Abstract
At high bacterial cell density the gene expression program of Pseudomonas aeruginosa is regulated by quorum sensing. Among the gene products highly up-regulated by this system is an exoprotease, leucine aminopeptidase (PA-LAP), which is coexpressed with several known virulence factors and secreted as a proenzyme. We undertook a study of its activation by expressing the full-length proform of PA-LAP recombinantly in Escherichia coli (here termed, rLAP55) and characterizing individual steps in its conversion to an active enzyme. Activation is initiated with the proteolytic removal of a C-terminal prosequence. Removal of ∼20 amino acids is accomplished by Pseudomonas elastase, which is also positively regulated by quorum sensing. Activation is also mediated by other proteases that cleave rLAP55 near its C terminus. The importance of the C terminus was confirmed by showing that C-terminal deletions of 1-24 amino acids produce a fully active enzyme. The removal of C-terminal prosequences either by proteolysis or deletion leads to an unusual autoprocessing event at the N terminus. Autoprocessing is apparently an intramolecular event, requires the active site of LAP, and results in the removal of 12 N-terminal amino acids. Furthermore, a detailed analysis of the C-terminal prosequence suggests that the proenzyme state is dependent on the presence of a basic side chain contributed by the last amino acid, lysine 536. Our data support a model whereby full-length PA-LAP is activated in a two-step process; proteolytic cleavage at the C terminus is followed by an intramolecular autocatalytic removal of a 12-amino acid propeptide at the N terminus.
Pseudomonas aeruginosa is an opportunistic pathogen of humans causing acute life-threatening infections in immunocompromised and burned patients and chronic infections in individuals affected by cystic fibrosis (1). To cause cell and tissue damage, P. aeruginosa secretes several well characterized virulence factors, many of which are released to the growth media via the general secretion pathway (also called the Type II system) (1). These include 10 or more well characterized enzymes and toxins, including exotoxin A, elastase, alkaline protease, and hemolytic phospholipase (2-6). Secreted toxins and enzymes frequently assemble in the periplasm as proenzymes and are exported to the growth media as inactive proteins. This strategy has two potential purposes, 1) to prevent damage to self and 2) to facilitate activation at either a later time or a distant location. By studying the conversion of a proenzyme to its active form, it may be possible to gain further insights into the role the local environment plays in the activation of virulence factors and the possible interactions of two or more factors.
In low density cultures, P. aeruginosa grows in a planktonic or free swimming mode and usually exhibits sensitivity to antibiotic treatment. However, once a culture reaches high density, a quorum-sensing (QS)2 system is triggered, and this directs the expression of approximately 500 gene products (7, 8). QS in Pseudomonas, which is mediated by the accumulation of homoserine lactones, has been intensively studied because chronic high density infections of cystic fibrosis-affected individuals are frequently antibiotic-resistant and are associated with extensive damage to airway tissue. In fact, many virulence factors are expressed preferentially at high density. Among the products exported via the Type II system and highly up-regulated by the QS system is leucine aminopeptidase (PA-LAP), the product of gene PA2939 (7). It is noteworthy that a prominent virulence factor of P. aeruginosa, elastase (9), is likewise highly up-regulated during QS and also secreted as a proenzyme via the Type II system (7, 10).
Aminopeptidases are exoproteases that remove specific amino acids from the N termini of proteins and peptides (11). In mammalian cells aminopeptidases are found within the cytosol or are membrane-bound and associated with particular subcellular compartments (11). In bacteria, aminopepidase enzymes are found in roughly equivalent locations, and depending on the export system of the organism, some bacterial aminopeptidases are secreted to the growth media (12). Although aminopeptidases play major roles in normal mammalian cell physiology, they have also been implicated in cancer cell invasion via the degradation of tissue matrix proteins (13-15). In bacterial systems, however, the contribution of aminopeptidases to virulence has not been widely investigated. Because many aminopeptidases are synthesized as proenzymes, we decided to investigate the activation steps of the secreted PA-LAP with a particular focus on the role of prosequences. It is potentially noteworthy that the only other organisms that express LAPs with a similar arrangement of prosequences have been isolated from marine environments.
At high density, PA-LAP is positively regulated (∼30-fold) by the QS system (7, 16) and, thus, may be of some advantage to a biofilm community. Cahan et al. (17) purified PA-LAP from high density culture supernatants of P. aeruginosa and characterized some of its biochemical features including its preference for leucine and methionine over other substrates and its requirement for Zn2+. From their data 17 and that of Braun et al. (9), at least two N-terminal sequences have been reported for PA-LAP isolated from culture supernatants; 1 beginning at amino acid 25, indicating the removal of the signal sequence, and 1 at amino acid 39, suggesting the removal an N-terminal prosequence (9, 17). Here we investigate the activation of PA-LAP by expressing a full-length version of the enzyme (without the signal sequence) and various recombinant variants of the protein (recombinant LAP (rLAP) in Escherichia coli and measuring enzyme activity against a small molecular weight reporter substrate. After refolding and purification, full-length rLAP (rLAP55) is enzymatically inactive and, thus, represents an appropriate species to study the process of activation. Activation was investigated using protease treatments and deletion mutagenesis. Data from activity assays suggest a complex model of activation whereby a “third party” protease initiates activation via cleavage at the C terminus followed by the autocatalytic removal of a 12-amino acid N-terminal prosequence.
