Abstract
After endocytic uptake by mammalian cells, the heterodimeric plant toxin ricin is transported to the endoplasmic reticulum (ER), where the ricin A chain (RTA) must cross the ER membrane to reach its ribosomal substrates. Here, using gel filtration chromatography, sedimentation, fluorescence, fluorescence resonance energy transfer, and circular dichroism, we show that both fluorescently labeled and unlabeled RTA bind both to ER microsomal membranes and to negatively charged liposomes. The binding of RTA to the membrane at 0-30 °C exposes certain RTA residues to the nonpolar lipid core of the bilayer with little change in the secondary structure of the protein. However, major structural rearrangements in RTA occur when the temperature is increased. At 37 °C, membrane-bound toxin loses some of its helical content, and its C terminus moves closer to the membrane surface where it inserts into the bilayer. RTA is then stably bound to the membrane because it is nonextractable with carbonate. The sharp temperature dependence of the structural changes does not coincide with a lipid phase change because little change in fluorescence-detected membrane mobility occurred between 30 and 37 °C. Instead, the structural rearrangements may precede or initiate toxin retrotranslocation through the ER membrane to the cytosol. The sharp temperature dependence of these changes in RTA further suggests that they occur optimally in mammalian targets of the plant toxin.
Ricin is a potent A-B cytotoxin composed of an RNA-specific N-glycosidase (A chain or RTA)4 disulfide bonded to a cell binding lectin (B chain or RTB). The interaction of holotoxin with galactosylated surface components of mammalian cells is mediated by RTB and is followed by endocytic uptake (reviewed in Ref. 1). There is evidence that a tiny fraction of toxin then reaches the endoplasmic reticulum (ER) lumen (2) where it can be reduced to liberate RTA (3) in preparation for retrotranslocation across the membrane. However, RTA is not thought to penetrate the ER membrane directly. Instead, it appears to exploit protein-conducting translocons (4) as a non-native species (5, 6) in a manner akin to misfolded ER proteins that are dispatched by proteasomal degradation via the ER-associated degradation (ERAD) pathway (7, 8). There is evidence to suggest that once released to the cytosol, non-native RTA can uncouple from the ERAD pathway by virtue of its low lysine content (9). This would reduce the chance for polyubiquitylation and subsequent proteasomal degradation and thereby provide opportunities for refolding in a way that is not normally possible for terminally misfolded ERAD substrates whose dislocation is inextricably linked to degradation. Experimental evidence that toxins exploit various components of the ERAD pathway to reach the cytosol has been provided for ricin (9, 10), cholera toxin (11-14), pertussis toxin (15, 16), Shiga toxin (17, 18), and Pseudomonas exotoxin (19-21).
It is generally assumed that RTA must make specific interactions with ER components to accomplish the unfolding that is required for translocation to the cytosol. However, in contrast to the PDI-dependent unfolding of cholera toxin A (22), ricin is only reduced by PDI (3, 23). Other than the known RTA interactions with PDI and thioredoxin reductase (23), there is a reported nonglycan-mediated interaction of RTA with the ER degradation enhancing α-mannosidase-like protein (EDEM) (24) and an interaction of ricin holotoxin with calreticulin that appears to facilitate toxin trafficking from the Golgi to the ER (25). Of course, ER chaperone interactions act downstream of any unfolding step because ricin presumably arrives by endocytosis to this compartment as a native protein.
The question therefore arises: how is RTA perceived as a potential substrate for the ERAD/retrotranslocation machinery? Unlike the processed and reduced A1 chain of cholera toxin that is conformationally unstable at 37 °C in aqueous solution (26), RTA at pH 7.0 does not become structurally unstable at temperatures below 42 °C. However, above this nonphysiological temperature, RTA can unfold noncooperatively to generate a molten globule (27). Hence, at 37 °C, RTA must employ a different tack to be recognized as unfolded/misfolded by ER quality control systems. One possibility is that the hydrophobic C terminus of RTA (28), newly exposed following reduction and dissociation of RTB (3), may directly promote the kind of chaperone interaction that ultimately leads to membrane translocation. Alternatively, this region may trigger membrane lipid interactions in a process that induces specific or random structural change in the toxin. Indeed, a previous study has shown that upon mixing RTA with liposomes containing a negatively charged phospholipid (POPG), the toxin underwent major structural changes while bound to the bilayer and was rendered sensitive to protease (29). This suggestive result led us to examine the interaction of RTA with liposomes containing phosphatidylserine (POPS), a negatively charged lipid that, in contrast to POPG, is a significant component of biological membranes (30). In addition, because negatively charged lipids such as PS are synthesized on, and can be detected within, the cytosolic leaflet of the ER membrane (30, 31), we have characterized RTA binding to the surface of intact canine pancreatic ER microsomes to assess RTA affinity for a natural membrane while precluding potential complications from lumenally exposed proteins that may interact with RTA. We show, using chromatography, fluorescence spectroscopy and circular dichroism, that RTA binds to such bilayers in a temperature-independent manner but that the physiologically relevant temperature of 37 °C is required for specific changes in toxin structure and exposure to the nonpolar lipid core. The implications of these findings for RTA retrotranslocation are discussed.
EXPERIMENTAL PROCEDURES
Modification of RTA with NBD—RTA and RTA variants (RTA(C171A)259-NBD, RTA(E177D,C259S,I249C)249-NBD, and RTA(C171A,E177D); the following variants each contain K4R, C171A, E177D, and K239R mutations: RTA(R31K)31-NBD, RTA(E61K)61-NBD, RTA(Q98K)98-NBD, RTA(R114K)114-NBD, RTA(Q128K)128-NBD, and RTA(E135K)135-NBD) were created, expressed, and purified using published methods (9). Because this study focuses on NBD dyes attached to single residues within RTA, NBD-labeled RTA variants are designated RTA#-NBD, and # indicates the amino acid position to which NBD was attached. Cysteines in RTA259-NBD and RTA249-NBD were labeled with the thiol-reactive dye N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine (IANBD; Molecular Probes), whereas lysines in RTA31-NBD, RTA61-NBD, RTA98-NBD, RTA114-NBD, RTA128-NBD, and RTA135-NBD were labeled with the amine-reactive dye succinimidyl 6-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoate (NBD-X; Molecular Probes). In a typical labeling reaction, 2-5 ml of an RTA variant (0.5-2 mg/ml) was thawed and dialyzed overnight against 2 L of labeling buffer (20 mm HEPES, pH 8.0, 50 mm NaCl, 2 mm EDTA). IANBD or NBD-X in Me2SO was then added to provide final concentrations of 4-10-fold molar excess of reagent over the RTA and a maximum of 5% (v/v) Me2SO. After incubation overnight at 4 °C, the reaction was quenched for 1 h at 4 °C by the addition of dithiothreitol to a final concentration of 10 mm (thiol-reactive probes) or freshly prepared hydroxylamine (pH 8.5) to a final concentration of 150 mm (amine-reactive probes). The mixture was then passed over a Sephadex G-50 gel filtration column (30 × 1.5-cm inner diameter) equilibrated in buffer H (50 mm HEPES, pH 7.5). The fractions containing the NBD-labeled protein were pooled, sucrose was added to 0.25 m final concentration, and aliquots were quick-frozen in liquid nitrogen prior to storage at -80 °C. The extent of covalent reaction with NBD was estimated using a molar extinction coefficient of 25,000 m-1 cm-1 for IANBD or 22,000 m -1 cm-1 for NBD-X (32).
Preparation of Microsomes and Liposomes—Salt-washed rough microsomal membranes (KRMs) were prepared from dog pancreas as described previously (33). When KRMs needed to be further purified, a sample was layered on a sucrose step gradient and sedimented (Beckman SW55 rotor; 40,000 rpm; 2 h; 4 °C; ½ × 2 inch polyallomer centrifuge tube with 2 ml of 0.8 m sucrose and 2.5 ml of 2 m sucrose in buffer H). The purified KRMs were located at the 0.8-2 m sucrose interface. Liposomes were prepared as before (see supplemental material).
