thioredoxin and mutants, insulin reduction activity using DTT as the reducing source. C, in vitro reductase activity of thioredoxin and mutants, insulin reduction activity using NADPH with thioredoxin reductase as the reducing source. D, in vivo reductase activity of thioredoxin and mutants as measured by copper sensitivity. The gradient indicates a serial dilution of trxA null cells harboring thioredoxin and its variants (from 1 to 10-6 OD).
The crystal structures of thioredoxin fold proteins show that the cis-proline loop and the X-X dipeptide interact with the active site cysteines from opposite directions (Fig. 1), raising the possibility that they exert their influence independently of each other. To explore this question, we combined two of the most oxidizing thioredoxin mutations: the cisPro-minus1 I75T mutation (Table 2) with the CPHC DsbA-like active site sequence mutation (11) to generate the double mutant. The double mutant has a redox potential of -180 mV, 91 mV more oxidizing than the wild type and thus by far the most oxidizing thioredoxin mutant that we know. Significantly, the increase in the redox potential in the double mutant of 91 mV is almost additive of the increases conferred by the individual mutations, I75T (45 mV, Table 2) and CPHC (42 mV from supplemental Fig S1C or 60 mV using the value from Ref. 10). These results raise the interesting possibility that the majority of the redox properties of thioredoxin fold proteins are determined by a very small number of residues.
To further investigate what makes the thioredoxin I75T mutant more oxidizing than WT thioredoxin, we solved the crystal structure of the oxidized form of the protein to a resolution of 2.0 Å (supplemental Table S3). The thioredoxin I75T structure shows that the mutation has little effect on the overall structure of the protein; the root mean square deviation to the wild type protein structure is 0.51 Å (Fig. 1B, supplemental Fig. S4). However, the I75T substitution positions the hydroxyl group of Thr-75 within hydrogen bond distance (3.4 Å) of the nucleophilic Cys-33 sulfur (Fig. 1C). This hydrogen bond is absent in WT thioredoxin, as the isoleucine side chain at position 75 lacks an electronegative oxygen or nitrogen atom. Interestingly, hydrogen bonds with the equivalent nucleophilic cysteines are observed in DsbC (39) and DsbG (40) structures (Fig. 1C) and have been proposed to be partially responsible for the oxidizing redox potential of these isomerases (40). These observations suggest that a small change in the local environment of the CXXC motif contributes to the dramatic change in the redox properties of the variant. This is substantiated by our analysis of the pKa values of the active site nucleophilic cysteine of the thioredoxin fold variants. In most cases, mutation of the cisPro-minus1 residue resulted in changes in the pKa values of the nucleophilic cysteine, and in some cases these changes were substantial (Table 2). In general our data agree with previous observations suggesting that as the pKa of the nucleophilic cysteine is lowered the disulfide bond becomes easier to reduce, increasing the intrinsic redox potential of the protein (38). In the case of the oxidase DsbA, replacement of WT Val by Thr had little effect on the pKa (3.3 ± 0.1 and 3.5 ± 0.1, respectively), despite the considerable difference in redox potential of the two proteins (Table 2, supplemental Fig. S1G). A value of 3.3-3.5 for the pKa may represent a lower limit of measurement when using this method.
The cisPro-minus1 Residue Influences in Vitro Enzyme Activity—Redox potential is a thermodynamic measure of the redox properties of a protein. To determine whether the cisProminus1 residue contributes to redox function, we analyzed the ability of the thioredoxin fold mutants to participate in thiol exchange reactions by measuring their ability to reduce disulfides, oxidize cysteines, and isomerize incorrect disulfide bonds.