EXPERIMENTAL PROCEDURES
rLAP Constructs
rLAP clones were generated from genomic DNA prepared from P. aeruginosa (strain PAO1). Primers used for the relevant constructs and mutants are listed on Table 1. PCR was performed using standard reaction conditions: a 50 μm dNTP mix, 10 ng of purified plasmid DNA template (∼30 fmol), 1× PCR buffer (PCR Optimizer kit; Invitrogen), 100 ng each of forward and reverse oligonucleotide primers (∼6 pmol), and 2.5 units of EASY-A polymerase (Stratagene) in a total volume of 50 μl. Reaction temperatures differed depending on oligonucleotides used. All PCR products were cloned directly into a TOPO-TA vector (pCR2.1). TA clones were digested with appropriate restriction endonucleases and separated by 0.9% agarose gel electrophoresis, and fragments were isolated using a Qiaquick gel extraction kit (Qiagen) or PureLink gel extraction kit (Invitrogen). Ligations of fragments were performed using an ∼3:1 insert to vector molar ratio with T4 DNA ligase (New England Biolabs) for 1.5 h at 37 °C or 16 h at 16 °C. The base expression vector, pRS10, was a pCAC19-pCAC7-based plasmid (22) that has an inducible T7 promoter and carries chloramphenicol resistance (25 μg/ml in E. coli).
TABLE 1.
Oligonucleotides used to make rLAP constructs
APΔsig seq forward (FOR), 5′-CATATGCCTTCGGAAGCGCAACAGT-3′; AP39 forward, 5′-CATATGAAACCCAACCCGTCGAT-3′; AP537 reverse (REV), 5′-GAATTCTTACTTGATGAAGT-3′; AP512K reverse, 5′-ATGAATTCAAGC-TTTTATTTCTGGCCGGCGGCGGCGATCTC-3′; AP529R reverse, 5′-ATGA-ATTCAAGCTTTTAGCGTTCGATCTGGCTGGCGCTC-3′; APΔ536 reverse, 5′-ATGAATTCAAGCTTTTAGATGAAGTCGTGACCCCAGC-3′; D308A forward, 5′-GCGCGCACCTCGACTCGGTGTTCGAAGGCCCCGGTATCAACG-CCAACGGTTCGGGCAGC-3′; SPH wild type (WT) reverse, 5′-AATAAGTGA-TAATAAGCGGACCTGCA-3′; D370A-1 forward, 5′-AAGAAGAAGATCAAG-GCCTACCTGAACTTCGCCATGATCGGCTCG-3′; D370-2 WT reverse, 5′-AAGTTCAGGTAGGCCTTGATCTTCTTC-3′.
| Construct name | Forward primer | Reverse primer |
|---|---|---|
| rLAP55 | APΔsig seq FOR | AP537 REV |
| rLAP53 | AP39 FOR | AP537 REV |
| rLAP55Δ513-536 | APΔsig seq FOR | AP512K REV |
| rLAP55Δ530-536 | APΔsig seq FOR | AP529R REV |
| rLAP55Δ536 | APΔsig seq FOR | APΔ536 REV |
| rLAP53Δ513-536 | AP39 FOR | AP512K REV |
| rLAP53Δ530-536 | AP39 FOR | AP529R REV |
| rLAP53Δ536 | AP39 FOR | APΔ536 REV |
| rLAP55D308AΔ536 | D308A FOR | SphI WT REV |
| rLAP55D369AΔ536 | D370A-1 FOR | D370A-2 WT REV |
The expression of recombinant rLAP variants was carried out in BL21-Star(DE3) E. coli cells (Invitrogen) grown at 37 °C in Superbroth supplemented with chloramphenicol at 25 μg/ml. Cells were diluted to A600 ∼ 0.1 from overnight cultures in fresh broth plus antibiotics and grown to A600 ∼ 0.5 when target protein expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 2 h. Cells were harvested by centrifugation and stored frozen at -80 °C. Expressed rLAP proteins were recovered from the insoluble material of the cell (i.e. inclusion bodies). Cell pellets (∼2 g wet weight) were thawed and resuspended in lysis buffer containing 18 ml of 50 mm Tris, 20 mm EDTA (Tris:EDTA (TE) 50/20; pH 8.0), 0.8 ml of lysozyme (5 mg/ml in water), 2.0 ml of 5 m NaCl, and 2.0 ml of Triton X-100 (25% v/v solution). Bacterial suspensions were further disrupted using a Tissuemizer. Inclusion bodies were recovered and washed (as noted immediately below) via repeated (×9) centrifugations at 12,000 × rpm in an SS34 rotor; the first 3× washes were in TE 50/20 plus Triton X-100 (1% v/v), the second 3× washes were in TE 50/20, and the final 3× washes were in Tris 50 mm. The final inclusion body pellet was solubilized in 6 m guanidine HCl, 50 mm Tris, pH 8.0, in approximately a 5-ml final volume. After solubilization, insoluble material was removed by a final centrifugation.