To prepare KRMs for nitroxide quenching experiments, 1-palmitoyl-2-stearoyl-(5-doxyl)-sn-glycero-3-phosphocholine (5NOPC; Avanti Polar Lipids) or POPC in chloroform was first dried under a stream of nitrogen and further dried under vacuum for an additional 3 h. The phospholipid film was then hydrated in buffer H by vortexing (final lipid concentration, 10 mm). Thereafter, lipid micelles were created by sonication for 30 min at 25 °C in a Bransonic 12 bath sonicator (Branson). Lipid micelles were mixed with an equal volume of microsome buffer (50 mm triethanolamine, pH 7.5, 200 mm sucrose, 1 mm dithiothreitol) containing 250-400 eq KRMs and fused by incubation for 1 h at 25 °C. The nitroxide-labeled (5NOPC) and control (POPC) KRMs were further purified as described above.
Membrane Binding Assays—RTA interactions with membranes were assayed nonspectroscopically using gel filtration chromatography, sedimentation, or alkaline extraction. KRMs (40 eq) were incubated with 2.5 μm RTA259-NBD in buffer H for 30 min at 4 or 26 °C, after which free RTA259-NBD was separated from KRM-bound protein by Sepharose CL-2B (8 × 0.5-cm inner diameter) gel filtration chromatography in buffer H at 4 °C. The fractions (350 μl) were scanned for the presence of RTA259-NBD by NBD emission (λex = 468 nm, λem = 530 nm) or for the presence of KRMs by performing right angle light scattering measurements (λex = 405 nm, λem = 420 nm). When unlabeled 7 μm RTA samples were used, the KRMs were replaced by liposomes (200 μm), the mixture was incubated in buffer C (20 mm sodium phosphate, pH 7.1) for 30 min at the indicated temperature, and RTA in gel filtration fractions was detected by intrinsic fluorescence (λex = 280 nm, λem = 322 nm).
For sedimentation assays, KRMs (20 eq) were incubated with 250 nm RTA259-NBD in buffer H for 30 min at 4 °C or 26 °C. KRMs were collected by sedimentation through a 0.5 m sucrose cushion in buffer H (Beckman TLA100 rotor; 100,000 rpm; 5 min; 4 °C), washed, collected again, and finally resuspended in buffer H. Bovine serum albumin was added as carrier (final concentration, 0.2 mg/ml) to both the combined supernatants of both centrifugation steps and also the resuspended KRM pellet. The proteins were then precipitated with 12.5% (w/v) trichloroacetic acid and analyzed by SDS-PAGE. The bands were visualized by NBD emission using a Pharos FX Plus fluorescence imager (Bio-Rad). For some experiments, microsomal or liposomal membranes were purified by sedimentation and then carbonate extracted (supplemental material) to assess the extent of protein integration into the lipid bilayer (34).
Fluorescence Spectroscopy—Steady-state fluorescence of NBD-labeled RTA samples was measured using an SLM 8100 photon-counting spectrofluorimeter as described previously (35). At low temperatures, the cuvette chamber was continuously flushed with N2 to prevent condensation of water on the microcells. The excitation wavelength (λex) for NBD was 468 nm, and the emission wavelength (λem) was 530 nm; the band-pass was 4 nm. Emission intensity was scanned at 1-nm intervals between 500 and 600 nm, and typically three to five spectra were averaged. The measurements were taken in four 4-mm quartz microcells that were coated with POPC vesicles to minimize protein adsorption (36) at the indicated temperature. In all cases, the samples containing or lacking NBD-labeled proteins were examined in parallel, and the net NBD intensity was determined by subtracting the signal of an equivalent NBD-free sample prepared with unmodified RTA from the RTA#-NBD sample signal. The final RTA concentration was set to 450 nm in all samples using unlabeled RTA as necessary to reach that value. When excess KRMs or liposomes (determined by titration) were added to a sample in microcells, the contents were mixed thoroughly with a 2 × 2-mm magnetic stirring bar as described previously (37), and the resulting sample was incubated at the indicated temperature for 10 (KRMs) or 20 min (liposomes) before the intensity was measured again. Association of RTA#-NBD with KRMs or liposomes was detected by an increase in NBD emission intensity or anisotropy (r, measured as before) (38). For samples that lacked NBD, tryptophan emission of 7 μm RTA samples was scanned using a 4-mm-path length cuvette in a Photon Technology International fluorimeter at 37 °C with 2-nm slit widths, 0.5-s integration time, and 1-nm step size. After the appropriate blank subtraction, four spectra were typically averaged.
Quenching of NBD Emission Intensity by Spin-labeled Phospholipids—The emission spectra were obtained as above using λex = 468 nm and 1-nm intervals between 520 and 550 nm. The net NBD intensities at λem max were determined and normalized to account for differences in the NBD content in each sample. Thereafter, excess PCPS liposomes, either with or without NOPC, were added to parallel samples to a final concentration of 600 μm. When KRMs were used for the quenching experiments, 20 eq of KRMs containing either 5NOPC or POPC were added to each sample. After 20 min of incubation with the membranes at the indicated temperature, the emission spectra were recorded, and the net NBD intensities at λem max were determined and normalized using the membrane-free intensities. F0 was the net intensity of the nitroxide-free sample, and FNO was the net intensity of the sample containing NOPC after the signals of equivalent NBD-free samples prepared with unmodified RTA were subtracted from the NBD sample intensities.
Circular Dichroism—Far-UV CD was measured on a Jasco J-715 spectrapolarimeter at 37 °C using a 1-mm-path length cuvette. Typically six spectra were recorded from 190 to 260 nm with 1-nm step size and averaged. The protein concentrations were 5 μm. The spectra were corrected for background by subtraction of appropriate blanks.
RESULTS
Experimental Strategy—RTA, but not ricin holotoxin, interacts with negatively charged phospholipid vesicles (29). Because RTA is released from RTB within the lumen of the ER, it is conceivable that RTA binds or even inserts into the ER membrane during its passage from the lumen to the cytosol. To examine membrane-protein interactions, we used a spectroscopic approach that has many advantages, including the ability to monitor those interactions in real time with intact membranes under native aqueous conditions and to selectively focus on one or more residues or domains in the protein while doing so (39). Fluorescence spectroscopy is particularly useful because the fluorescence signal can be analyzed using multiple independent techniques.
A water-sensitive fluorescent dye was covalently attached to a single cysteine or lysine residue within RTA to monitor directly the environment of that residue. Each fluorescently labeled RTA was then incubated with either liposomal or purified microsomal ER membranes, and any changes in its spectral properties were characterized. NBD was chosen as the fluorescent probe because its emission properties change dramatically upon moving from an aqueous to a hydrophobic environment (35, 40). Moreover, NBD has a relatively small size for a dye, it is uncharged, it has sufficient polar character to be soluble in an aqueous milieu, and it is not so hydrophobic that it aggressively buries itself in the nonpolar interior of a membrane. Therefore, NBD is a minimally intrusive probe that acts as a sensitive and stable reporter group in both aqueous and nonaqueous environments. By using this experimental approach, many different structural and mechanistic aspects of protein-membrane interactions can be characterized using functional proteins and membranes in aqueous conditions that mimic the physiological (39).