Perhaps the most widely used kinetic assay for thiol-disulfide oxidoreductases is measurement of the rate at which these proteins accelerate the reduction of disulfides in insulin (27). We initially used DTT as the electron donor in this assay, which allowed a direct comparison of the reductase activity of the cisPro-minus1 variants in DsbA, DsbG, DsbC, and thioredoxin. As shown in Figs. 3A and 2B and supplemental Fig. S2A, mutation of the cisPro-minus1 residue in DsbA, DsbC, DsbG, and thioredoxin significantly affects the reductase activity of these proteins, confirming the key role this residue plays in redox enzymatic activity. In the case of Dsb proteins, the cisPro-minus1 modification affected the reductase activity in ways that reflected changes in the redox potential (see Table 2). Thus, DsbA V150T has a substantially more oxidizing redox potential and, not surprisingly, is less efficient in reducing insulin. The cisPro-minus1 mutants of DsbC and DsbG are more efficient in reducing insulin, in keeping with their more reducing redox potentials.
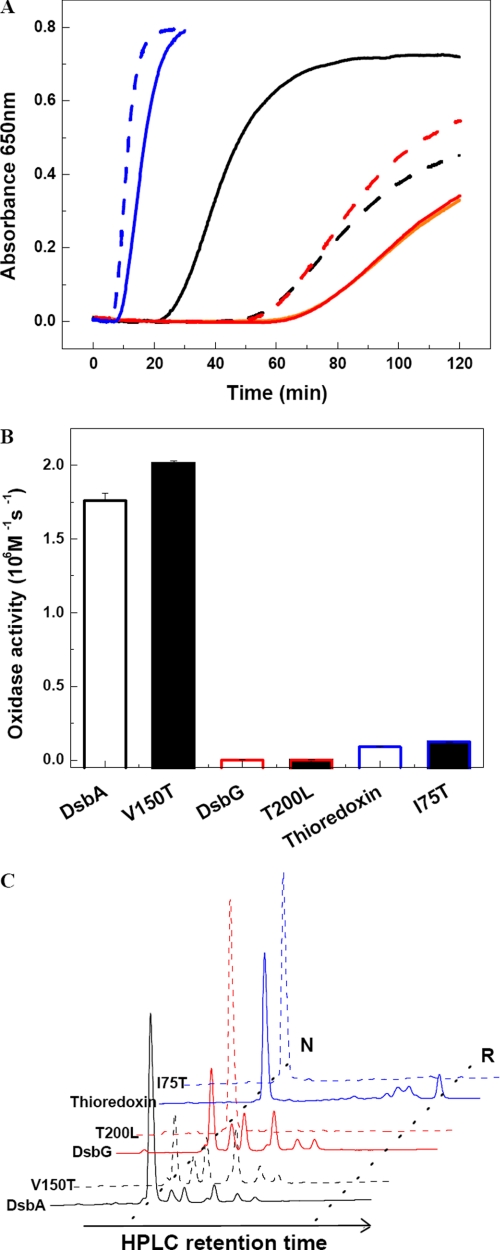
FIGURE 3.
In vitro activity assays for DsbA, DsbG, and thioredoxin cisPro-1 variants. A, reductase activity was assayed using insulin and DTT as substrates for DsbA (black), DsbG (red), and thioredoxin (blue) (same in B and C). The orange line shows the DTT-only control. WT proteins are shown with solid lines and cisPro-1 mutants with dashed lines, sample names are as indicated in the panels. B, oxidase activity was assayed using reduced hirudin as a substrate. C, isomerase activity was assayed with scrambled hirudin as a substrate. The scrambled hirudin starting material is shown in Fig. S2C. Equimolar quantities of reduced proteins and scrambled hirudin were incubated for 30 min for the DsbA and thioredoxin samples and 18 h for the DsbG samples. The reactions were then acid-quenched, and samples were analyzed by reverse-phase HPLC on a Vydac™218TP54 C18 column. N and R indicate native and reduced hirudin, respectively.
