Abstract
MspA is the major porin of Mycobacterium smegmatis and mediates diffusion of small and hydrophilic solutes across the outer membrane. The octameric structure of MspA, its sharply defined constriction zone, and a large periplasmic loop L6 represent novel structural features. L6 consists of 13 amino acids and is directly adjacent to the constriction zone. Deletion of 3, 5, 7, 9, and 11 amino acids of the L6 loop resulted in functional pores that restored glucose uptake and growth of a porin mutant of M. smegmatis. Lipid bilayer experiments revealed that all mutant channels were noisier than wild type (wt) MspA, indicating that L6 is required for pore stability in vitro. Voltage gating of the Escherichia coli porin OmpF was attributed to loops that collapse into the channel in response to a strong electrical field. Here, we show that deletion mutants Δ7, Δ9, and Δ11 had critical voltages similar to wt MspA. This demonstrated that the L6 loop is not the primary voltage-dependent gating mechanism of MspA. Surprisingly, large deletions in L6 resulted in 3-6-fold less extractable pores, whereas small deletions did not alter expression levels of MspA. Pores with large deletions in L6 were more permissive for glucose than smaller deletion mutants, whereas their single channel conductance was similar to that of wt MspA. These results indicate that translocation of ions through the MspA pore is governed by different mechanisms than that of neutral solutes. This is the first study identifying a molecular determinant of solute translocation in a mycobacterial porin.
Mycobacteria have a unique outer membrane composed of covalently attached mycolic acids and a variety of other loosely associated lipids. Visualized by cryo-electron tomography (1), the outer membrane is an efficient permeability barrier that renders mycobacteria intrinsically resistant to many drugs and toxic compounds. As is the case for Gram-negative bacteria, this permeability barrier must be functionalized by outer membrane proteins. Although over 60 outer membrane proteins are known to exist in Escherichia coli (41), very few have been described in mycobacteria to date.
The major protein of Mycobacterium smegmatis is MspA (42), which is the primary member of the nearly identical Msp (M. smegmatis porin) proteins. Msp proteins account for ∼80% of detergent-extractable proteins at 100 °C (2) in wt4 M. smegmatis mc2155. MspA represents the main pathway for small and hydrophilic solutes and nutrients across the outer membrane (3). One hallmark of MspA is its extremely high stability. Functional pores are detectable at temperatures up to 100 °C, a pH range from 0-14, and resist the action of denaturing agents such as 2% SDS (4), thus providing the utility of MspA for nanotechnological applications. However, it is unknown how M. smegmatis utilizes Msp porins to functionalize the outer membrane in varying conditions or what parts of the channel determine substrate translocation. It is our goal to understand the translocation determinants of the MspA channel.
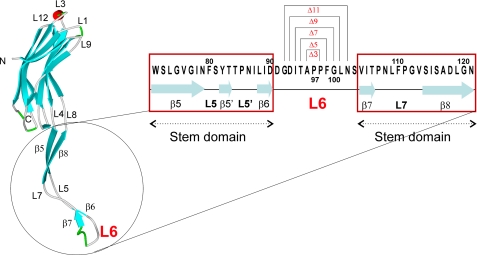
The crystal structure revealed that eight monomers comprise one functional MspA pore (5). Like all other known outer membrane proteins (6), MspA contains a β-barrel that spans the outer membrane. A novel feature of MspA is the constriction zone where the region of the channel with the smallest diameter is lined by at least 16 aspartic acid residues, likely explaining the cation preference of MspA (7). Most porins of Gram-negative bacteria display large, extracellular loops and much smaller periplasmic loops (8-11). In contrast, the single 13-amino acid periplasmic loop of MspA, herein referred to as L6, is larger than the extracellular loops (see Fig. 1). Additionally, the L6 loop is positioned adjacent to the 1-nm-wide constriction zone, whereas the smaller extracellular loops surround a vestibule opening of 4.8 nm in diameter (5). The L6 loop was also found to be exposed to the periplasmic space via biotinylation of cysteine mutants (12).
FIGURE 1.
Schematic representation of the deletions in the periplasmic loop 6 of MspA. The assignment of the secondary structural elements was taken from the crystal structure of MspA (5). The arrows represent the β-sheets, lines represent the loops, and the red cylinder represents the L3 helix. The periplasmic loop is marked as L6, and the primary sequence of the loop 6 and stem domain is shown to the right. Deletion of amino acids 96-98, 95-99, 94-100, 93-101, and 92-102 generated the mspA mutants Δ3, Δ5, Δ7, Δ9, and Δ11, respectively.
It is unknown what the role of this unusual periplasmic loop is. It was shown that MspA exhibited voltage gating (13), suggesting that, in response to a strong electric field, the channel undergoes a conformational change that restricts ionic conductance through the pore. Although the rest of MspA is primarily comprised of a rigid β-barrel, the periplasmic loop L6 appeared to be a likely candidate for mediating such channel gating.
Many outer membrane proteins have also been shown to utilize soluble periplasmic domains for functional purposes. For example, OmpA of E. coli contains a C-terminal domain that binds peptidoglycan (14), whereas TolC links to inner membrane efflux pumps via its large periplasmic domain (15). It may be that the loop L6, because of its larger size, also serves as a binding site for proteins or other effector molecules that aid in transport processes across the cell envelope of M. smegmatis.
To examine the role of the periplasmic loop of MspA, we constructed five mutants with symmetric deletions of 3, 5, 7, 9, or 11 amino acids in L6. The function of these mutants was analyzed both in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Chemicals and Enzymes—Chemicals were of the highest purity available from Merck, Roth (Karlsruhe, Germany), Invitrogen, or Sigma unless otherwise noted. The detergent n-octylpolyoxyethylene (n-octyl-POE) was from Alexis (Plymouth Meeting, PA). The oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA).
Bacterial Strains and Growth Conditions—M. smegmatis ML16, which lacks the porin genes mspA, mspC, and mspD (16) was grown at 37 °C in 7H9 liquid medium (BD Biosciences) supplemented with 0.2% glycerol and 0.05% Tween 80 or on 7H10 agar (BD Biosciences) supplemented with 0.2% glycerol, unless otherwise indicated. E. coli DH5α was used for cloning experiments and was routinely grown in Luria-Bertani broth at 37 °C. Hygromycin was used in concentrations of 50 and 200 μg/ml for M. smegmatis and E. coli, respectively.