Fluorescently Labeled RTA259-NBD—Wild type RTA contains two cysteine residues at positions 171 and 259. Only 8 amino acids from the C terminus of RTA, Cys259 forms the interchain disulfide bond that links RTA and RTB in the ricin holotoxin. Because this disulfide bond is cleaved after the holotoxin reaches the ER lumen, Cys259 of RTA will normally be reduced in the lumen. NBD was therefore reacted with Cys259 (Cys171 in native RTA does not react because it is deeply buried within the protein; data not shown). However, labeling at Cys171 was further prevented by converting this residue to an Ala (C171A) using site-directed mutagenesis of RTA cDNA. Replacement of Cys171 was shown previously to yield a catalytically active RTA protein with unaffected rRNA depurination and almost unaffected in vitro protein synthesis inhibition activity (5). After purification of the overexpressed recombinant RTAC171A protein, NBD reacted covalently with more than 60% of the Cys259, and further purification yielded the NBD-labeled RTA sample we designated RTA259-NBD.
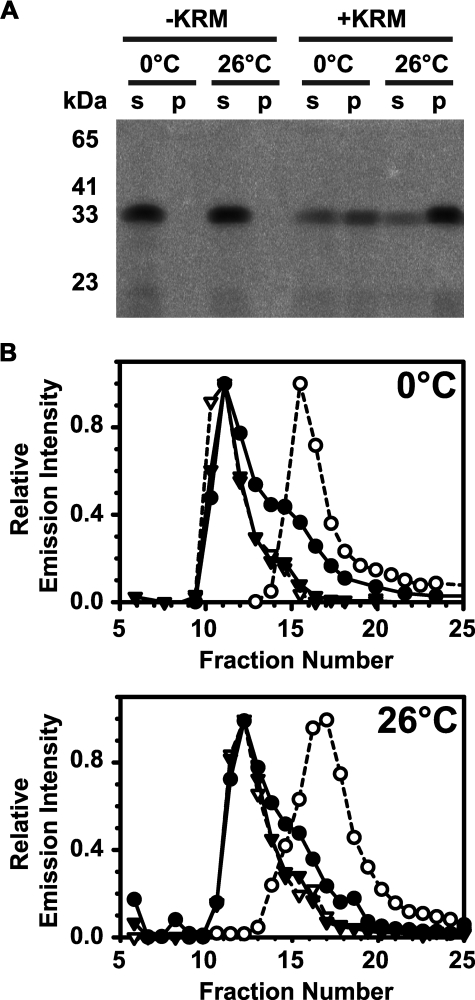
RTA259-NBD Binds to Microsomal ER Membranes—Day et al. (29) showed that RTA binds to liposomes containing negatively charged phospholipids, but RTA binding to natural membranes has not been examined. We therefore investigated whether RTA binds to purified KRMs. Although ricin is exposed first to the lumenal leaflet of the ER membrane after endocytosis, we chose to examine the binding of RTA to the outer surface of ER microsomes because the cytoplasmic leaflet is always exposed in the sealed vesicles. This approach allowed us to focus on RTA with genuine bilayer lipids (including cytoplasmically exposed phosphatidylserine) in the absence of lumenal proteins that might interact with or intercept RTA to complicate interpretation. After incubation of RTA259-NBD with KRMs at 0 or 26 °C, membrane-bound RTA was separated from free protein either by sedimentation or by gel filtration. KRM-exposed RTA259-NBD was found in the pellet following ultracentrifugation, whereas RTA259-NBD remained in the supernatant in the absence of membranes (Fig. 1A). Also, when a sample of RTA259-NBD and KRMs was analyzed by gel filtration, the RTA co-eluted with KRMs in the void volume of the column, whereas KRM-free RTA259-NBD eluted in later fractions (Fig. 1B). Thus, both sedimentation and gel filtration assays demonstrated that RTA259-NBD binds to microsomal membranes with significant affinity. Moreover, this binding occurs at both 0 and 26 °C.
FIGURE 1.
RTA binds to ER microsomal membranes. 0.25 μm (A) 2.5 μm (B) RTA259-NBD were incubated in buffer H for 30 min on ice (0 °C) or at 26 °C with either microsomal membranes (+KRM; 20-40 eq) or an equal amount of buffer without microsomes (-KRM). Free RTA259-NBD was then separated from KRM-bound RTA259-NBD either by sedimentation (A) or by gel filtration chromatography (B). A, following sedimentation, the supernatant (s) and the microsomal pellet (p) were analyzed by SDS-PAGE. NBD-labeled proteins were visualized using a fluorescence imager. B, following mixing, free and KRM-bound RTA259-NBD were separated by gel filtration chromatography in buffer H at 4 °C using a Sepharose CL-2B column (8 × 0.5-cm inner diameter). Each fraction was scanned for the presence of RTA259-NBD (•; λex = 468 nm; λem = 530 nm) and KRMs (▾; λex = 405 nm; λem = 420 nm). As controls, only RTA259-NBD (○) or only KRMs (▿) were run and analyzed separately.
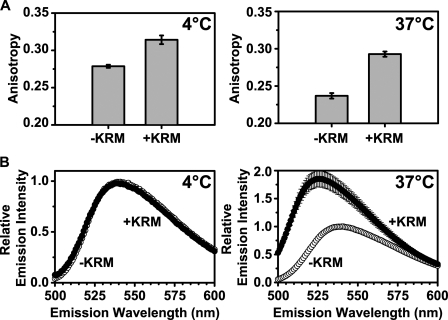
To determine whether RTA259-NBD binding to KRMs can be detected spectroscopically, the membrane dependence of both NBD anisotropy and emission intensity was measured. After determining the anisotropy of free RTA259C-NBD, KRMs were added at either 4 °C or 37 °C. The resulting increases in anisotropy (Fig. 2A) showed that the slowly rotating microsomes significantly slowed the rotational rate of the protein and its NBD, thereby demonstrating that RTA259-NBD bound to the KRMs at both 4 and 37 °C. No change in the NBD emission spectrum of RTA259-NBD was observed when KRMs were added to the protein at 4 °C (Fig. 2B). However, upon RTA259-NBD incubation with KRMs at 37 °C, a substantial increase in fluorescence intensity was observed and also a significant blue shift in the wavelength of maximum emission intensity (λem max) (Fig. 2B). Thus, the NBD at residue 259 moved from an aqueous environment to a more hydrophobic milieu when RTA was exposed to microsomal membranes at 37 °C.
FIGURE 2.
RTA259-NBD binds to microsomes differently at 4 °C and 37 °C. Anisotropy measurements (A) and emission scans (B; λex = 468 nm) of RTA259-NBD (450 nm) were performed before (-KRM) and immediately after the addition of ER microsomal membranes (+KRM; 15-20 eq) in buffer H. Emission intensity and anisotropy data were corrected by the subtraction of the signal obtained from an equivalent NBD-free RTA sample. The averages of at least three independent experiments are shown, and the error bars indicate the S.D. of the experiments.