Surprisingly, about half of the 19 thioredoxin variants are more active than WT thioredoxin in accelerating DTT-mediated insulin reduction, some spectacularly so. Yet all thioredoxin mutants have more oxidizing redox potentials than the WT enzyme. Generally, mutations have a detrimental effect on activity of proteins; sophisticated directed evolution experiments are generally required to isolate proteins with enhanced activity (41, 42). Thus the observation that many of the mutations in the thioredoxin cisPro-minus1 residue have improved activity in DTT-mediated insulin reduction is striking. Mutants that exhibit higher activity than WT thioredoxin can be divided into two groups according to the properties of their cisProminus1 side chains. The mutants in group I, where Ile-75 is exchanged for another large hydrophobic group such as Phe, Leu, Met, or Val have redox potentials similar to WT (-257 to -270 mV versus -271 mV) (supplemental Table S2) but show very large increases in DTT-mediated insulin reduction. The mutants in group II (I75C, I75S, and I75T) have a free hydroxyl or thiol group in the side chain of the cisPro-minus1 residue and so can potentially interact with the active site cysteines. These group II variants also show large increases in DTT-mediated insulin reduction activity, despite their more oxidizing redox potentials (supplemental Table S2). Interestingly, the thioredoxin variant I75Y, which has an aromatic ring and a free hydroxyl, apparently shows the effects of both groups I and II, as it has a 30-fold increase in DTT-mediated insulin reduction activity compared with WT thioredoxin. In all, seven substitutions at the cisPro-minus1 position had activity >250% of WT thioredoxin in this assay. On the other hand, charged residues at this position had decreased activity, with I75D and I75E completely inactive.
We also measured the oxidase activity of the proteins by looking at the rate at which the variant proteins donate their disulfide bond to substrates like hirudin. A disulfide donation step is one of several reactions that need to occur during enzymatic thiol-disulfide oxidoreductase reaction cycles. None of the mutants significantly altered the rate of this reaction step (Fig. 3B), which implies that alterations in the kinetics of other steps in the reaction cycle, such as the oxidation of DsbA by insulin, are more important in explaining the changes in enzymatic activity seen in DTT-mediated insulin reduction.
Isomerase activity requires the ability of enzymes to reduce mismatched protein disulfides and subsequently form correct disulfides; this requires both reductase and oxidase activity. As the oxidase activity of the cisPro-minus1 variants was essentially unaltered compared with the WT thioredoxin fold proteins, it is not surprising that the isomerase activity of the variants was altered in the same way as their reductase activity (Fig. 3C and supplemental Fig. S2). DsbC T182V showed a slightly increased reductase activity but showed greatly decreased isomerase activity (supplemental Fig. S2B). We predicted that T182V would have decreased oxidase activity compared with WT based on its redox potential and pKa. This case provides a further indication that both reductase and oxidase activity contribute to the ability of a protein to function as an isomerase.
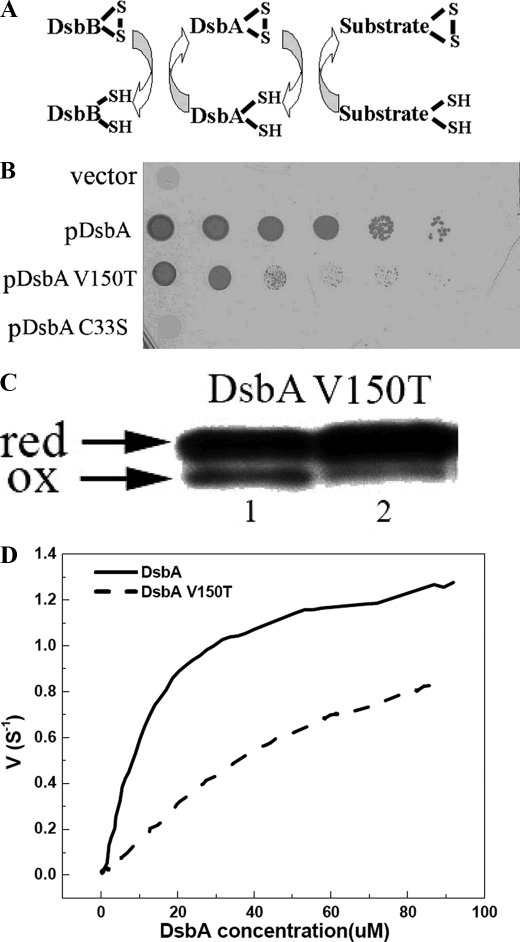