Construction of MspA Loop 6 Deletion Mutants—The L6 deletion mutants were constructed using the plasmid pMN016, which carries the psmyc-mspA transcriptional fusion (16) as a template. Psmyc1 and pMS-seq1 were used as end primers along with appropriate mutagenesis primers to perform a two-step PCR. Separate PCR amplifications of the upstream and downstream portions of the gene flanking the deletion were performed. Then the two purified PCR products were amplified and ligated together in the same reaction using ampliligase (Epicenter). Mutated mspA genes were then ligated into SphI and HindIII double digested pMN016 to generate the plasmids pML905-pML909. Primers and resulting plasmids are listed in Table 1. All of the plasmids were verified by sequencing the entire mspA gene before they were transformed into the porin mutant M. smegmatis ML16 for protein production.
TABLE 1.
Oligonucleotides used in this work
| Mutant | Plasmid | Primer pair for fragment A (5′ → 3′) | Primer pair for fragment B (5′ → 3′) |
|---|---|---|---|
| Δ3 | pML905 | TTCGGCCTGAACTCGGTCATCACC | GGTGATGTCACCGTCGTCGATCAGG |
| CGACCAGCACGGCATACATC (psmyc1) | CGTTCTCGGCTCGATGATCC (pMS-seq1) | ||
| Δ5 | pML906 | GGCCTGAACTCGGTCATCACCCCG | GATGTCACCGTCGTCGATCAGGATG |
| CGACCAGCACGGCATACATC | CGTTCTCGGCTCGATGATCC | ||
| Δ7 | pML907 | CTGAACTCGGTCATCACCCCGAACC | GTCACCGTCGTCGATCAGGATGTTC |
| CGACCAGCACGGCATACATC | CGTTCTCGGCTCGATGATCC | ||
| Δ9 | pML908 | AACTCGGTCATCACCCCGAACCTG | ACCGTCGTCGATCAGGATGTTCGG |
| CGACCAGCACGGCATACATC | CGTTCTCGGCTCGATGATCC | ||
| Δ11 | pML909 | TCGGTCATCACCCCGAACCTGTTC | GTCGTCGATCAGGATGTTCGG |
| CGACCAGCACGGCATACATC | CGTTCTCGGCTCGATGATCC |
Expression, Extraction, and Purification of MspA Mutants—The wild type (wt) mspA gene (pMN016) and the mutated mspA genes (pML905-pML909) were constitutively expressed in M. smegmatis ML16. This strain contains at least 15-fold less Msp porins in the outer membrane (16). To examine whether the five L6 deletion mutants were expressed in ML16, a selective extraction procedure was employed that yields predominantly MspA when whole cells of M. smegmatis are heated in POPO5 buffer (300 mm NaH2PO4/Na2HPO4, 0.3 mm Na2EDTA, 150 mm NaCl, 0.5% (w/v) n-octyl-POE) at a cell density of 100 μl/10 mg cells (wet weight) to 100 °C for 30 min (2). Protein extracts were separated on a denaturing 10% polyacrylamide gel and stained with Coomassie Blue. Quantitative image analysis of protein gel bands by pixel densitometry was performed using Labworks 4.6 (UVP, Inc.) software. wt and mutant MspA proteins were purified as previously described (2). Briefly, protein extracts were precipitated in acetone and resuspended in AOPO5 buffer (25 mm HEPES/NaOH, 10 mm NaCl, 0.5% n-octyl-POE, pH 7.5). The proteins were then loaded onto a 1.7-ml POROS 20HZ anion exchange column (Perceptive Biosystems) and eluted off in a salt concentration gradient ranging from 0.76 to 1.02 m NaCl. The buffers were then exchanged by gel filtration in AOPO5.
Lipid Bilayer and Voltage Gating Experiments—The channel activity of the proteins was measured in lipid bilayer experiments as described (2). In all single channel conductance experiments, 1 m KCl was used as the electrolyte. The single channel conductances of at least 100 pores reconstituted from a freshly diluted solution of purified proteins were analyzed in several diphytanoyl phosphatidylcholine (Avanti Polar Lipids) membranes with a constant applied voltage of -10 mV. Where applicable, the cis side of the cuvette was defined as the side with the positive electrode. In voltage gating experiments, the applied voltage increased from 0 to -60 mV or +60 mV in 10-mV increments. Using Ag/AgCl electrodes, the membrane current under the voltage clamp was measured with a current amplifier (Keithley 428 current amplifier) and digitized by a desktop computer equipped with Keithley Metrabyte STA 1800U interface. The data were recorded using a macro and the software Test Point 4.0 (Keithley). Analysis of the current traces was carried out using a macro based on the Igor Pro 5.03 software (written by Dr. Harald Engelhardt). For all measurements where the conductance fluctuations (noise) caused by individual pore insertion were negligible, the entire current trace with 50-100 pores was analyzed. However, for analysis of the pores where insertion of an individual pore created fluctuations (noise) in the recording, only the initial 10-20 pores were used. The steps recorded in four to six membranes were pooled to generate the final distribution plots (SigmaPlot 9.0). The noise suppression factor was constant for all measurements.
Growth Assays in Minimal Media—Growth on solid media was performed by inoculating primary cultures in Hartmans's-de Bond (HdB) liquid medium with 0.05% Tween 80 and supplemented with 1% glucose. The cultures were filtered through a 5-μm filter and grown overnight in beveled flasks on a shaker at 37 °C. The cultures were then filtered again through a5-μm filter, serially diluted in HdB with 0.05% Tween 80 and no glucose, and plated onto HdB solid medium supplemented with 1% glucose. Pictures at 12.5× magnification were taken after 5 days of incubation in a sealed container at 37 °C.
Growth in liquid medium was performed by inoculating 5 ml of primary cultures in HdB with 0.025% tyloxapol and 0.2% glucose. The cultures were filtrated with a 5-μm filter and grown overnight. The cultures were then diluted in triplicate in 30 ml of HdB with 0.025% tyloxapol and 0.2% glucose in beveled flasks and incubated on a shaker at 37 °C. At indicated time points, samples of each culture were taken and the optical density at 600 nm was recorded using a photometer and cuvettes with a 1-cm path length.