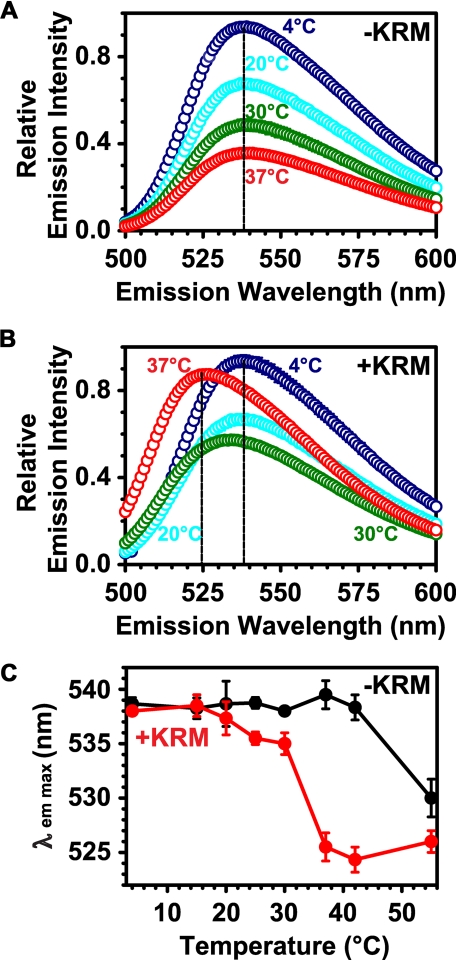
The NBD Emission Spectrum of Membrane-bound RTA259-NBD Is Temperature-dependent—To determine at which temperature the NBD environment of membrane-exposed RTA changes, we compared the emission spectra of free RTA with those of membrane-bound RTA as a function of temperature. When incubated without KRMs, the fluorescence intensity of RTA259-NBD declined with increasing temperature (Fig. 3A) because of the reduced quantum yield of the NBD dye at higher temperatures. In the presence of membranes, the same effect was observed until the temperature reached 30 °C. However, when the temperature was further increased to 37 °C, a substantial increase in RTA259-NBD fluorescence intensity was observed, as well as a significant blue shift in λem max (Fig. 3, B and C). Thus, a relatively small increase in temperature from 30 to 37 °C caused a dramatic spectral change and hence a highly temperature-dependent transition from an aqueous to a hydrophobic environment for the NBD dye at the C terminus of RTA. To determine whether this change in RTA environment originated from a phase change in the lipid bilayer, 1,6-diphenyl-1,3,5-hexatriene was incorporated into KRMs to monitor bilayer fluidity. Because no sudden change in the fluidity or phase of the ER lipids occurred between 20 and 37 °C (supplemental Fig. S1), the sharp temperature-dependent transition from an aqueous to a hydrophobic environment for the C terminus of RTA most likely results from a conformational change in the membrane-bound RTA protein that occurs between 30 and 37 °C.
FIGURE 3.
Temperature dependence of the RTA259-NBD probe environment. Net emission scans (λex = 468 nm) of RTA259-NBD (450 nm) are shown in the presence of either microsome buffer (A) or of KRMs (B; 15-20 eq) at increasing temperatures in buffer H (A and B: only 4, 20, 30, and 37 °C are shown). The λem max at different temperatures is shown in (C). The averages of three independent experiments are shown, and the error bars indicate the S.D. of the experiments. However, most of the error bars in A and B are smaller than the circles on the graph.
RTA Binding to Membranes Detected by Intrinsic Fluorescence—To exclude the possibility that the covalently attached NBD dye is responsible for the binding of RTA259-NBD to ER microsomal membranes, we monitored the intrinsic fluorescence of unlabeled RTA in the absence or presence of membranes. Wild type RTA contains one Trp (Trp211) and 14 Tyr residues. Conformational changes in RTA can therefore be detected by changes in Trp or Tyr emission without introducing an extrinsic fluorescent dye into the protein. However, to focus solely on RTA fluorescence, we had to use liposomes to avoid the substantial Trp emission from membrane proteins in KRMs.
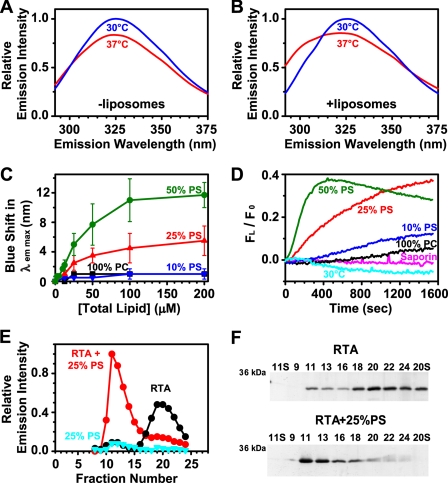
When the temperature of parallel RTA samples was increased from 30 to 37 °C, a change in Trp emission (aside from the usual temperature-dependent decrease in intensity) was observed only in the sample containing excess liposomes (Figs. 4, A and B). Thus, RTA binding to the membrane can be detected spectroscopically by the lowering of the λem max that occurs when the Trp environment becomes more nonpolar. In fact, by monitoring the temperature change at higher resolution, it is clear that RTA binding to membranes is initiated when the temperature is between 33 and 35 °C (Fig. S2). This effect was membrane-dependent because there was very little change in RTA Trp fluorescence upon incubation with PC liposomes (Figs. 4, C and D). But increasing the concentration of the liposomes and the molar ratio of PS within them markedly increased the blue shift in λem max (Fig. 4C). Kinetically, fluorescence changes occurred more rapidly as the mole fraction of PS in the liposomes increased (Fig. 4D). The Trp fluorescence of saporin, a structurally related type-I ribosome-inactivating protein that lacks both a C-terminal hydrophobic region and the ability to internalize to the ER of cells, was not affected by the addition of negatively charged liposomes (Fig. 4D).
FIGURE 4.
Temperature and PS dependence of RTA binding to liposomes. Trp emission spectra (λex = 280 nm) of 7 μm RTA are shown in the absence (A) and presence (B) of 200 μm PCPS liposomes at 30 °C and 37 °C in buffer C. The λem max of RTA Trp emission was determined as a function of phospholipid concentration and composition (C). The error bars show S.D. from two or three experiments. D, after the addition of liposomes at 0 s, Trp emission intensity at 300 nm (FL) was monitored over time relative to the initial liposome-free intensity (F0). Emission intensity data were corrected by both subtraction of the signal obtained from samples lacking RTA and subtraction of the signal obtained by RTA or saporin alone. The average of at least two different experiments is shown. E and F, RTA was incubated for 30 min at 37 °C with 200 μm PCPS liposomes, the mixture was subjected to gel filtration on a Sepharose CL-2B column (18 × 0.5-cm inner diameter), and 250-μl fractions were collected. The samples were analyzed for RTA emission intensity (λex = 280 nm; λem = 322 nm; E) and protein content by SDS-PAGE followed by silver staining (F). The lanes are labeled in fraction numbers with S indicating the loaded supernatant of given fractions following a 10-min microcentrifuge centrifugation at 14,000 rpm.
To confirm binding, we incubated RTA with liposomes at 37 °C and separated membrane-bound RTA from free protein by gel filtration. RTA and liposomes co-eluted in the void volume from a gel filtration column (Fig. 4E; light scattering data not shown). In this case, RTA remained bound to lipids even after gel filtration and sedimentation of the liposomes (Fig. 4F). In contrast free RTA eluted in the later liposome-free fractions (Figs. 4, E and F). This can be seen from a gel analysis of the fractions (Fig. 4F). It should be noted that a small amount of RTA was observed in the early fractions (11-13) of the RTA-only separation, most likely because of a small amount of aggregation. From this, it can be seen that the binding of RTA to lipids is driven by the RTA protein itself, not by any attached NBD dye.
The RTA C Terminus Is Exposed to the Nonpolar Core of the Bilayer—When RTA binds to a microsome, the NBD at residue 259 could move into a more hydrophobic environment by inserting into the nonpolar interior of either the membrane or the protein. These two possibilities can be distinguished directly by using a technique based on the collisional quenching of fluorescence (39, 41). We therefore integrated 5NOPC as a lipophilic quenching agent into KRMs and liposomes. We chose to use 5NOPC because the nitroxide moiety (NO) is covalently attached to a phospholipid acyl chain and hence is restricted to the nonpolar interior of the membrane. Although the NO is covalently attached partway into the nonpolar membrane core, the flexibility and dynamic motion of the acyl chain allows essentially all NBD probes facing the bilayer to contact the nitroxide moiety and hence be quenched, although to different extents (41, 42). This approach unambiguously distinguishes between nonpolar environments formed by the bulk lipid of the membrane and those formed at protein-protein interfaces or by protein folding because only NBD dyes exposed to the interior of the lipid bilayer can collide with NO and be quenched.