Refolding of rLAP
Denatured rLAP was refolded by adding guanidine-solubilized protein to the following solution: 0.5 m Arg, 50 mm Tris, pH 8.0, 1 mm CaCl2, and 50 μm ZnCl2, to a final protein concentration of ∼50 μg/ml. Refolding was for 48 h at 10 °C. After refolding, rLAP was dialyzed against 20 mm Tris, pH 8.0, 50 mm NaCl, 1 mm CaCl2, 50 μm ZnCl2. A rapid renaturation assay was also used for small scale assessment of rLAP activity. Inclusion bodies were washed ∼3 times in 1.5-ml microcentrifuge tubes, denatured, and renatured into refolding solution as indicated above. After 1 h, samples were diluted 100-fold and assayed for LAP activity.
Ion Exchange Chromatography
Dialyzed rLAP was adsorbed to Q-Sepharose Fast Flow (GE Healthcare) layered on a sintered glass funnel. Proteins were eluted with step increases of NaCl: 0.1 m NaCl, 0.35 m NaCl, and 1.0 m NaCl. Most rLAP eluted at 0.35 m NaCl, and this fraction was used exclusively for further analysis and most enzyme assays.
Rapid Refolding Assay
When evaluating potential active site mutants of various rLAPs, recombinant proteins were denatured in guanidine and then refolded in 1) Tris (50 mm, pH 8.0) supplemented with arginine (0.5 m), calcium (1 mm), zinc (50 μm), in 2) Tris (50 mm, pH 8.0), with arginine (0.5 m) and DTT (1 mm), or in 3) Tris (50 mm, pH 8.0) with arginine (0.5 m) and EDTA (1 mm). After refolding, the arginine was removed via dialysis, but in those samples where DTT or EDTA had been added, these compounds were retained in the dialysis step. Furthermore, when assayed for activity, the usual AP buffer containing Tris, calcium, and zinc (see below) was replaced with only Tris (50 mm, pH 8.0). Thus, samples could not acquire cations from the assay buffer.
Antibodies
Rabbit Antibodies to LAP—Rabbit antisera were raised to the last 15 amino acids, KSASQIERWGHDFIK, of rLAP55 (underlined in Fig. 1A). The peptide was conjugated to keyhole limpet hemocyanin via an N-terminal cysteine, injected with complete Freund's adjuvant, and boosted with incomplete Freund's adjuvant.
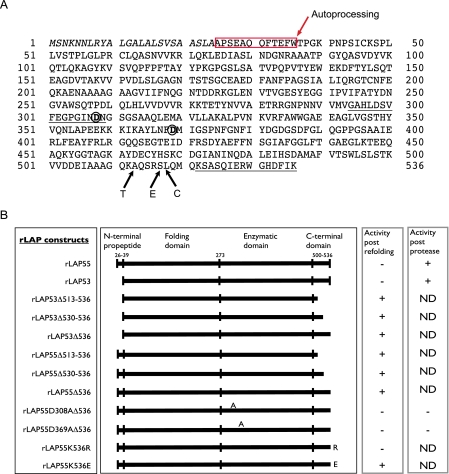
FIGURE 1.
Sequence and construct information for LAP. A, the primary sequence of PA-LAP, the product of gene PA2939, is provided along with certain features related to this study. The signal sequence (1-24) is shown in italics. The N-terminal prosequence is boxed in red. The putative autoprocessing site is shown between tryptophan 36 and threonine 37. The underlined sequence from 294 to 309 corresponds to the peptide used to generate monoclonal antibody 2C3; a second underlined sequence from 522 to 536 shows the peptide used to generate a polyclonal rabbit anti-C terminus antibody. Putative active site aspartic acid residues are shown in bold at positions 308 and 369. Primary cleavage sites for trypsin, elastase, and chymotrypsin are shown with arrows near the C terminus. B, constructs used in this study. Amino acid numbering is from the primary sequence of Fig. 1A. Putative domain boundaries are provided. All constructs were confirmed by DNA sequencing, and the expressed proteins were checked for reactivity by Western blots using the antibody 2C3. Only the D308A active site mutant failed to react with this antibody because the mutation eliminated the epitope. For expression in BL21-STAR cells, each rLAP gene construct was preceded by a T7 promoter. A summary of aminopeptidase activity is provided for each protein. ND, not determined.