Enzyme-linked Immunosorbent Assay—To confirm expression and localization of MspA mutants, a sandwich-type enzyme-linked immunosorbent assay (ELISA) using whole cells of M. smegmatis ML16 was adapted from a previously described whole cell ELISA protocol (3). Cells were grown to an A600 of ∼0.8, harvested by centrifugation, washed with TBST (0.01 m Tris-HCl, pH 8, 0.15 m NaCl, 0.05% Tween 20), and resuspended in coating buffer (50 mm NaHCO3, pH 9.6) to an A600 of ∼10. Wells of a MaxiSorp plate (NUNC-Immuno™ MaxiSorp™ surface; Nalge Nunc International) were coated with 100 μl of a 1:2,000 dilution of MspA polyclonal antibody pAK813 (7) for 1 h at 37 °C to trap cells. After blocking with 5% powdered skim milk in TBST for 1 h, ∼108 cells were loaded into each well and incubated at 37 °C for 1 h. Unbound cells were removed by washing three times with TBST, and bound cells were incubated with 100 μl of a 1:20 dilution of MspA mAb P2 (unpublished results) for 1 h at 37 °C. The cells were washed three times and then incubated with a 1:2,000 dilution of horseradish peroxidase-conjugated goat anti-mouse antibody (Sigma) for 1 h at 37 °C. After washing three times, 50 μl of O-phenylenediamine substrate (Sigma) were added to each well, and the plate was incubated at room temperature in the dark until a yellow color developed (∼20 min). The reaction was then stopped with 50 μl of 1 m H2SO4. The plate was centrifuged (3,000 rpm, 5 min), the supernatant was removed to a new microtiter plate, and the absorbance was read at 490 nm using a microplate reader (Synergy HT, Bio-TEK Instrument Inc.).
Glucose Uptake Measurements—Glucose uptake measurements were carried out as previously described (3, 17). To reduce aggregation and clumping, all M. smegmatis cells were filtered through a 5-μm-pore size filter (Sartorius) and grown to an A600 of ∼0.8 in the presence of 1 mm glucose. The cells were harvested (3000 × g at 4 °C for 10 min), washed once in uptake buffer (2 mm PIPES, pH 6.5, 0.05 mm MgCl2), and resuspended in the same buffer to an A600 of ∼0.4. Radiolabeled [14C]glucose and unlabeled glucose were mixed and added to the cell suspension to a final concentration of 20 μm. The cells were then incubated at 37 °C for 5 min. At the indicated time points, 1-ml samples were removed to 3 ml of kill buffer (0.1 m LiCl, 6.7% formalin). The cells were captured on a 0.45-μm-pore size filter (Sartorius) and counted in a liquid scintillation counter. All of the experiments were performed in triplicate. The mean dry weight of the cells in these samples was 0.6 ± 0.2 mg. The glucose uptake rate was expressed as nmol/mg cells/min and was determined by fitting a straight line to the first four data points (from 1 to 4 min). Uptake rates at the individual concentrations and Km and vmax values for the overall transport and a minimal estimate of the permeability coefficient were determined as described previously (3, 18).
RESULTS
Construction of mspA L6 Deletion Mutants—The crystal structure of MspA (5) revealed a single loop (L6) at the periplasmic end of the monomer (Fig. 1). To determine whether L6 contributes to the translocation of small and hydrophilic compounds through the MspA channel, symmetric deletions centered around Pro97 were constructed in this loop. The resulting mspA mutant genes were cloned into pMN016, which contains a psmyc-mspA transcriptional fusion (16). Using appropriate mutagenesis primers (Table 1), Ala96-Pro98, Thr95-Phe99, Ile94-Gly100, Asp93-Leu101, and Gly92-Asn102 were constructed (Fig. 1). Henceforth, these mutants are referred to as Δ3, Δ5, Δ7, Δ9, and Δ11, respectively.
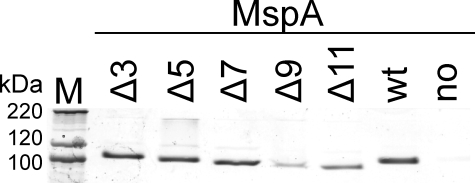
Expression of L6 Deletion Mutants in M. smegmatis—For expression of the mspA genes encoding the loop deletion mutants, the porin triple mutant M. smegmatis ML16, which lacks the mspA, mspC, and mspD genes, was used. The amount of porins in the outer membrane is reduced by ∼15-fold in this strain (16). To examine whether the loop deletion mutants were expressed in ML16, a selective extraction procedure was utilized that yields predominantly MspA when whole cells of M. smegmatis are heated with 0.5% n-octyl-POE to 100 °C (2). Whole cell extracts of these mutants were separated on a denaturing 10% polyacrylamide gel and stained with Coomassie Blue. Almost no Msp porins were detected in extracts of M. smegmatis ML16 carrying the empty vector pMS2 (19) (Fig. 2, no MspA lane), indicating a very low background expression of mspB. Expression of wt mspA in ML16 using the vector pMN016 (Fig. 2, wt lane) yields MspA levels similar to that in wt M. smegmatis mc2155 (12, 16). Mutants with the smallest deletions in the periplasmic loop (Δ3, Δ5, and Δ7) had expression levels similar to that of wt mspA (Fig. 2). However, expression of mutants harboring larger deletions was significantly reduced in comparison with wt mspA (Fig. 2). Quantitative image analysis of the bands representing the MspA octamer showed that Δ3, Δ5, and Δ7 had expression levels that were 104, 94, and 91% of wt levels, whereas Δ9, Δ11, and the empty vector had expression levels that were 17, 40, and 0.2% of wt (Fig. 2). It should be noted that the intensity of the monomeric band was similar for all mutants and the wt. The same quantitative expression patterns were obtained in several other Coomassie Blue-stained protein gels. Further, expression of all loop deletion mutants was confirmed via Western blot using the Msp-specific antiserum pAK813 (not shown). These results demonstrated that all mutants were expressed in M. smegmatis ML16 and were stable during heat extraction. The much lower expression of Δ9 and Δ11 indicated the importance of the periplasmic loop for MspA expression in the outer membrane.
FIGURE 2.