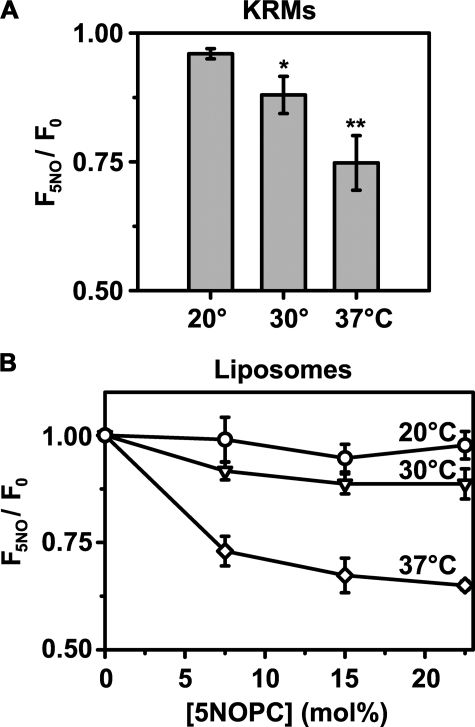
Upon binding to KRMs containing 5NOPC at 37 °C, RTA259-NBD emission intensity was quenched by the membrane-integrated nitroxide moiety (Fig. 5A). Thus, the dye attached to 259 is exposed to the membrane interior when RTA binds to the microsomal membrane at 37 °C. When incubated at 30 °C, the quenching was much less efficient, and almost no quenching of RTA259-NBD was detected at 20 °C. Because we could not quantify the concentration of 5NOPC within the KRMs, we next prepared PCPS liposomes containing increasing concentrations of 5NOPC. Concentration-dependent quenching curves for the liposomes containing 5NOPC are shown in Fig. 5B. Thus, RTA appears to bind similarly to both KRMs and PCPS liposomes. At 37 °C, maximal quenching efficiency was achieved with liposomes containing about 22.5 mol% 5NOPC. But at the same 5NOPC concentration, quenching was less efficient at 30 °C and was almost undetectable at 20 °C. These data therefore strongly indicate that exposure of the NBD of RTA259-NBD to the nonpolar lipid core of ER microsomes and of PCPS vesicles is strongly temperature-dependent. RTA does not bind to PC liposomes (Fig. 4, C and D), and as expected, no quenching was observed with liposomes containing only POPC and 5NOPC at either 20 °C (F5NO/F0 = 1.0 ± 0.1) or 37 °C (F5NO/F0 = 1.1 ± 0.1).
FIGURE 5.
Exposure of RTA259-NBD to the membrane interior. The emission intensity of RTA259-NBD (450 nm in buffer H) was measured before and after the addition of either KRMs (20 eq; A) or 600 μm PCPS liposomes (B). Emission intensities of parallel samples containing either 5NOPC (F5NO) or an equal mol% of POPC (F0) were compared. The net emission intensities are shown as a function of the mol% of 5NOPC in the liposomes (B), but the final mol% of 5NOPC/POPC in the bulk lipid of natural ER membranes (A) cannot be quantified. The averages of at least three independent experiments are shown, and the errorbars indicate the S.D. of the experiments. *, p = 0.208; **, p = 0.006 compared with the quenching efficiency at 20 °C, respectively (Student's t test).
Collisional quenching experiments were also done with iodide ions, quenchers that are restricted to the aqueous phase. As expected, the temperature dependence of NBD accessibility to I- was the reverse of that to 5NOPC; the extent of I- quenching decreased substantially when the temperature was raised to 37 °C in the presence of KRMs (supplemental Fig. S3). The data obtained with an aqueous and a lipophilic quencher therefore complement each other and strongly indicate that residue Cys259 at the C terminus of membrane-bound RTA is exposed to the aqueous medium at low temperatures but is inserted and exposed to the nonpolar lipid core at 37 °C.
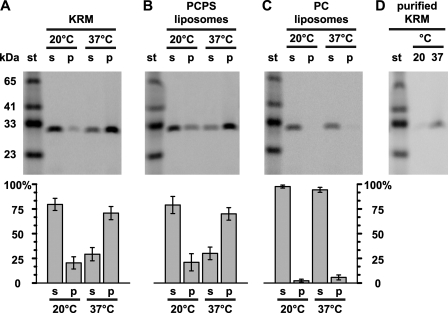
Membrane-bound RTA259-NBD Is Resistant to Alkaline Extraction—To determine whether or not RTA is stably embedded in the bilayer core or is bound peripherally to the membrane surface, we examined whether RTA became resistant to alkaline carbonate extraction following its incubation with membranes at various temperatures. After RTA259-NBD was preincubated with KRMs or PCPS liposomes at either 20 or 37 °C, membrane-bound RTA was separated from free protein by sedimentation through a sucrose cushion, and the membranes were treated with sodium carbonate (pH 11.5). The majority of RTA259-NBD incubated at 37 °C was retained in the membrane pellet fraction (KRMs: 71 ± 7%; liposomes: 70 ± 6%; Fig. 6, A, B, and D), thereby indicating that RTA259-NBD was stably integrated into the bilayer. In contrast, at 20 °C, much less RTA259-NBD was found in the carbonate pellet (KRMs: 20 ± 6%; liposomes: 21 ± 9%), suggesting that most membrane-bound RTA was carbonate-extractable (Fig. 6, A, B, and D).
FIGURE 6.
Some membrane-bound RTA is stably embedded in the bilayer. RTA259-NBD (500 nm) was incubated with either 40 eq of KRMs (A and D), or 2.5 mm PCPS liposomes (B) in buffer H for 30 min at 20 °C or 37 °C. Membrane-bound RTA was purified by centrifugation and then extracted with alkaline sodium carbonate. The protein contents of the supernatant (s), the membrane pellet (p) fractions, and a molecular weight standard (st) were then analyzed by SDS-PAGE. C, RTA259-NBD (1 μm) was incubated with 5 mm PC liposomes in 10 mm HEPES (pH 7.5) for 30 min at 20 °C or 37 °C. The samples were then treated as in B. D, KRMs were incubated with RTA259-NBD, treated with carbonate, and then purified using a sucrose step gradient as described under “Experimental Procedures.” NBD-labeled proteins were visualized and quantified using a fluorescence imager. Representative gels from a set of at least three independent experiments are shown. Histograms show the average fraction of the total protein in the supernatant and membrane pellet fractions, respectively. The error bars indicate the S.D. of the experiments.
As a control for this unexpected result, we incubated RTA259-NBD with PC liposomes that do not bind to the protein either at 20 or 37 °C and then subjected them to treatment with alkaline carbonate. The fact that RTA259-NBD was exclusively found in the supernatant (99 and 97% after incubation at 20 or 37 °C, respectively) excludes the possibility that RTA aggregates and then precipitates with membranes after the carbonate treatment (Fig. 6C). To further exclude the possibility of temperature-dependent RTA259-NBD precipitation, RTA259-NBD-exposed and carbonate-treated KRMs were purified using a sucrose step gradient in 0.1 m Na2CO3, pH 11.5. RTA259-NBD was then recovered only in the purified KRMs that had been incubated at 37 °C and not those incubated at 20 °C (Fig. 6D). Altogether, these data strongly indicate that RTA259-NBD is stably inserted into the nonpolar cores of both KRMs and liposomes containing negatively charged lipids. Furthermore, this insertion is temperature-dependent; a significant portion of RTA is embedded in the membrane only when the temperature rises to 37 °C.