Mouse Monoclonal Antibodies to LAP—The following peptide, GAHLDSVFEGPGINDN (underlined in Fig. 1A), was conjugated to keyhole limpet hemocyanin via an N-terminal cysteine residue and injected into mice at A&G Pharmaceutical Inc., Columbia MD, to generate monoclonal antibodies. Bleeds and hybridomas were screened using a biotinylated version of the same peptide immobilized on streptavidin-coated microtiter plates (Pierce). Monoclonal antibodies 1F12 and 2C3 were highly reactive for the peptide and were used in Western blot analysis of LAP. Neither monoclonal reacted with soluble forms of rLAP.
Proteases
Sequencing grade trypsin and chymotrypsin were from Roche Applied Science. Pseudomonas elastase, human neutrophil elastase, and carboxypeptidase B were from Calbiochem. Generally, proteases were used in a 50:1 molar substrate-to-enzyme ratio.
Aminopeptidase Assay
Aminopeptidase activity was assayed in the following AP buffer: 50 mm Tris, pH 8.0, 1 mm CaCl2, 50 μm ZnCl2. Assays were carried out at room temperature for 0-30 min. To assess the role of enzyme-bound cations, some samples were assayed in 50 mm Tris only (without added cations). Samples were assayed in 96-well microtiter plates and read at 405 nm. A molar extinction coefficient of 10,400 for para-nitroaniline was used to calculate the amount of product released from nitroanilide (NA)-based substrates (18). Typical assays were carried out with 120 nmol of Leu-NA-HCl as the substrate and 20-200 pmol of rLAP in a final volume of 200 μl. Results are reported either as nmol of nitroanilide/μg of rLAP or in absorbance units at 405 nm.
Gels and Western Blots
Routinely, 8-16% precast Tris-glycine gels (Invitrogen) were used to analyze LAP samples. Gels were stained for proteins using Microwave Blue (Protega). For Western blots, samples were transferred to polyvinylidene difluoride membranes and blocked overnight with 2% nonfat milk. Primary antibodies, either rabbit or mouse, were used at ∼1-2 μg/ml followed by extensive washing in Tris-buffered saline with 0.1% Tween 20 and followed by donkey anti-rabbit or donkey anti-mouse secondary antibodies conjugated to horseradish peroxidase (Jackson ImmunoResearch) and used at a dilution of 1:15,000. Bands were detected on x-ray film using a chemiluminescence substrate detection kit (KPL).
Samples for N-terminal Sequencing
Gels were transferred to polyvinylidene difluoride membranes, and specific bands were cut from the membrane and sequenced at the Johns Hopkins University Sequencing facility, Baltimore, MD.
Samples for Mass Spectrometry
Samples were analyzed at the Mass Spectrometry/Proteomics Facility at Johns Hopkins School of Medicine, Baltimore, MD. Samples were run by electrospray liquid chromatography/mass spectroscopy on an Applied Biosystems QSTAR quadru-pole-TOF mass spectrometer. Mass values are reported with a confidence of ±2 daltons.
Gel Filtration Chromatography
rLAP55 that eluted from Q-Sepharose in 0.35 m NaCl (see above) was concentrated to ∼400 μg/ml using Centriprep-30 concentrators and then loaded (∼0.5 ml) onto a Superdex™200, 10/300 GL column (GE Healthcare). An identical sample that was treated with trypsin (50:1 substrate to enzyme) for 30 min at room temperature was loaded in a separate run. The mobile phase was Na3PO4, 0.2 m, pH 7.0, and the column was calibrated using gel filtration standards from Bio-Rad. Absorbance at 280 nm was used to detect proteins, and the column was run at 0.3 ml/min. Chromatograms were recorded and analyzed using the software supplied with the AKTA™ chromatography system (GE Healthcare).
Bioinformatics
Blast search of ∼900 microbial genomes was performed via the NCBI website. ClustalX alignments were performed with ClustalX2.09 operating locally on a desktop computer.
RESULTS
Activation of Recombinant rLAP Correlates with Proteolytic Processing—PA-LAP is secreted to the growth media as a proenzyme (the primary sequence of 536 amino acids is provided in Fig. 1A). Features of the enzyme include a signal sequence (italics, amino acids (aa) 1-24), an N-terminal prosequence (boxed and including aa 25-36, described below), an epitope near the active site (underlined aa 294-309), active site residues Asp-308 and Asp-369, and a C-terminal epitope (underlined residues aa 521-536). Also shown are primary cleavage sites for Pseudomonas elastase, trypsin, and chymotrypsin (arrows E, T, and C; see below).