Expression of MspA deletion mutants in the porin mutant M. smegmatis ML16. Comparison of detergent extracts by SDS-polyacrylamide gel electrophoresis is shown. MspA proteins were selectively extracted from the porin mutant M. smegmatis ML16 (ΔmspA, ΔmspC, and ΔmspD) (16) at 100 °C in a buffer containing 0.5% n-octyl-POE, separated on a 10% polyacrylamide gel, and stained with Coomassie Blue. Lane M contains the protein mass marker (Mark12; Invitrogen). The expression vector pMN016 (wt mspA, wt lane), the empty vector pMS2 (no MspA lane), and the mutated mspA genes (pMN016 derivatives, lanes Δ3--Δ11) were constitutively expressed in ML16. The part of the gel containing the bands of octameric MspA is shown. The intensity of the monomeric band was identical for all MspA proteins. The amount of the loop deletion mutants was quantified by image analysis and normalized to wt MspA. For mutants Δ3, Δ5, Δ7, Δ9, and Δ11, expression was 104, 94, 91, 17, and 40% of wt, respectively. The background signal (no MspA) was 0.2% of that of wt MspA.
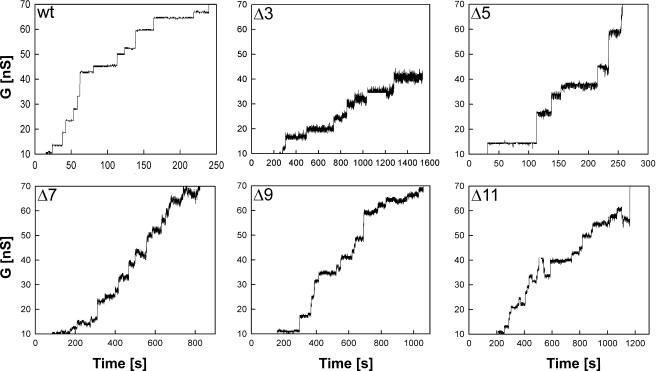
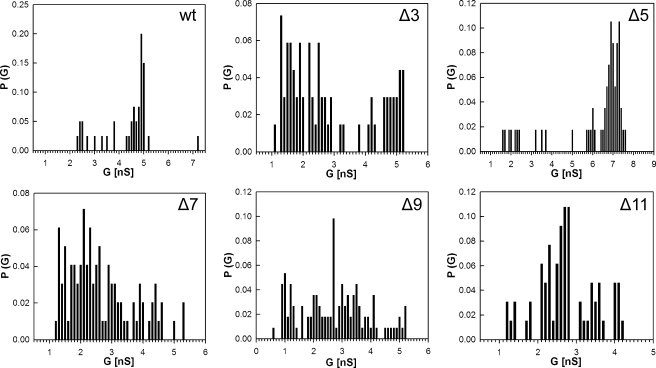
Analysis of Channel Activity in Artificial Lipid Bilayers—Lipid bilayer experiments provide direct evidence of whether a particular protein forms channels within a lipid membrane (7). To determine whether the mutated MspA proteins formed functional pores, the genes were overexpressed in M. smegmatis ML16, and proteins were purified by a two-step chromatographic protocol as previously described (2). No pores were recorded in control experiments when only detergent-containing buffer was added to the lipid bilayer (not shown). As can be seen in Fig. 3, all of the mutants formed open channels in artificial lipid bilayers. All loop deletion mutants produced noisier channels. In contrast to wt and the other deletion mutants, the loss of conductance events were much more frequent in Δ11. Individual conductance steps were plotted to determine the probability of conductance for single channels. Significant changes in the single channel conductance were observed for all mutants. A reduced ability to conduct ions was apparent for Δ7, Δ9, and Δ11, which were determined to be 2.1, 2.6, and 2.8 nS in 1 m KCl, respectively (Fig. 4). In contrast, the ion conductance of Δ5 was much higher with a peak at 7.0 nS. The Δ3 mutant displayed a bimodal distribution of ionic conductances with one species conducting ions near wt levels and the other reduced to levels similar to Δ7, Δ9, and Δ11. These experiments showed that L6 has a considerable effect on the single channel conductance of purified MspA and the stability of the pores in artificial bilayers.
FIGURE 3.
Single channel recordings of purified MspA loop deletion mutants in lipid bilayers. Single-channel recordings of a diphytanoyl phosphatidylcholine membrane in the presence of 0.02, 500, or 0.1 ng/ml of wt, Δ3, and Δ5-Δ11 purified MspA protein, respectively. Protein solutions were added to both sides of the membrane, and data were collected from at least five different membranes. The membrane current was measured in an aqueous solution of 1 mKCl with an applied transmembrane potential of -10 mV.
FIGURE 4.
Analysis of single channel conductances of purified MspA deletion mutants. Analysis of the probability P of a conductance step G for single channel events. The average single channel conductances were 4.9, 1.3, and 5.1 (bimodal), 7.0, 2.1, 2.6, and 2.8 nS for wt, Δ3, Δ5, Δ7, Δ9, and Δ11, respectively.
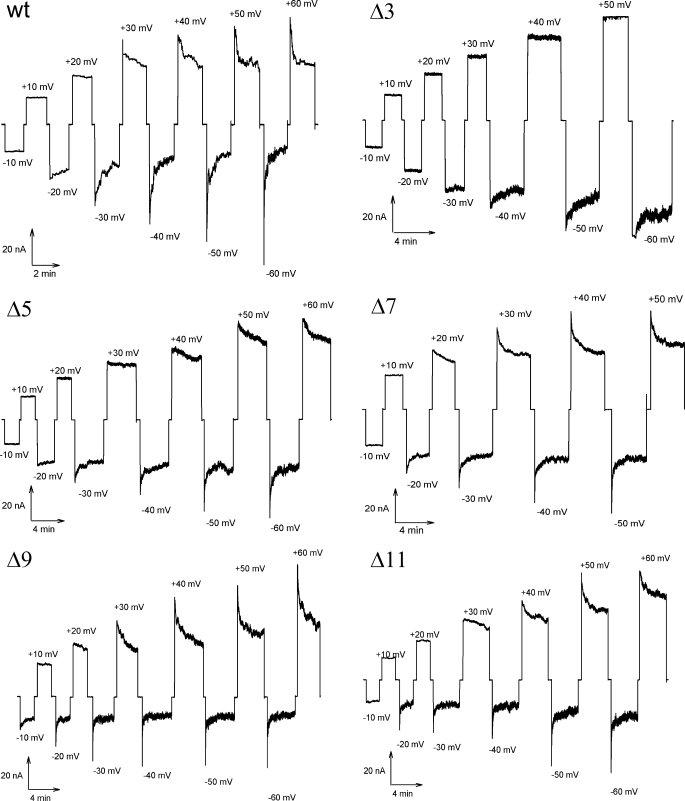
Voltage-dependent Channel Closure—It was shown that MspA inserts unidirectionally into lipid membranes and that the MspA channel closes at voltages exceeding -20 and +40 mV (13). To investigate the role of the periplasmic loop in voltage gating of MspA, ∼100 pores of purified MspA were reconstituted into artificial lipid bilayers after the addition of protein in the cis side of the cuvette. Ionic conductance through the channels was measured at increasing voltage magnitudes and alternating polarities. In this experiment, wt MspA channels closed starting at voltages of +30 and -20 mV (Fig. 5). These values are consistent with those previously published (13). Upon deletion of the periplasmic loop, different voltage gating responses were observed. Δ3 formed stable pores, did not gate at positive voltages up to +50 mV, and was less sensitive to negative voltages, gating at -40 mV. Voltages above +50 mV could not be used because no membrane was obtained under in those conditions. Δ5 was also less sensitive to positive voltages, undergoing closure events only after +40 mV were applied. However, negative voltage resulted in gating similar to wt.