The cisPro-minus1 Residue Is Involved in Interactions with Upstream Partner Proteins—We also investigated the role of the cisPro-minus1 residue in contributing to interactions with partner proteins. One way to do this is by studying the in vivo phenotype of DsbA V150T and its ability to be reoxidized by the membrane protein DsbB (Fig. 4) (43). Negatively influencing the interaction between DsbA and its downstream substrates (partially folded proteins) or its upstream partner (DsbB) should impair disulfide bond formation in vivo so that the in vivo effects would be similar to those seen in dsbA- strains. For example, dsbA- strains are sensitive to cadmium because of the high affinity of Cd2+ for protein thiol groups (11, 44). Similarly, E. coli-expressing DsbA V150T is cadmium-sensitive (Fig. 4B) showing that the cisPro-minus1 residue is important for in vivo DsbA function. Given that DsbA V150T is equivalent to WT DsbA (Fig. 4B) in oxidizing downstream substrates such as hirudin, at least in vitro, it is possible that the in vivo phenotype is a result of reduced affinity of DsbA V150T for its upstream partner DsbB. Supporting this explanation, the Km for interaction with DsbB is 5-fold higher for the V150T variant than for WT DsbA (98 versus 16 μm) (Fig. 4D). Similarly, the variant DsbA V150G is 40-fold slower than WT DsbA in the rate at which it is reoxidized by DsbB (10). Moreover, the DsbG T200L mutant is reduced only very slowly by its upstream partner, DsbD (supplemental Fig. S3C). Thus, mutation of the cisPro-minus1 residue in both DsbA and DsbG appears to result in a defect in the interaction with their respective upstream partners.
FIGURE 4.
Characterization of DsbA and mutant V150T-DsbB interaction in vivo and in vitro. A, E. coli disulfide bond formation pathway in the periplasm. B, DsbA null mutants are very sensitive to cadmium. In vivo DsbA oxidase activity was examined by measuring the cadmium resistance of DsbA null mutant strains expressing various mutants of DsbA on a plasmid. Spot titers were performed on LB plates with 40 μm cadmium. C33S, represents the DsbA active site mutant of CXXS. C, an acid trapping assay shows the slow oxidation of DsbA V150T by DsbB in vivo. Expression of DsbA and mutants was induced 10 min before AMS acid trapping. Reduced DsbA shows a 1-kDa upshift on SDS-PAGE. D, in vitro measurement of Km and kcat of DsbB catalyzing the oxidation of DsbA and variants by multi-turnover assay. The curves were fit from the average of three independent experiments. Original data are shown in supplemental Fig. S3A.
We also investigated the interaction of the thioredoxin cis-Pro-minus1 variants with their upstream partner by testing the ability of the variants to reduce insulin in the presence of their physiological reductant, thioredoxin reductase, rather than DTT. Once again, the cisPro-minus1 residue strongly affected the rates of insulin reduction (Fig. 2C and supplemental Fig. S3B). In many cases the activities paralleled those obtained using DTT as the reductant (compare Fig. 2, panels B and C). However, in some cases the variants have the opposite effect on activity. For instance, the thioredoxin I75Y variant, which was so spectacularly active in DTT-dependent insulin reduction (Fig.2B), was almost completely in active in thioredoxin reductase-dependent insulin reduction (Fig. 2C), suggesting that the cis-Pro-minus1 residue affects the interaction with the reductant. E. coli thioredoxin null mutant strains are Cu2+-sensitive due to their inability to keep DsbD reduced (31, 45). Thus, copper sensitivity can be used to monitor in vivo reductase activity of thioredoxin and its variants. We examined the in vivo activity of the thioredoxin cisPro-minus1 variants by measuring copper sensitivity. Cells expressing these mutants are all more copper-sensitive than cells expressing WT thioredoxin (Fig. 2D). These results suggest that in thioredoxin, as well as in DsbA and DsbG, the nature of the cisPro-minus1 residue affects interaction with upstream partner proteins and that this residue has been optimized by evolution.