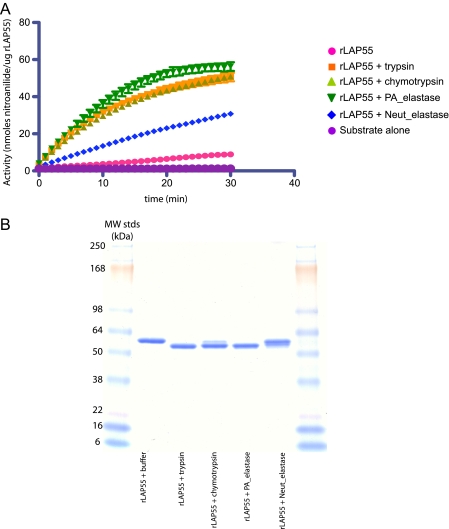
To study the steps of activation and gain mechanistic insights, we expressed aminopeptidase variants in E. coli. All constructs (outlined in Fig. 1B) were expressed without a signal peptide. Recombinant proteins accumulated in inclusion bodies and were denatured and refolded as indicated under “Experimental Procedures.” The longest rLAP variant, rLAP55, contained all wild type amino acids (26-536) with alanine 25 replaced with an initiating methionine. Despite refolding in buffers of various compositions (data not shown), rLAP55 failed to exhibit enzyme activity when assayed with Leu-NA as a substrate. This was consistent with the supposition that rLAP55 had refolded to a proenzyme state. To investigate protease-mediated activation, rLAP55 was incubated with a variety of proteases and re-assayed for activity. The addition of Pseudomonas elastase, trypsin, or chymotrypsin each converted rLAP55 to an active enzyme (Fig. 2A). However, the addition of neutrophil elastase activated rLAP relatively poorly (Fig. 2A and Table 2). Activation was accompanied by a change in migration on SDS-PAGE gels. Regardless of the activating protease, the cleaved rLAP migrated ∼3-4 kDa faster than untreated rLAP55, suggesting the loss of ∼35 amino acids (Fig. 2B). Proteases that failed to activate rLAP55 (data not shown) or activated it poorly (neutrophil elastase) resulted in little or no change in migration, confirming that the removal of ∼35 residues correlates with activation (Fig. 2B).
FIGURE 2.
Activation of rLAP55 via limited proteolysis. rLAP55 was incubated in a molar ratio of 50:1 for 30 min with either trypsin, chymotrypsin, elastase from P. aeruginosa, or neutrophil elastase. A, protease treatment activated the aminopeptidase activity of rLAP55. After the 30-min incubation with each of the proteases mentioned above, samples were removed and assayed for aminopeptidase activity (as described under “Experimental Procedures”). Results are expressed as nmol of nitroanilide/μg of rLAP over a 30-min period. B, SDS-PAGE analysis of rLAP55 after protease incubations. Untreated rLAP55 migrates at 55 kDa. Each protease treatment produced a product that migrated at ∼52 kDa. In a parallel experiments the bands were transferred to polyvinylidene difluoride membranes and submitted for N-terminal sequence analysis; results are reported in Table 3. Samples were also sent for mass spectroscopy time of flight analysis (results are reported in Table 3).
TABLE 2.
Specific activity of protease-treated rLAP proteins
Activity was calculated from initial rates of Leu-NA hydrolysis over a period of 10 min.
| Activity | |
|---|---|
| nmol/μg/min | |
| rLAP55 + trypsin | 1.4 |
| rLAP55 + chymotrypsin | 1.5 |
| rLAP55 + elastase | 1.7 |
| rLAP55 + neutrophil elastase | 0.95 |
| rLAP53 + trypsin | 0.43 |
| rLAP53 + elastase | 0.61 |
N-terminal Sequence and Mass Spectrometry Analysis of rLAP55—To understand the basis for the change in size after protease activation, we performed both N-terminal sequence and mass spectrometry TOF determinations on rLAP55 before and after treatment with the three activating proteases. Untreated rLAP55 (amino acids 25-536) began with serine 27, indicating the loss of the initiating methionine and proline 26 (the loss of the initiating methionine was universal; however, the loss of proline 26 was variable from preparation to preparation). And the TOF mass for untreated rLAP55 was 54,925 Da, indicating the presence of all amino acids from 27-536 (Table 3). Remarkably, the N terminus of each of the three protease-activated rLAP55s began with threonine 37 regardless of the protease used for activation (Table 3). This was not the result anticipated from a direct cleavage at this location because the three proteases exhibit distinct substrate specificities, and the result was especially puzzling for trypsin where the scissile bond typically follows immediately after a basic residue. The amino acid immediately preceding Thr-37 is Trp-36 (Fig. 1A). Clarification was sought using mass spectrometry. The TOF analysis was consistent with two cleavage events, one direct and one potentially “indirect” (see below). Results are reported in Table 3 and reflect two processing events after treatment with trypsin, one between Lys-512 and Ala-513 (direct) and the other between Trp-36 and Thr-37 (indirect), producing a fragment with a mass of 50,853 (Fig. 1A and Table 3). For elastase, the direct cleavage was between Ser-517 and Leu-518, and the indirect cleavage was between Trp-36 and Thr-37, producing a fragment with a mass of 51,382 Da. Cleavage by chymotrypsin produced two products; one reflected cleavages between Leu-518—Gln-519 and Trp-36 -Thr-37, whereas a less abundant product was the result of a single cleavage at Trp-36 -Thr-37; see Table 3. Although there was no initial explanation for the cleavages between Trp-36 and Thr-37, it was clear that activation correlated with the removal of both N- and C-terminal sequences. This was investigated further using a systematic deletion analysis.