FIGURE 5.
Voltage gating properties of purified MspA L6 deletion mutants. Purified MspA was added to the cis-side of a diphytanoyl phosphatidylcholine membrane. Increasingly positive (upper traces) and negative (lower traces) voltages were applied to the membrane when ∼100 channels were reconstituted into the membrane. The membrane current was recorded at each applied voltage. The critical voltage at which the channels began to close (Vc) was determined to be the voltage where conductance decreased after an initial spike. The wt was measured to have a Vc of +30 and -20 mV. For the L6 mutants Δ5, Δ7, Δ9, and Δ11, Vc was approximately +40/-30, +20/-20, +20/-20, and +30/-20 mV, respectively. The Δ3 mutant did not gate at positive voltages before membrane breakage, whereas gating occurred at -40 mV.
Conversely, both Δ7 and Δ9 were slightly more sensitive and gated at lower voltages. Δ7 began to close at 20 mV in both polarities. Δ9 was also affected by voltages of both polarities, undergoing gating events with voltages of +20 and -10 mV.
Most importantly, the gating properties of Δ11 were similar to that of wt MspA with evidence of gating events at +30 and -20 mV. This mutant has lost almost the entire L6 loop. These data show that L6 affects the voltage-dependent gating properties of MspA but is not the primary gating mechanism.
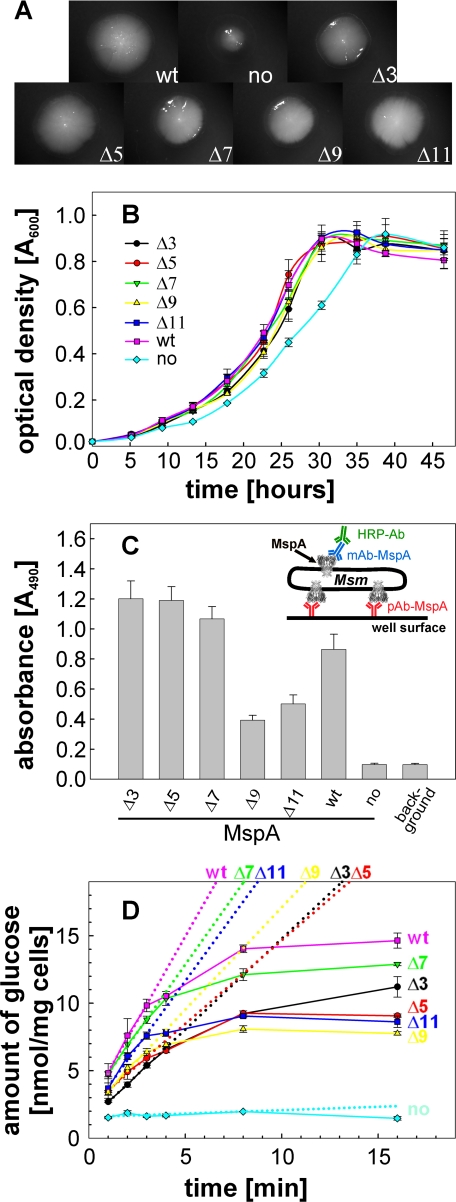
Activity of the Loop Deletion Mutants in Vivo—The periplasmic L6 loop of MspA has been shown to affect the conductance properties and stability of the pore in artificial membranes. To determine whether the loop alterations influenced the channel function in vivo, the growth defect of the triple porin mutant M. smegmatis ML16 was exploited in growth complementation assays. Strains of ML16 complemented with MspA loop deletion mutants were grown to mid log phase, filtered to obtain single cell suspensions, and plated onto minimal HdB media supplemented with 1.0% glucose as the sole carbon source. Representative colony pictures showed that wt mspA gave rise to full size colonies in contrast to the empty vector (Fig. 6A). Colonies formed by M. smegmatis ML16 containing L6 deletion mutants had a similar size as those containing wt MspA, indicating functional expression of the L6 mutants. To quantify the growth rates, the same strains were grown in HdB liquid medium containing 0.025% tyloxapol and supplemented with 0.2% glucose as the sole carbon source. All of the mutants complemented the growth defect of ML16 with similar growth kinetics as wt MspA (Fig. 6B). These results are consistent with the growth experiments on agar plates and show that the L6 loop is dispensable for porin function in vivo. Thus, all of the L6 deletion mutants appear to be fully functional in vivo despite their different in vitro properties.
FIGURE 6.
Surface accessibility and in vivo function of MspA L6 deletion mutants. A, growth on agar plates and colony morphology. M. smegmatis ML16 complemented with MspA L6 deletion mutants were grown on HdB minimal medium supplemented with 1% glucose. After 5 days of incubation at 37 °C, pictures of representative colonies were taken at 12.5× magnification. B, growth in liquid medium. Triplicate cultures in beveled flasks were inoculated to an A600 of 0.02 in HdB liquid medium, 0.025% tyloxapol, and supplemented with 0.2% glucose. The optical density at 600 nm was recorded at the indicated time points during growth at 37 °C. C, surface accessibility of MspA mutants by whole cell capture ELISA. The experimental set-up is drawn as an insert. Wells of a Maxisorp 96-well microtiter plate were coated with an MspA antiserum to which ML16 cells expressing MspA were then “captured.” The anti-MspA mAb P2 was used for detection of surface-exposed MspA in combination with a secondary antibody conjugated with horseradish peroxidase. D, uptake of glucose. ML16 complemented with MspA mutants were grown to an A600 of ∼0.8 and incubated with a 20-μm mixture of nonlabeled and [14C]glucose. At the indicated time points, 1 ml of the cell suspensions were removed, filtered, and counted on a scintillation counter. The dotted lines represent the best fit for the first four time points (1-4 min). Uptake rates for wt, no MspA, Δ3, Δ5, Δ7, Δ9, and Δ11 were determined to be 2.5, 0.05, 1.3, 1.2, 2.0, 1.5, and 2.0 nmol/mg cells/min, respectively.