DISCUSSION
Nature often uses the same scaffold in functionally related proteins and modifies the common architecture to give rise to different functionalities. One case that exemplifies this process is the thioredoxin fold protein family. Proteins within this family share only limited sequence homology, yet they have a remarkable structural resemblance (1) and a broad range of redox activities. This is achieved in part through the identity of residues in the X-X dipeptide between the catalytic cysteines of the conserved CXXC motif (13, 46).
The redox modulating effect of the X-X residues prompted us to investigate another highly conserved motif: the cis-proline loop, which is distant in linear sequence but very close in space to the CXXC active site motif. We found that the residue that precedes the cis-proline is at least as important in determining the redox properties of thioredoxin fold proteins as the well studied CXXC motif. In addition, the cisPro-minus1 residue seems to be vital for regulating the ability of the proteins to interact with both upstream and downstream substrates. The choice of the cisPro-minus1 residue in any specific thioredoxin fold protein appears to represent an evolutionary compromise across all of these factors, allowing optimal activity of an individual thioredoxin-family member.
On the basis of the comprehensive study reported here, we can conclude that the cisPro-minus1 residue plays a key role in controlling the activity of thioredoxin fold proteins in part by modifying the active site cysteine reactivity. In all of the known structures of thioredoxin fold proteins, this residue appears to interact with the active site cysteine either via hydrogen donation or hydrophobic interaction. This is best illustrated when the cisPro-minus1 residue is threonine. Thus, the crystal structure of thioredoxin I75T indicates the presence of a hydrogen bond between the threonine and the active site cysteine that is absent in the WT structure (Fig. 1). This hydrogen bond is predicted to be stronger in the reduced form because the distance between cisPro-minus1 and the active site cysteine (as determined from the structures of oxidized and reduced DsbG) is optimal in the reduced form. The presence of the Thr likely stabilizes the reduced form of the protein by formation of a hydrogen bond with the thiolate and destabilizes the oxidized form through electrostatic and steric interactions with the cystine. This is measurable as a decrease in the pKa of the nucleophilic active site cysteine and an increase in the redox potential, thereby favoring oxidase activity (26, 38, 47). At the same time, threonine may favor more rapid kinetics, as shown by the substantial increase in the rate at which threonine mutation at cisProminus1 in thioredoxin catalyzes DTT-dependent insulin reduction. This may explain why threonine is so commonly found in prokaryotic disulfide isomerases yet is absent in reductases (Table 1). Similar effects on redox properties have been reported previously for Staphylococcus aureus DsbA, which naturally contains a threonine in the cisPro-minus1 position; mutation of this Thr to Val dramatically increases the pKa of the nucleophilic cysteine from 3.4 to 5.2 (48). Moreover, a recent bioinformatics study highlighted the potential importance of threonines in modifying the pKa of nearby cysteines in proteins (49).
Thioredoxin fold proteins transfer disulfide bonds from one substrate to another. These proteins often have a wide range of partially folded proteins that serve as downstream substrates and usually just one upstream partner. For instance, the upstream partner of thioredoxin is thioredoxin reductase; for DsbA it is DsbB; and for DsbC (and DsbG) it is DsbD. Several crystal structures of disulfide-linked complexes of thioredoxin fold proteins with downstream and upstream partners have recently been reported. These include thioredoxin·BASI, DsbA·DsbB, and thioredoxin·thioredoxin-reductase·ferredoxin (20-22, 50). In all of these studies, the side chains of the cis-proline loop residues are oriented toward the substrate, implying a role in substrate interaction. By altering the cisPro-minus1 residue, we have shown that it is surprisingly easy to improve the activity of thioredoxin fold proteins toward their downstream substrates. However, none of the variants improved the interaction with specific upstream partners, and most of them negatively affected this interaction. This finding implies that the cisPro-minus1 residue is important in the interaction of thioredoxin fold proteins with upstream partners and that the selection of this residue may represent a compromise between optimal redox properties and optimal substrate interactions. For example WT E. coli DsbA has a valine at the cisPro-minus1 residue, even though a threonine at this position would make it more oxidizing. Valine may allow a balance between the ability of E. coli DsbA to oxidize substrates and to be oxidized by E. coli DsbB.