TABLE 3.
Characterization of rLAP55 before and after protease treatment
| Treatment of rLAP55 | N terminus by Edman | Mass by TOF | Amino acids present | Mass by calculation | Enzyme activity after treatment |
|---|---|---|---|---|---|
| None | SEAQQF... | 54,923 | 27-536 | 54,925 | None |
| Trypsin | TPGKPN.. | 50,853 | 37-512 | 50,856 | ++++ |
| PA elastase | TPGKPN | 51,382 | 37-517 | 51,386 | ++++ |
| Chymotrypsin | TPGKPN | 51,498 | 37-518 | 51,499 | +++ |
| Chymotrypsin | TPGKPN | 53,671 | 37-536 | 53,671 | ?? |
| Neutrophil elastase | SEAQQF | 54,923 | 27-536 | 54,925 | +/− |
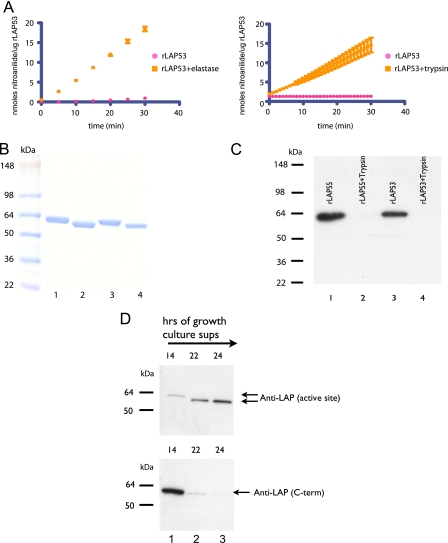
Deletions at the C Terminus Resulting in Active rLAP—Cahan et al. (17) reported the existence of a processed form of PA-LAP beginning at Gly-39. We, therefore, attempted to recapitulate this protein via expression of a construct we termed rLAP53 (all wild type amino acids from 40 to 536 were included, with an initiating methionine in place of Gly-39; see Fig. 1, A and B). However, like rLAP55, rLAP53 exhibited no enzyme activity after refolding (Fig. 3A). From this we concluded that the removal of the N-terminal peptide alone (amino acids 26-39) was not sufficient to achieve activation. Therefore, we investigated the activation of rLAP53 via proteolysis. Like rLAP55, enzyme activity was noted after incubations with elastase or trypsin (shown) or chymotrypsin (not shown) (Fig. 3A), and in each instance treatment produced a large fragment that migrated several kilodaltons faster on SDS-PAGE than untreated rLAP53 (Fig. 3B). When the specific activities of protease-activated rLAP55 and rLAP53 were compared, the rLAP55 version was ∼3-fold more active (Table 2). We do not yet understand the basis of this difference.
FIGURE 3.
Protease treatment activates rLAP53 via limited proteolysis. rLAP53 was incubated in a molar ratio of 50:1 for 30 min with either trypsin, elastase (shown), or chymotrypsin (not shown). A, protease-treated rLAP53 was assayed for aminopeptidase activity and compared with rLAP53 incubated in buffer alone. Results are expressed as nmol of nitroanilide/μg of rLAP over a 30 min period. B, trypsin treatment of rLAP55 or rLAP53 produced similar sized stable products. Untreated rLAP55 and rLAP53 are shown (lanes 1 and 3), whereas trypsin-treated samples are shown (lanes 2 and 4). In a parallel experiment the product of trypsin-mediated cleavage of rLAP53 was submitted for N-terminal sequence determination. C, Western blot analysis of rLAP55 and rLAP with either a monoclonal antibody (2C3) to the active site (lanes 1 and 3) or a polyclonal antibody to the C-terminal last 15 amino acids (lanes 2 and 4). D, P. aeruginosa strain PAO1 was grown in LB broth overnight, and samples were taken at various time points. Cells were removed by centrifugation. Supernatants were analyzed via SDS-PAGE and Western blots. Blots were probed with either the 2C3 monoclonal antibody (upper panel) or the polyclonal antibody to the C terminus of LAP (lower panel).
N-terminal sequence analysis was performed on the trypsin-generated fragment of rLAP53 (Fig. 3B, lane 4). Results showed that only the initiating methionine was absent, indicating a cleavage event at the C terminus. This confirmed that activation required the removal of C-terminal sequences.