Surface Accessibility of Loop Deletion Mutants—As shown in Fig. 2, large deletions in L6 resulted in a reduced number of extractable pores in M. smegmatis ML16. To determine whether L6 deletion mutants were efficiently inserted into the outer membrane of M. smegmatis and that loop alterations did not result in pores more resistant to extraction, the surface accessibility of each mutant was examined by whole cell capture ELISA. Cells expressing MspA were bound to wells of a microtiter plate by a polyclonal MspA antibody. Surface-exposed MspA was detected with a monoclonal MspA antibody. Because ML16 complemented with the empty vector expresses no MspA in the outer membrane, no capture above background was detected. Conversely, wt MspA was clearly detectable (Fig. 6C). Mutant pores with smaller deletions in L6 (Δ3-Δ7) were better detected on the cell surface of M. smegmatis than wt MspA. In contrast, larger deletions of the periplasmic loop resulted in decreased surface detection. These results reflect the expression pattern in Fig. 2, indicating that the L6 deletion mutants are localized in the outer membrane and are extractable.
Uptake of Glucose—To determine whether the periplasmic loop of MspA is important for the translocation of small molecules in vivo, we examined the accumulation of glucose by whole cells as a reference solute for porin activity (3, 16, 20). M. smegmatis ML16 strains expressing MspA L6 deletion mutants were incubated with a 20 μm mixture of unlabeled and 14C-labeled glucose. Accumulation of glucose was measured over time and normalized to the dry weight of the cells. As previously shown, the porin mutant ML16 was almost completely deficient for glucose uptake (16). Expression of wt mspA in ML16 increased glucose uptake rates by 50-fold (Fig. 6D). L6 deletion mutants partially complemented the porin defect of ML16, with uptake rates ranging from 50 to 80% of that mediated by wt MspA. These results are consistent with the growth experiments and showed that all L6 mutants were functionally expressed in the outer membrane and enabled diffusion of glucose in vivo.
DISCUSSION
This study marks the first analysis of a molecular determinant of channel activity for a mycobacterial porin. The large periplasmic loop L6 is directly adjacent to the constriction zone of MspA (Fig. 1) and might influence the channel conductance of the pore and its gating properties by dynamic motions. Indeed, deletions in L6 resulted in altered single channel conductances of the purified proteins reconstituted in artificial bilayers. All of the mutants with the exception of Δ5 had a reduced single channel conductance in 1 m KCl compared with wt MspA (Fig. 4). It appears to be counter-intuitive that deletion of a possibly occluding loop resulted in decreased conductance. One explanation might be that residues lining the entire barrel lumen from the vestibule to the periplasmic tip of MspA are required for an optimal, ordered flow of ions through the channel as previously shown for OmpF in a molecular dynamics study (21). An alternative explanation is that the L6 loop is required for stabilizing the constriction zone. The observation that all channels of the loop deletion mutants are much more noisy than wt MspA is consistent with both mechanisms and supports the conclusion that the L6 loop is required for a stable MspA pore. In light of this interpretation, the larger channel conductance of the Δ5 mutant (Thr95-Phe99) might result from a different stabilization of the constriction zone by sequence-specific effects that are not present in the other loop deletion mutants. A simple alternative explanation is the increased propensity of Δ5 to aggregate as indicated by a high molecular weight band above that of the octamer (Fig. 2). Thus, more than one functional pore of the Δ5 mutant might simultaneously insert into the artificial lipid bilayer and cause a higher conductance value. It should be noted that bi- or multimodal distributions of apparent single channel conductances have also been observed for other porins such as PhoE of E. coli (22).
Voltage gating is a fundamental feature of β-barrel membrane channels (23). This phenomenon was observed for porins in Gram-negative bacteria and has received a lot of attention because the elucidation of its molecular mechanism helps to understand the translocation of ions through these pores (24). The deletion mutants Δ7, Δ9, and Δ11, in which the L6 loop was mostly or completely removed, had critical voltages (Vc) very similar to wt MspA (Fig. 5). This clearly demonstrated that the L6 loop is not the primary voltage-dependent gating mechanism of MspA. This observation rules out the motion of loops that fold back to gate the pore as discussed for OmpF of E. coli as a mechanism underlying voltage gating (25). In the aforementioned study, large extracellular loops were found to collapse in response to high voltage and block the OmpF pore (25). However, deletion of any single of the seven extracellular loops of OmpF only affected the pH sensitivity of the pores but did not significantly alter voltage gating (26). These experiments did not rule out redundancy of extracellular loops for channel closure. A similar argument does not apply to MspA because MspA has only one periplasmic loop/monomer. A voltage-induced conformational change of the smaller extracellular loops (3-6 amino acids) as an alternative mechanism cannot block the wide vestibule opening with a diameter of 4.8 nm (5). Several reports suggested that individual residues close to the constriction zone of the OmpF pore are involved in voltage gating (27-29). This may hold true also for MspA because minor changes in voltage-dependent gating were observed for the loop deletion mutants, which have either altered spacing or numbers of charged residues (Fig. 1). These alterations may also explain why the Δ3 mutant closed at significantly higher positive voltages than wt MspA (Fig. 5). The discovery of voltage gating in MspA as another β-barrel protein with no structural similarities to either porins of Gram-negative bacteria nor β-barrel toxins such as α-hemolysin of Staphylococcus aureus (30, 31) supports the hypothesis that voltage gating might indeed be an intrinsic property of β-barrel proteins as first described by Lakey and co-workers (23). Hence, our finding favors a mechanism in which an externally applied voltage perturbs the electric field in β-barrel pores that is required for ion translocation (23). It should be noted that it is unclear both for porins of Gram-negative bacteria (32) and for mycobacteria (13) whether voltage-dependent gating is physiologically relevant.