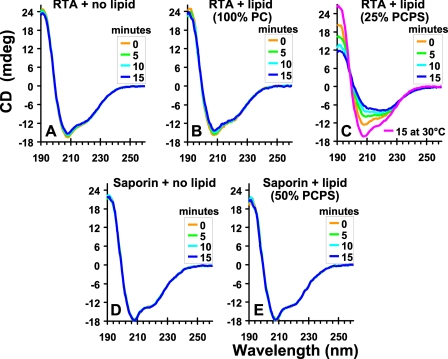
The Secondary Structure of RTA Is Altered upon Binding to Lipids—To determine whether the binding of RTA to membranes introduces a change in its secondary structure, we compared the CD spectra of unmodified RTA in the absence and presence of liposomes at 37 °C. The CD spectrum of RTA indicates a significant amount of α-helix with minima at 208 and 222 nm (Fig. 7A), consistent with the published crystal structure (43). Upon exposure to PCPS liposomes, the conformation of RTA changes and its α-helical structure is reduced (Fig. 7C) concomitant with an increase in β sheet. The CD data therefore show that some α-helical structure is lost upon RTA binding to membranes. In contrast, the secondary structure of saporin was not altered by PCPS liposomes (Fig. 7, compare D and E). The RTA secondary structure was unchanged by incubation at 30 °C in the presence of PCPS liposomes (Fig. 7C), although increasing the proportion of PS in the liposomes promoted the loss of α-helical structure of RTA at lower temperatures (supplemental Fig. S4). Thus, the secondary structural change detected by CD requires both binding to a PS-containing membrane and a temperature of 37 °C.
FIGURE 7.
Secondary structural changes in RTA and saporin. Recordings of the far-UV CD spectra of RTA and saporin, each 5 μm in buffer C at 37 °C, in the absence or presence of 200 μm liposomes containing different percentages of PC or PS. The scans were corrected by the subtraction of blanks containing only buffer and/or liposomes. The panels show the averaged spectra of at least two experiments.
Topographical Rearrangements of Membrane-bound RTA—Changes in the topography of the membrane-bound RTA were detected by distance-of-closest-approach fluorescence resonance energy transfer measurements between RTA259-NBD and the cytoplasmic leaflet of the microsomal membrane. When the temperature was increased to 37 °C, the spatial separation between the C terminus of RTA and the membrane surface decreased (see supplemental material).
Exposure of Specific RTA Residues to Bilayer Lipids—Above, we have documented a temperature-dependent exposure of RTA259-NBD to the nonpolar lipid core of the membrane that coincides with a conformational change in RTA. Does this conformational change constitute the partial unfolding of RTA that has long been postulated to precede its retrotranslocation through the ER (5, 6)? If so, the unfolding might proceed randomly to create a collection of different partially unfolded RTA molecules or via an ordered pathway of specific structural rearrangements that reproducibly create a specific membrane-exposed topography. We therefore examined the membrane exposure of various sites within RTA (Fig. 8A). Each of these derivatives bound to KRMs as shown by increases in NBD anisotropy upon KRM exposure (supplemental Tables S1 and S2).
FIGURE 8.
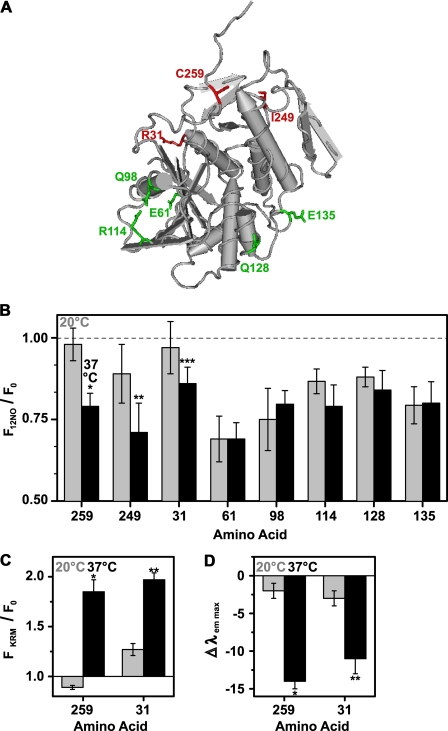
Exposure of different RTA residues to the membrane interior. A, the tube worm representation of the α-carbon backbone of the RTA crystal structure is shown. Amino acids that are exposed to the membrane at 20 °C are shown in green, whereas amino acids that are exposed to the nonpolar lipid core only at 37 °C are indicated in red. B, the emission intensities of NBD-labeled RTA mutants (450 nm in buffer H) were measured before and after the addition of PCPS liposomes. Emission intensities of parallel samples containing either 22.5 mol% 12NOPC (F12NO) or 22.5 mol% of PC (F0) were compared at 20 °C (gray bars) or at 37 °C (black bars), respectively. The averages of at least three independent experiments are shown, and the error bars indicate the S.D. of the experiments. Sequence numbers of NBD-labeled amino acids are shown on the x axis. *, p = 0.0004; **, p = 0.04; ***, p = 0.07 when compared with the corresponding quenching efficiency at 20 °C (Student's t test). C and D, the ratio of RTA mutant (450 nm in buffer H) NBD emission intensity (C) and the change in λem max (D) are shown before (F0) and after (FKRM) binding to KRMs (20 eq), either at 20 °C (gray bars) or at 37 °C (black bars). The average of at least three independent experiments is shown, and the error bars indicate the S.D. of the experiments. A, *, p < 0.00004; **, p < 0.0002. B, *, p < 0.00001; **, p < 0.001 when the measurements at 20 °C were compared with those at 37 °C (Student's t test).
Because NBD exposure to the nonpolar lipid core is revealed directly by 5NOPC quenching, each RTA-NBD mutant was bound to and quenched by liposomes containing 22.5 mol% 12NOPC (Fig. 8B). The NO is located deeper in the bilayer with 12NOPC than with 5NOPC, so the extent of RTA259-NBD quenching was lower for 12NOPC than for 5NOPC at 37 °C (compare Figs. 5B and 8B). NBD probes at RTA residues 259, 249, and 31 were quenched more efficiently at 37 °C than at 20 °C (Fig. 8B), thereby showing that the membrane- and temperature-induced structural transitions in RTA have the greatest effect on residues 259, 249, and perhaps 31 in terms of exposing them to the bilayer interior. The probe environment at residue 31 was characterized further after binding to natural KRMs at 20 °C and at 37 °C, and the temperature-induced changes in emission intensity and λem max were similar for the probes at positions 259 and 31 (Fig. 8, C and D).
In contrast, probes at position 61, 98, 114, 128, and 135 of RTA were quenched at both 37 and 20 °C (Fig. 8B). Hence, each of these residues is exposed to the bilayer interior when RTA binds to the membrane, regardless of the temperature. Furthermore, the extents of quenching do not change significantly as the temperature is increased, which suggests that the location of each probe in the bilayer interior is not altered much, if at all.
Thus, two groups of RTA residues have been identified; one group is membrane-exposed only at the physiological relevant temperature of 37 °C, whereas the other is membrane-exposed even at 20 °C. Interestingly, the residues in each group localize on different sides of RTA (Fig. 8A). Thus, at 20 °C one half of membrane-bound RTA appears to be in an aqueous environment, whereas the other half of the protein is exposed to the membrane interior. However, at 37 °C, much of RTA is exposed to the lipid core of the bilayer.
DISCUSSION
It is generally assumed that after separation from RTB in the ER lumen, free RTA initiates a series of specific interactions that facilitate its translocation through the ER membrane to reach its ribosomal substrates in the cytosol. The biochemical and spectroscopic evidence provided here strongly suggest that RTA binding directly to a membrane surface may constitute an early, and perhaps obligatory, early step in this pathway. Furthermore, the extent of RTA interaction with the bilayer is strikingly temperature-sensitive (Fig. 3). At temperatures of 30 °C or less, RTA appears to bind peripherally to both natural ER membranes (Figs. 1 and 2) and liposomes (Fig. 4) because the bound protein is carbonate-extractable (Fig. 6) and because only certain RTA residues are exposed to the nonpolar lipid core of the membrane (Fig. 8B). However, at the physiologically relevant temperature of 37 °C, membrane-bound RTA loses some of its helical content (Fig. 7), undergoes a conformational change that exposes C-terminal residues to the membrane interior (Figs. 5 and 8B), is no longer extracted from the membrane by carbonate (perhaps because of the additional hydrophobic interactions) (Fig. 6), and hence is apparently embedded in the bilayer. The temperature dependence of these structural alterations correlate remarkably well with the growing temperature of Ricinus communis and the physiological 37 °C temperature of the mammalian “target” organisms of ricin holotoxin. These results therefore suggest that RTA inserts into and is optimally transported across mammalian ER membranes by a mechanism that is potentially unavailable in the plant itself because of its generally lower growing temperature.