To track the loss of C-terminal residues, we generated an antibody to the last 15 amino acids of LAP and performed Western blot analysis (Fig. 3C). Blots confirmed that untreated rLAP55 and rLAP53 had an intact C terminus before activation. Activation via proteolysis resulted in the removal of a substantial number of amino acids (at least enough to remove the reactive epitope) from the C terminus of both rLAP55 and 53 (Fig. 3C). This was in full agreement with the mass spectrometry data (see above). The removal of C-terminal amino acids is apparently also relevant to the activation of native PA-LAP. A Western blot analysis of culture supernatants of P. aeruginosa grown in LB confirmed that PA-LAP is initially secreted with its C terminus intact (Fig. 3D; see the 14-h sample, lower panel) and is then processed to a smaller version (Fig. 3D, upper panel) lacking some or all of the last 15 amino acids at later times (22 or 24 h) (Fig. 3D).
To characterize the role of C-terminal amino acids, we performed a deletion analysis. Either 24, 7, or 1 amino acid was removed from the C terminus of rLAP53, and the three mutant proteins are expressed as outlined under “Experimental Procedures.” Initially, a rapid renaturation approach was used to screen for activity against Leu-NA. Proteins were scored as active or inactive. Results indicated that each deletion, including rLAP53ΔK536 lacking only the terminal lysine but not full-length rLAP53, exhibited enzyme activity (Fig. 1B). Thus, we concluded that the removal of a C-terminal propeptide was necessary for activation of rLAP, and activation could be achieved with the removal of one or more amino acids from the C terminus.
The deletion of the C-terminal lysine, leading to activation, prompted us to probe structural features of the C terminus of rLAP53. Carboxypeptidase B, which removes basic amino acids from the C termini of proteins, was added to test for the accessibility of the terminal lysine. No activation was noted (data not shown), suggesting that the peptide bond linking isoleucine 535 with lysine 536 was not available and/or not susceptible to cleavage.
Because each of the rLAP53 deletion mutants began at amino acid 39, it was not possible to assess the role of the N-terminal propeptide in these constructs. Therefore, each C-terminal deletion was recapitulated in the longer rLAP55 construct (Fig. 1B). Expressed proteins were again collected as inclusion bodies, denatured, rapidly renatured, and assayed for enzymatic activity. As with rLAP53, each C-terminal deletion mutant, but not full-length rLAP55, exhibited enzyme activity. When these deletion proteins were subjected to N-terminal sequence analysis, we were surprised to note that each began at threonine 37. Thus, the refolding of rLAP lacking a C-terminal prosequence resulted in processing at the N terminus. We considered it highly significant that the elimination of C-terminal sequences via deletion resulted in the same N terminus (Thr-37) as was noted with the activation of full-length rLAP via proteolytic processing. This suggested a potential auto-processing step at the N terminus.
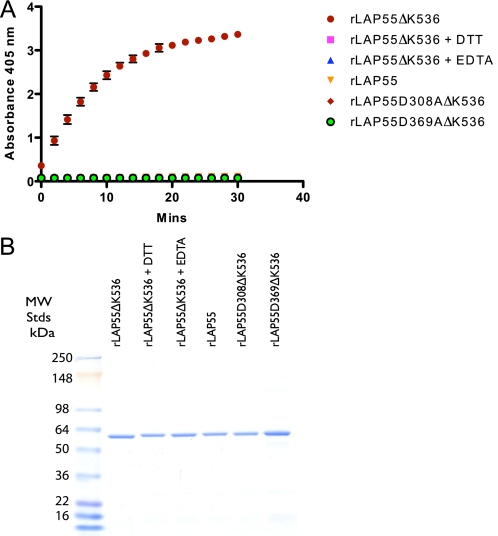
Active Site Mutants—Because autoprocessing could be mediated theoretically by either the active site responsible for aminopeptidase activity, another portion of the molecule, or possibly by a contaminating protease, we generated active site mutants and re-assessed processing at the N terminus. PA-LAP is a zinc-dependent enzyme and exhibits high sequence homology in its activity domain to LAP from Streptomyces griseus (17). Sequence alignments of PA-LAP with the Streptomyces enzyme and other LAPs suggested that zinc ions coordinated at the active site likely involve the side chains of the amino acids His-296, Asp-308, Glu-341, Asp-369, and His-467 (17). As noted above, the refolding of rLAP55ΔK536 (with the active site intact) in a calcium- and zinc-containing buffer resulted in an active enzyme and an N terminus that had been processed to threonine 37. Therefore, to investigate the participation of the active site, two mutants (D308A and D369A) were generated within the construct rLAP55ΔK536 (Table 1 and Fig. 1B). Both mutant proteins were refolded in the Ca2+/Zn2+ buffer and found to have no aminopeptidase activity (with or without trypsin treatment), thus confirming them as active site mutants. The refolding of either active site mutants in a Ca2+/Zn2+ buffer followed by SDS-PAGE and N-terminal sequence determination revealed that both mutant proteins began with proline 26. This result supports a role for the active site in the processing of the N-terminal propeptide, here termed “autoprocessing.” To expand and confirm the study, we devised a refolding experiment whereby rLAP55ΔK536 (active when refolded in Ca2+/Zn2+) was refolded with either EDTA (1 mm) or DTT (1 mm) present (to prevent zinc binding at the active site (17). Results from these experiments indicated that no aminopeptidase activity was detected with the addition of either EDTA or DTT (Fig. 4A). As expected, no activity was detected either with rLAP55 or the active site mutants (Fig. 4A). SDS-PAGE analysis indicated that rLAP55ΔK536 migrated slightly faster than the other proteins and also slightly faster than rLAP55ΔK536 treated with EDTA or DTT (Fig. 4B). N-terminal sequence analysis revealed that there was no autoprocessing when rLAP55ΔK536 was treated with EDTA or DTT. From these data we conclude that autoprocessing is mediated by the same catalytic site used in the exoproteolytic removal of leucine from substrates.