Structural data (20-22) indicate that thioredoxin fold proteins recognize their partner proteins in part by forming a transient antiparallel β-sheet interaction with residues of the cis-Pro loop. To allow this interaction to occur, the cis-Pro loop must be able to adopt a β-strand conformation, and steric clashes between the residues of the two strands must be avoided. Independent biophysical modeling studies have consistently shown that Asp is a nonfavored residue for β-strand structure (51, 52), so this residue in the cisProminus1 position would be predicted to be deleterious for activity of thioredoxin fold proteins. Consistent with this prediction, we found that introducing Asp or Glu at the cis-Pro-minus1 position of thioredoxin (I75D and I75E) completely abolished oxidoreductase activity in vivo and in vitro (Fig. 2, C and D). Similarly, DsbAV150E had no significant in vivo oxidase activity (data not shown). These results are also consistent with sequence alignments of thioredoxin fold proteins showing the complete absence of charged residues at this cisPro-minus1 position.
In conclusion, the present study finds that the cisPro-minus1 residue is a general activity regulator of thioredoxin fold proteins. It appears to control the activity of thioredoxin-family proteins by affecting both their redox properties and their ability to interact with partner proteins. These findings provide insight into the relationship between the thioredoxin fold structure and its function. They also highlight a functional regulator that is distinct from the well studied CXXC redox rheostat. This knowledge expands our scientific understanding of this important class of proteins. Furthermore, modification of the cisPro-minus1 residue could lead to the development of thioredoxin fold proteins with a range of redox activities that will be applicable in industrial or academic settings, for example, in the in vitro oxidative folding of protein substrates.
Supplementary Material
Acknowledgments
We thank members of the Bardwell, Jakob, and Martin laboratories for helpful discussions. We also thank Tim Tapley and Shu Quan for supplying protein samples for this study.
The atomic coordinates and structure factors (code 3DYR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by a grant from the National Institutes of Health (to J. C. A. B.). This work was also supported by the Australian Research Council (to B. H. and J. L. M.) and a University of Queensland Early Career Research Award (to B. H.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. S1-S4, and Tables S1-S3.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: TRX, thioredoxin; WT, wild type; AMS, 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid; cisPro, cis-proline; cis-Pro-minus1, residue preceding cisPro; PDB, Protein Data Bank; DTT, dithiothreitol.