Partial unfolding of RTA has long been postulated to precede its retrotranslocation through the ER (5, 29). Here, we show that upon exposure to membranes containing negatively charged phospholipids at 37 °C, RTA undergoes significant conformational changes, including the fluorescence resonance energy transfer-detected movement of the C terminus of RTA closer to the membrane surface. Previous work has shown that modifications to the C terminus of RTA, although neutral to ricin trafficking and catalytic activity, had a profound affect on some aspect of the membrane translocation process and has implicated the C terminus in translocation (28). Because this major spatial movement is not likely to be explained by a small localized conformational change in RTA, this observation may indicate that RTA (partially) unfolds upon membrane exposure at 37 °C. Although these changes cannot yet be assigned solely to RTA unfolding, there is a substantial reduction in the secondary structure of RTA (Fig. 7) that coincides with RTA exposure to and stable insertion into to the nonpolar lipid core of the membrane (Figs. 5, 6, and 8).
This transition from a folded, soluble protein to a partly unfolded, membrane-exposed state might trigger the recognition of RTA as a misfolded protein substrate by the ERAD machinery. RTA would then presumably cross the ER membrane via a protein translocon, as do other misfolded proteins. The potential participation of such a translocon in RTA retro-translocation was suggested by the co-immunoprecipitation of RTA with Sec61p (4). Recent studies also indicated that EDEM, which seems to be crucial for extracting terminally misfolded proteins from the calnexin cycle (44, 45), is involved in RTA retrotranslocation to the cytosol (24). In those studies, RTA was co-immunoprecipitated with both EDEM and Sec61α, and EDEM promoted RTA retrotranslocation to the cytosol when the interaction between EDEM and other misfolded proteins was disrupted. Because these experiments were done at 37 °C, it is likely that RTA was in a membrane-inserted state prior to its interaction with EDEM and hence had lost some secondary structure (Fig. 7) and had become more exposed to the bilayer lipid core (Figs. 5 and 8). However, the identity and role of any protein cofactors involved in RTA retrotranslocation in a mammalian system have yet to be clarified.
Also, other mechanisms of RTA translocation through the ER membrane cannot yet be ruled out. For example, RTA may pass through the membrane without the involvement of a translocon, or alternatively, RTA may form a pore itself. Partly unfolded RTA has been described to resemble a molten globule species (27). Such potential insertion intermediates may be capable of membrane transversal (46) and might be involved in the insertion of the pore-forming domain of colicin A into membranes (47). Moreover, interactions with negatively charged membranes have been reported to induce a transition from a folded protein to a molten globule (48). The interaction with negatively charged membranes at the physiological temperature of 37 °C might induce similar changes in RTA as well, although it is beyond the scope of the present study to determine whether membrane-exposed RTA at 37 °C resembles a real molten globule. However, we have clearly shown that, at 37 °C, membrane-bound RTA is exposed to the nonpolar lipid core (Fig. 8) and is embedded in the membrane because of its resistance to alkaline extraction (Fig. 6). It remains to be seen whether membrane-inserted RTA is a self-sufficient insertion intermediate en route through the ER membrane or whether RTA mimics a misfolded protein that is subsequently recognized by the ERAD machinery prior its passage through a protein translocon.
The goal of this study was to characterize the interaction of RTA with the lipid components of a natural membrane. Because the same results were obtained with both natural ER microsomes and simple artificial liposomes that contained only PC and PS and lacked any integral membrane proteins, it appears that RTA binding to membranes primarily requires a membrane surface with negatively charged phospholipids (Fig. 4). Yet this study would have ideally involved the lipids that comprise the lumenal leaflet of the ER membrane, because that is what RTA initially faces after separation from RTB. However, interpretation of the lipid dependence of RTA binding to the lumenal surface would be complicated by RTA binding to various lumenal proteins and protein domains. Thus, to avoid interference from protein-protein interactions, we chose to examine RTA binding to the best natural membrane alternative, the cytoplasmic surface of purified and sealed ER microsomes. Negatively charged lipids such as PS can be detected within both leaflets of the ER membrane (30, 31), although the asymmetric distribution of some lipid species and the precise rate of lipid flipping is poorly understood. However, a difference between cytosolic and lumenal ER lipids may stimulate rather than inhibit RTA binding. For example, PS has been found in higher concentrations in the lumenal than in the cytosolic leaflet of the ER membrane (30, 31). It would therefore appear, given the PS dependence of RTA binding (Fig. 4), that RTA binding to a lumenal ER membrane surface would be even higher than we observe at the cytosolically exposed microsomal surface.
The most important observation of this study is that binding to a membrane surface is an intrinsic and highly temperature-dependent property of RTA. Upon binding to a membrane surface at 37 °C, RTA undergoes reproducible changes in state that substantially alter both its conformation and its exposure to the nonpolar membrane interior. It will now be important to determine how, and if, RTA-membrane interactions and topography are influenced by the presence of any soluble lumenal proteins and/or lumenally exposed membrane protein domains.
Supplementary Material
Acknowledgments
We are grateful to Philip Robinson (University of Warwick) for helpful advice regarding circular dichroism, Y. Miao and Y. Shao for outstanding technical assistance, and B. Hou for providing rhodamine-phosphatidylethanolamine·KRMs. Saporin was a gift from Dr. David Flavell (Southampton General Hospital).
This work was supported, in whole or in part, by National Institutes of Health Grant GM26494 (to A. E. J.). This work was also supported by Wellcome Trust Program Grant 080566Z/06/Z (to J. M. L. and L. M. R.) and by Robert A. Welch Foundation Chair Grant BE-0017 (to A. E. J.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1-S4.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: RTA/RTB, ricin A/B chain; ER, endoplasmic reticulum; ERAD, ER-associated degradation; EDEM, ER degradation enhancing α-mannosidase-like protein; NBD, 7-nitrobenz-2-oxa-1,3-diazole; KRM, salt-washed rough microsomal membrane; POPS, 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-l-serine]; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; POPG, 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]; PC liposomes, 100 mol% POPC; PCPS liposomes, 75 mol% POPC and 25 mol% POPS; IANBD, N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine; NBD-X, succinimidyl 6-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoate; 5NOPC, 1-palmitoyl-2-stearoyl-(5-doxyl)-sn-glycero-3-phosphocholine; NO, nitroxide moiety; 12NOPC, 1-palmitoyl-2-stearoyl-(12-doxyl)-sn-glycero-3-phosphocholine.