Expression of MspA in M. smegmatis was also affected by mutations in L6 (Fig. 2). However, all of the mutants were still extracted from the outer membrane, albeit at differing levels, indicating that the localization of the protein was not altered. This conclusion is confirmed by the observation that all of the loop deletion mutants were functionally active and complemented an Msp porin triple mutant of M. smegmatis similar to wt MspA (Fig. 6). Although small deletions in L6 did not alter expression levels and surface accessibility of MspA, large deletions in L6 (Δ9 and Δ11) resulted in 3-6-fold less extracted pores. A reduced number of pores in the outer membrane for Δ9 and Δ11 was confirmed by whole cell ELISA (Fig. 6C), excluding the possibility that these mutants were less extractable by our standard method. It is unlikely that reduced transcription of the genes accounts for this observation because all mutants were expressed from the same genetic constructs. Hence, these experiments demonstrated that deletion of the L6 loop reduced the levels of MspA in the outer membrane of M. smegmatis. In Gram-negative bacteria, insertion of proteins into the outer membrane is accomplished by a machinery composed of an assembly factor such as Omp85 of Neisseria meningitidis and YaeT of E. coli and other proteins (33, 34). This assembly complex inserts OmpF of E. coli in a reversed orientation compared with in vitro experiments (35). It has recently been shown that the assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif (36). A similar mechanism might exist in mycobacteria and may involve interactions of the periplasmic loop of MspA with a yet unknown outer membrane protein assembly machinery of M. smegmatis.
All of the loop deletion mutants complemented the growth defect of the porin triple mutant M. smegmatis ML16 on solid and in liquid HdB minimal media supplemented with glucose (Fig. 6, A and B), demonstrating functional pores in vivo. It is concluded that the L6 loop is dispensable for porin function in M. smegmatis. However, larger deletions of L6 resulted in MspA proteins that enable a faster uptake of glucose than smaller loop deletion mutants (Fig. 6D). This is in apparent contrast to the expression levels because much fewer pores are found in the outer membrane for these mutants (Fig. 6C). Normalization to the amount of extractable protein resulted in uptake rates for glucose of 2.5, 1.3, 1.3, 2.2, 8.8, and 5.0 nmol/mg cells/min for wt, Δ3, Δ5, Δ7, Δ9, and Δ11, respectively. This shows that large deletions in L6 result in pores 2-3-fold more permissive for translocation of glucose. It might be speculated that although large deletions in L6 do not increase the conductance of small ions in vitro, they may remove a blockade to neutral and larger solutes such as glucose. These results indicate that the molecular mechanisms governing the translocation of ions are different from those of neutral solutes. It should be noted that the lipid environment in the outer membrane of M. smegmatis and in the lipid bilayer experiments are very different and likely to influence the translocation of solutes as shown for the porin OmpF of E. coli (37).
The periplasmic portions of many outer membrane proteins of Gram-negative bacteria, such as TolC (15) and OmpA (14), have been shown to interact with periplasmic and/or inner membrane effectors. Demonstration that deletion of almost the entire periplasmic loop in Δ11 results in a pore still capable of complementing a porin mutant growth defect shows that the periplasmic loop is not essential for docking of periplasmic or inner membrane effectors required for growth.
In conclusion, the L6 loop affects expression of MspA in the outer membrane and its permeability to both ions and larger, neutral solutes in vitro. It is not, however, required for channel function in whole cells, nor is it the primary mechanism for voltage-dependent gating. The role of the periplasmic loop L6 in substrate translocation is not surprising given its large size and proximity to the constriction zone. MspA is the prototype of a new family of porins with more than 30 members identified solely in mycolic acid-containing bacteria (38) and has excellent perspectives in nanotechnological applications because of its extraordinary biophysical and biochemical properties (39, 40). In addition to the periplasmic loop L6, the constriction zone represents another structural novelty of MspA. Hence, work is underway to elucidate the role of the constriction zone in translocation of solutes through the MspA pore.
Acknowledgments
We gratefully acknowledge Ying Wang for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant HG004145 (to M. N.).
Footnotes
The abbreviations used are: wt, wild type; n-octyl-POE, n-octylpolyoxyethylene; ELISA, enzyme-linked immunosorbent assay; PIPES, 1,4-piperazinedi-ethanesulfonic acid; HdB, Hartman's-de Bond.
References
- 1.Hoffmann, C., Leis, A., Niederweis, M., Plitzko, J. M., and Engelhardt, H. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 3963-3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinz, C., and Niederweis, M. (2000) Anal. Biochem. 285 113-120 [DOI] [PubMed] [Google Scholar]
- 3.Stahl, C., Kubetzko, S., Kaps, I., Seeber, S., Engelhardt, H., and Niederweis, M. (2001) Mol. Microbiol. 40 451-464 [DOI] [PubMed] [Google Scholar]
- 4.Heinz, C., Engelhardt, H., and Niederweis, M. (2003) J. Biol. Chem. 278 8678-8685 [DOI] [PubMed] [Google Scholar]