References
- 1.Heras, B., Kurz, M, Shouldice, S. R., and Martin, J. L. (2007) Curr. Opin. Struct. Biol. 17 691-698 [DOI] [PubMed] [Google Scholar]
- 2.Martin, J. (1995) Structure 3 245-250 [DOI] [PubMed] [Google Scholar]
- 3.Pan, J., and Bardwell, J. C. (2006) Protein Sci. 15 2217-2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmgren, A., Johansson, C., Berndt, C., Lönn, M. E., Hudemann, C., and Lillig, C. H. (2005) Biochem. Soc. Trans. 33 1375-1377 [DOI] [PubMed] [Google Scholar]
- 5.Gruber, C., Cemazar, M., Heras, B., Martin, J. L., and Craik, D. J. (2006) Trends Biochem. Sci. 31 455-464 [DOI] [PubMed] [Google Scholar]
- 6.Krause, G., Lundström, J., Barea, J. L., Pueyo de la Cuesta, C., and Holmgren, A. (1991) J. Biol. Chem. 266 9494-9500 [PubMed] [Google Scholar]
- 7.Lin, T., and Kim, P. S. (1989) Biochemistry 28 5282-5287 [DOI] [PubMed] [Google Scholar]
- 8.Zapun, A., Bardwell, J. C., and Creighton, T. E. (1993) Biochemistry 32 5083-5092 [DOI] [PubMed] [Google Scholar]
- 9.Appenzeller-Herzog, C., and Ellgaard, L. (2008) Biochim. Biophys. Acta 1783 535-548 [DOI] [PubMed] [Google Scholar]
- 10.Jonda, S., Huber-Wunderlich, M., Glockshuber, R., and Mössner, E. (1999) EMBO J. 18 3271-3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quan, S., Schneider, I., Pan, J., Von Hacht, A., and Bardwell, J. C. (2007) J. Biol. Chem. 282 28823-28833 [DOI] [PubMed] [Google Scholar]
- 12.Mössner, E., Huber-Wunderlich, M., and Glockshuber, R. (1998) Protein Sci. 7 1233-1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber-Wunderlich, M., and Glockshuber, R. (1998) Folding Des. 3 161-171 [DOI] [PubMed] [Google Scholar]
- 14.Woycechowsky, K., and Raines, R. T. (2008) Biochemistry 42 5387-5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA
- 16.Georgescu, R., Li, J. H., Goldberg, M. E., Tasayco, M. L., and Chaffotte, A. F. (1998) Biochemistry 37 10286-10297 [DOI] [PubMed] [Google Scholar]
- 17.Charbonnier, J., Belin, P., Moutiez, M., Stura, E. A., and Quéméneur, E. (1999) Protein Sci. 8 96-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadokura, H., Tian, H., Zander, T., Bardwell, J. C., and Beckwith, J. (2004) Science 303 534-537 [DOI] [PubMed] [Google Scholar]
- 19.Su, D., Berndt, C., Fomenko, D. E., Holmgren, A., and Gladyshev, V. N. (2007) Biochemistry 46 6903-6910 [DOI] [PubMed] [Google Scholar]
- 20.Maeda, K., Hägglund, P., Finnie, C., Svensson, B., and Henriksen, A. (2006) Structure (Lond.) 14 1701-1710 [DOI] [PubMed] [Google Scholar]
- 21.Inaba, K., Murakami, S., Suzuki, M., Nakagawa, A., Yamashita, E., Okada, K., and Ito, K. (2006) Cell 127 789-801 [DOI] [PubMed] [Google Scholar]
- 22.Dai, S., Friemann, R., Glauser, D. A., Bourquin, F., Manieri, W., Schürmann, P., and Eklund, H. (2007) Nature 448 92-96 [DOI] [PubMed] [Google Scholar]
- 23.Hiniker, A., Ren, G., Heras, B., Zheng, Y., Laurinec, S., Jobson, R. W., Stuckey, J. A., Martin, J. L., and Bardwell, J. C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11670-11675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bessette, P., Cotto, J. J., Gilbert, H. F., and Georgiou, G. (1999) J. Biol. Chem. 274 7784-7792 [DOI] [PubMed] [Google Scholar]
- 25.Collet, J., Riemer, J., Bader, M. W., and Bardwell, J. C. (2002) J. Biol. Chem. 277 26886-26892 [DOI] [PubMed] [Google Scholar]
- 26.Nelson, J. W., and Creighton, T. E. (1994) Biochemistry 33 5974-5983 [DOI] [PubMed] [Google Scholar]
- 27.Holmgren, A. (1979) J. Biol. Chem. 254 9627-9632 [PubMed] [Google Scholar]
- 28.Lundström, J., and Holmgren, A. (1990) J. Biol. Chem. 265 9114-9120 [PubMed] [Google Scholar]