References
- 1.Roberts, L. M., and Lord, J. M. (2004) Mini Rev. Med. Chem. 4 505-512 [DOI] [PubMed] [Google Scholar]
- 2.Rapak, A., Falnes, P. O., and Olsnes, S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3783-3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spooner, R. A., Watson, P. D., Marsden, C. J., Smith, D. C., Moore, K. A., Cook, J. P., Lord, J. M., and Roberts, L. M. (2004) Biochem. J. 383 285-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wesche, J., Rapak, A., and Olsnes, S. (1999) J. Biol. Chem. 274 34443-34449 [DOI] [PubMed] [Google Scholar]
- 5.Argent, R. H., Roberts, L. M., Wales, R., Robertus, J. D., and Lord, J. M. (1994) J. Biol. Chem. 269 26705-26710 [PubMed] [Google Scholar]
- 6.Beaumelle, B., Taupiac, M. P., Lord, J. M., and Roberts, L. M. (1997) J. Biol. Chem. 272 22097-22102 [DOI] [PubMed] [Google Scholar]
- 7.Romisch, K. (2005) Annu. Rev. Cell Dev. Biol. 21 435-456 [DOI] [PubMed] [Google Scholar]
- 8.Bukau, B., Weissman, J., and Horwich, A. (2006) Cell 125 443-451 [DOI] [PubMed] [Google Scholar]
- 9.Deeks, E. D., Cook, J. P., Day, P. J., Smith, D. C., Roberts, L. M., and Lord, J. M. (2002) Biochemistry 41 3405-3413 [DOI] [PubMed] [Google Scholar]
- 10.Di Cola, A., Frigerio, L., Lord, J. M., Ceriotti, A., and Roberts, L. M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14726-14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodighiero, C., Tsai, B., Rapoport, T. A., and Lencer, W. I. (2002) EMBO Rep. 3 1222-1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kothe, M., Ye, Y., Wagner, J. S., Deluca, H., Kern, E., Rapoport, T. A., and Lencer, W. I. (2005) J. Biol. Chem. 280 28127-28132 [DOI] [PubMed] [Google Scholar]
- 13.Teter, K., Allyn, R. L., Jobling, M. G., and Holmes, R. K. (2002) Infect. Immun. 70 6166-6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernardi, K. M., Forster, M. L., Lencer, W. I., and Tsai, B. (2007) Mol. Biol. Cell 19 877-884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carbonetti, N. H., Irish, T. J., Chen, C. H., O'Connell, C. B., Hadley, G. A., McNamara, U., Tuskan, R. G., and Lewis, G. K. (1999) Infect. Immun. 67 602-607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plaut, R., and Carbonetti, N. H. (2008) Cell Microbiol. 10 1130-1139 [DOI] [PubMed] [Google Scholar]
- 17.Sandvig, K., Garred, O., Prydz, K., Kozlov, J. V., Hansen, S. H., and van Deurs, B. (1992) Nature 358 510-512 [DOI] [PubMed] [Google Scholar]
- 18.Yu, M., and Haslam, D. B. (2005) Infect. Immun. 73 2524-2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida, T., Chen, C. C., Zhang, M. S., and Wu, H. C. (1991) Exp. Cell Res. 192 389-395 [DOI] [PubMed] [Google Scholar]
- 20.Jackson, M. E., Simpson, J. C., Girod, A., Pepperkok, R., Roberts, L. M., and Lord, J. M. (1999) J. Cell Sci. 112 467-475 [DOI] [PubMed] [Google Scholar]
- 21.Kreitman, R. J., and Pastan, I. (1995) Biochem. J. 307 29-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai, B., Rodighiero, C., Lencer, W. I., and Rapoport, T. A. (2001) Cell 104 937-948 [DOI] [PubMed] [Google Scholar]
- 23.Bellisola, G., Fracasso, G., Ippoliti, R., Menestrina, G., Rosen, A., Solda, S., Udali, S., Tomazzolli, R., Tridente, G., and Colombatti, M. (2004) Biochem. Pharmacol. 67 1721-1731 [DOI] [PubMed] [Google Scholar]
- 24.Slominska-Wojewodzka, M., Gregers, T. F., Walchli, S., and Sandvig, K. (2006) Mol. Biol. Cell 17 1664-1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day, P. J., Owens, S. R., Wesche, J., Olsnes, S., Roberts, L. M., and Lord, J. M. (2001) J. Biol. Chem. 276 7202-7208 [DOI] [PubMed] [Google Scholar]
- 26.Pande, A. H., Scaglione, P., Taylor, M., Nemec, K. N., Tuthill, S., Moe, D., Holmes, R. K., Tatulian, S. A., and Teter, K. (2007) J. Mol. Biol. 374 1114-1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Argent, R. H., Parrott, A. M., Day, P. J., Roberts, L. M., Stockley, P. G., Lord, J. M., and Radford, S. E. (2000) J. Biol. Chem. 275 9263-9269 [DOI] [PubMed] [Google Scholar]
- 28.Simpson, J. C., Lord, J. M., and Roberts, L. M. (1995) Eur. J. Biochem. 232 458-463 [DOI] [PubMed] [Google Scholar]
- 29.Day, P. J., Pinheiro, T. J., Roberts, L. M., and Lord, J. M. (2002) Biochemistry 41 2836-2843 [DOI] [PubMed] [Google Scholar]
- 30.Zachowski, A. (1993) Biochem. J. 294 1-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgins, J. A., and Dawson, R. M. (1977) Biochim. Biophys. Acta 470 342-356 [DOI] [PubMed] [Google Scholar]
- 32.Haugland, R. P. (2002) Handbook of Fluorescent Probes and Research Products, 9th Ed., Molecular Probes, Inc., Eugene, OR
- 33.Walter, P., and Blobel, G. (1983) Methods Enzymol. 96 84-93 [DOI] [PubMed] [Google Scholar]
- 34.Fujiki, Y., Hubbard, A. L., Fowler, S., and Lazarow, P. B. (1982) J. Cell Biol. 93 97-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shepard, L. A., Heuck, A. P., Hamman, B. D., Rossjohn, J., Parker, M. W., Ryan, K. R., Johnson, A. E., and Tweten, R. K. (1998) Biochemistry 37 14563-14574 [DOI] [PubMed] [Google Scholar]
- 36.Ye, J., Esmon, N. L., Esmon, C. T., and Johnson, A. E. (1991) J. Biol. Chem. 266 23016-23021 [PubMed] [Google Scholar]
- 37.Dell, V. A., Miller, D. L., and Johnson, A. E. (1990) Biochemistry 29 1757-1763 [DOI] [PubMed] [Google Scholar]
- 38.Johnson, A. E., Adkins, H. J., Matthews, E. A., and Cantor, C. R. (1982) J. Mol. Biol. 156 113-140 [DOI] [PubMed] [Google Scholar]
- 39.Johnson, A. E. (2005) Traffic 6 1078-1092 [DOI] [PubMed] [Google Scholar]
- 40.Crowley, K. S., Reinhart, G. D., and Johnson, A. E. (1993) Cell 73 1101-1115 [DOI] [PubMed] [Google Scholar]
- 41.Shatursky, O., Heuck, A. P., Shepard, L. A., Rossjohn, J., Parker, M. W., Johnson, A. E., and Tweten, R. K. (1999) Cell 99 293-299 [DOI] [PubMed] [Google Scholar]
- 42.London, E., and Feigenson, G. W. (1981) Biochemistry 20 1932-1938 [DOI] [PubMed] [Google Scholar]
- 43.Montfort, W., Villafranca, J. E., Monzingo, A. F., Ernst, S. R., Katzin, B., Rutenber, E., Xuong, N. H., Hamlin, R., and Robertus, J. D. (1987) J. Biol. Chem. 262 5398-5403 [PubMed] [Google Scholar]
- 44.Molinari, M., Calanca, V., Galli, C., Lucca, P., and Paganetti, P. (2003) Science 299 1397-1400 [DOI] [PubMed] [Google Scholar]
- 45.Oda, Y., Hosokawa, N., Wada, I., and Nagata, K. (2003) Science 299 1394-1397 [DOI] [PubMed] [Google Scholar]
- 46.Bychkova, V. E., Pain, R. H., and Ptitsyn, O. B. (1988) FEBS Lett. 238 231-234 [DOI] [PubMed] [Google Scholar]
- 47.van der Goot, F. G., Gonzalez-Manas, J. M., Lakey, J. H., and Pattus, F. (1991) Nature 354 408-410 [DOI] [PubMed] [Google Scholar]
- 48.de Jongh, H. H., Killian, J. A., and de Kruijff, B. (1992) Biochemistry 31 1636-1643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.