- 5.Faller, M., Niederweis, M., and Schulz, G. E. (2004) Science 303 1189-1192 [DOI] [PubMed] [Google Scholar]
- 6.Koebnik, R., Locher, K. P., and van Gelder, P. (2000) Mol. Microbiol. 37 239-253 [DOI] [PubMed] [Google Scholar]
- 7.Niederweis, M., Ehrt, S., Heinz, C., Klöcker, U., Karosi, S., Swiderek, K. M., Riley, L. W., and Benz, R. (1999) Mol. Microbiol. 33 933-945 [DOI] [PubMed] [Google Scholar]
- 8.Weiss, M. S., Wacker, T., Weckesser, J., Welte, W., and Schulz, G. E. (1990) FEBS Lett. 267 268-272 [DOI] [PubMed] [Google Scholar]
- 9.Schirmer, T., Keller, T. A., Wang, Y.-F., and Rosenbusch, J. P. (1995) Science 267 512-514 [DOI] [PubMed] [Google Scholar]
- 10.Cowan, S. W., Schirmer, T., Rummel, G., Steiert, M., Ghosh, R., Pauptit, R. A., Jansonius, J. N., and Rosenbusch, J. P. (1992) Nature 358 727-733 [DOI] [PubMed] [Google Scholar]
- 11.Basle, A., Rummel, G., Storici, P., Rosenbusch, J. P., and Schirmer, T. (2006) J. Mol. Biol. 362 933-942 [DOI] [PubMed] [Google Scholar]
- 12.Mahfoud, M., Sukumaran, S., Hülsmann, P., Grieger, K., and Niederweis, M. (2006) J. Biol. Chem. 281 5908-5915 [DOI] [PubMed] [Google Scholar]
- 13.Engelhardt, H., Heinz, C., and Niederweis, M. (2002) J. Biol. Chem. 277 37567-37572 [DOI] [PubMed] [Google Scholar]
- 14.Koebnik, R. (1995) Mol. Microbiol. 16 1269-1270 [DOI] [PubMed] [Google Scholar]
- 15.Koronakis, V., Li, J., Koronakis, E., and Stauffer, K. (1997) Mol. Microbiol. 23 617-626 [DOI] [PubMed] [Google Scholar]
- 16.Stephan, J., Bender, J., Wolschendorf, F., Hoffmann, C., Roth, E., Mailänder, C., Engelhardt, H., and Niederweis, M. (2005) Mol. Microbiol. 58 714-730 [DOI] [PubMed] [Google Scholar]
- 17.Stahl, C., Kubetzko, S., Kaps, I., Seeber, S., Engelhardt, H., and Niederweis, M. (2005) Mol. Microbiol. 57 1509. [DOI] [PubMed] [Google Scholar]
- 18.Jarlier, V., and Nikaido, H. (1990) J. Bacteriol. 172 1418-1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaps, I., Ehrt, S., Seeber, S., Schnappinger, D., Martin, C., Riley, L. W., and Niederweis, M. (2001) Gene (Amst.) 278 115-124 [DOI] [PubMed] [Google Scholar]
- 20.Mailaender, C., Reiling, N., Engelhardt, H., Bossmann, S., Ehlers, S., and Niederweis, M. (2004) Microbiology 150 853-864 [DOI] [PubMed] [Google Scholar]
- 21.Im, W., and Roux, B. (2002) J. Mol. Biol. 319 1177-1197 [DOI] [PubMed] [Google Scholar]
- 22.van Gelder, P., Saint, N., van Boxtel, R., Rosenbusch, J. P., and Tommassen, J. (1997) Protein Eng. 10 699-706 [DOI] [PubMed] [Google Scholar]
- 23.Bainbridge, G., Gokce, I., and Lakey, J. H. (1998) FEBS Lett. 431 305-308 [DOI] [PubMed] [Google Scholar]
- 24.Robertson, K. M., and Tieleman, D. P. (2002) Biochem. Cell Biol. 80 517-523 [DOI] [PubMed] [Google Scholar]
- 25.Müller, D. J., and Engel, A. (1999) J. Mol. Biol. 285 1347-1351 [DOI] [PubMed] [Google Scholar]
- 26.Basle, A., Qutub, R., Mehrazin, M., Wibbenmeyer, J., and Delcour, A. H. (2004) Protein Eng. Des. Sel. 17 665-672 [DOI] [PubMed] [Google Scholar]
- 27.van Gelder, P., Saint, N., Phale, P., Eppens, E. F., Prilipov, A., van Boxtel, R., Rosenbusch, J. P., and Tommassen, J. (1997) J. Mol. Biol. 269 468-472 [DOI] [PubMed] [Google Scholar]
- 28.Phale, P. S., Philippsen, A., Widmer, C., Phale, V. P., Rosenbusch, J. P., and Schirmer, T. (2001) Biochemistry 40 6319-6325 [DOI] [PubMed] [Google Scholar]
- 29.Bredin, J., Saint, N., Mallea, M., D, E., Molle, G., Pages, J. M., and Simonet, V. (2002) Biochem. J. 363 521-528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korchev, Y. E., Alder, G. M., Bakhramov, A., Bashford, C. L., Joomun, B. S., Sviderskaya, E. V., Usherwood, P. N., and Pasternak, C. A. (1995) J. Membr. Biol. 143 143-151 [DOI] [PubMed] [Google Scholar]
- 31.Song, L., Hobaugh, M. R., Shustak, C., Cheley, S., Bayley, H., and Gouaux, J. E. (1996) Science 274 1859-1866 [DOI] [PubMed] [Google Scholar]
- 32.Sen, K., Hellman, J., and Nikaido, H. (1988) J. Biol. Chem. 263 1182-1187 [PubMed] [Google Scholar]
- 33.Bos, M. P., Robert, V., and Tommassen, J. (2007) Annu. Rev. Microbiol. 61 191-214 [DOI] [PubMed] [Google Scholar]
- 34.Ruiz, N., Kahne, D., and Silhavy, T. J. (2006) Nat. Rev. Microbiol. 4 57-66 [DOI] [PubMed] [Google Scholar]
- 35.Hoenger, A., Pages, J. M., Fourel, D., and Engel, A. (1993) J. Mol. Biol. 233 400-413 [DOI] [PubMed] [Google Scholar]
- 36.Robert, V., Volokhina, E. B., Senf, F., Bos, M. P., Van Gelder, P., and Tommassen, J. (2006) PLoS Biol. 4 e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korkmaz, F., Koster, S., Yildiz, O., and Mantele, W. (2008) Biochemistry 47 12126-12134 [DOI] [PubMed] [Google Scholar]
- 38.Niederweis, M. (2008) in The Mycobacterial Cell Envelope (Daffe, M., and Reyrat, J.-M., eds), ASM Press, Washington, D. C.
- 39.Butler, T. Z., Pavlenok, M., Derrington, I. M., Niederweis, M., and Gundlach, J. H. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 20647-20652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wörner, M., Lioubashevski, O., Basel, M. T., Niebler, S., Gogritchiani, E., Egner, N., Heinz, C., Hoferer, J., Cipolloni, M., Janik, K., Katz, E., Braun, A. M., Willner, I., Niederweis, M., and Bossmann, S. H. (2007) Small 3 1084-1097 [DOI] [PubMed] [Google Scholar]
- 41.Molloy, M. P., Herbert, B. R., Slade, M. B., Rabilloud, T., Nouwens, A. S., Williams, K. L., and Gooley, A. A. (2000) Eur. J. Biochem. 267 2871-2881 [DOI] [PubMed] [Google Scholar]
- 42.Ojha, A., and Hatfull, G. F. (2007) Mol. Microbiol. 66 468-483 [DOI] [PMC free article] [PubMed] [Google Scholar]