- 29.Hillson, D. A., Lambert, N., and Freedman, R. B. (1984) Methods Enzymol. 107 281-294 [DOI] [PubMed] [Google Scholar]
- 30.Tapley, T., Eichner, T., Gleiter, S., Ballou, D. P., and Bardwell, J. C. (2007) J. Biol. Chem. 282 10263-10271 [DOI] [PubMed] [Google Scholar]
- 31.Hiniker, A., Collet, J. F., and Bardwell, J. C. (2005) J. Biol. Chem. 280 33785-33791 [DOI] [PubMed] [Google Scholar]
- 32.Brünger, A., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 33.Jones, T., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47 110-119 [DOI] [PubMed] [Google Scholar]
- 34.Murshudov, G., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 36.Adams, P., Grosse-Kunstleve, R. W., Hung, L. W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K., and Terwilliger, T. C. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1948-1954 [DOI] [PubMed] [Google Scholar]
- 37.Grimshaw, J. P., Stirmann, C. U., Brozzo, M. S., Malojcic, G., Grütter, M. G., Capitani, G., and Glockshuber, R. (2008) J. Mol. Biol. 380 667-680 [DOI] [PubMed] [Google Scholar]
- 38.Grauschopf, U., Winther, J. R., Korber, P., Zander, T., Dallinger, P., and Bardwell, J. C. (1995) Cell 83 947-955 [DOI] [PubMed] [Google Scholar]
- 39.McCarthy, A., Haebel, P. W., Törrönen, A., Rybin, V., Baker, E. N., and Metcalf, P. (2000) Nat. Struct. Biol. 7 196-199 [DOI] [PubMed] [Google Scholar]
- 40.Heras, B., Edeling, M. A., Schirra, H. J., Raina, S., and Martin, J. L. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 8876-8881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hibbert, E., and Dalby, P. A. (2005) Microb. Cell Fact. 4 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johannes, T., and Zhao, H. (2006) Curr. Opin. Microbiol. 9 261-267 [DOI] [PubMed] [Google Scholar]
- 43.Ritz, D., and Beckwith, J. (2001) Annu. Rev. Microbiol. 55 21-48 [DOI] [PubMed] [Google Scholar]
- 44.Vallee, B., and Ulmer, D. D. (1972) Annu. Rev. Biochem. 41 91-128 [DOI] [PubMed] [Google Scholar]
- 45.Rietsch, A., Belin, D., Martin, N., and Beckwith, J. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 13048-13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chivers, P. T., Laboissiere, M. C., and Raines, R. T. (1996) EMBO J. 15 2659-2667 [PMC free article] [PubMed] [Google Scholar]
- 47.Shaked, Z., Szajewski, R. P., and Whitesides, G. M. (1980) Biochemistry 19 4156-4166 [DOI] [PubMed] [Google Scholar]
- 48.Heras, B., Kurz, M., Jarrott, R., Shouldice, S. R., Frei, P., Robin, G., Cemazar, M., Thöny-Meyer, L., Glockshuber, R., and Martin, J. L. (2008) J. Biol. Chem. 283 4261-4271 [DOI] [PubMed] [Google Scholar]
- 49.Salsbury, F. R. Jr., Knutson, S. T., Poole, L. B., and Fetrow, J. S. (2008) Protein Sci. 17 299-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malojcić, G., Owen, R. L., Grimshaw, J. P., and Glockshuber, R. (2008) FEBS Lett. 582 3301-3307 [DOI] [PubMed] [Google Scholar]
- 51.Street, A., and Mayo, S. L. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9074-9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kallberg, Y., Gustafsson, M., Persson, B., Thyberg, J., and Johansson, J. (2001) J. Biol. Chem. 276 12945-12950 [DOI] [PubMed] [Google Scholar]
- 53.Hahn, J., and Pat, H. (1986) Syst. Appl. Microbiol. 7 178-183 [Google Scholar]
- 54.Guddat, L., Bardwell, J. C., Glockshuber, R., Huber-Wunderlich, M., Zander, T., and Martin, J. L. (1997) Protein Sci. 6 1893-1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katti, S. K., LeMaster, D. M., and Eklund, H. (1990) J. Mol. Biol. 212 167-184 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.