FIGURE 4.
Autoactivation and removal of the N-terminal propeptide are both dependent on the active site of rLAP. A, recombinant proteins were refolded, dialyzed, and then assayed for LAP activity. Recombinant LAP55ΔK536 was refolded in the presence of both calcium and zinc or in the presence of DTT or EDTA. The latter two treatments inactivate LAP via interference with cation binding at the active site. Active site mutants (rLAP55D308AΔK536 and rLAP55D369AΔK536) were refolded in the presence of calcium and zinc. As a negative control in the assay, rLAP55 was refolded in the presence of calcium and zinc. B, an SDS-PAGE analysis of the proteins used in these assays. Proteins following refolding and dialysis were run under reducing conditions (∼ 2 μg/lane).
Intramolecular Versus Intermolecular Autoprocessing—Because the active site is necessary for autoprocessing, it was of interest to determine whether this step was confined to individual molecules (the “cis” model) or whether one active LAP molecule could activate neighboring molecules (the “trans” model). To distinguish between these two possibilities, we incubated rLAP55 (the proenzyme) with a small amount of rLAP55ΔK536 (active enzyme) and looked for gain of aminopeptidase activity and/or removal of the N-terminal peptide (amino acids 26-39). Neither activity was noted. However, such an experiment was limited in scope because rLAP55 has an intact C-terminal propeptide, and if removal of this is required to allow access to the N terminus, then the experiment would not be informative. Therefore, we repeated the experiment with rLAP55ΔK536 as the potential activating enzyme and one of the active site mutants (lacking lysine 536) as the potential substrate. Results of N-terminal sequence analysis revealed that a small amount of active rLAP could not mediate autoprocessing at the N-terminal prosequence of rLAP55D308AΔK536. Therefore, we conclude that at the concentrations tested autoprocessing proceeds in cis but not in trans.
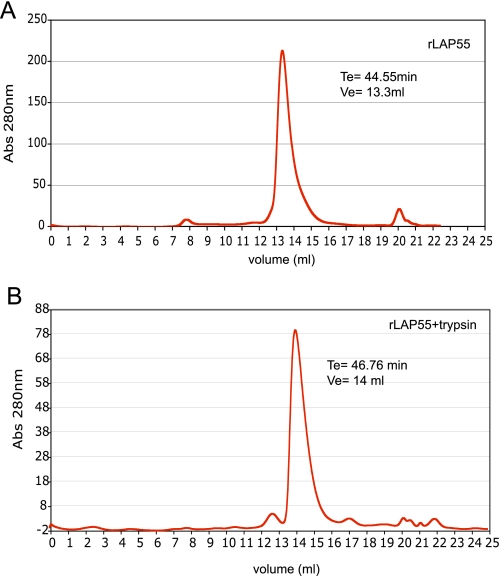
Recombinant LAP Appears to Be Active as a Monomer—The conversion of proenzymes to active ones can be associated with changes in quaternary structure. Dimers, hexamers, or multimers of LAPs are often associated with an active enzyme preparation (11, 19). Therefore, we performed gel filtration analysis on rLAP55 before and after activation with trypsin. Approximately 200 μg of rLAP55 that had been eluted from Q-Sepharose and concentrated to ∼400 μg/ml was loaded onto a Super-dex-200 column. When the mobile phase was Na3PO4 (0.2 m, pH 7.0), untreated rLAP55 migrated with an apparent size of 66 kDa (elution volume = 13.3 ml), and recovery was close to 100% (by absorbance at 280 nm) of the loaded sample (Fig. 5A). After treatment with trypsin, active rLAP eluted ∼0.7 ml later, indicating a more compact molecular size, calculated at 50 kDa (Fig. 5B). Recovery of trypsin-treated rLAP55 was ∼ 67% that of the loaded sample. However, we note that removal of the N- and C-terminal prosequences results in the loss of two of the proenzyme seven tryptophans, thus reducing the molar extinction coefficient (at 280 nm) and leading to an apparent reduction in recovery. Molecular sizes were calculated from plotting the molecular weight of known standards against elution volume. There was no evidence that activated rLAP formed dimers, multimers, or aggregates.
FIGURE 5.
Gel filtration analysis of rLAP55 (A)- and trypsin (B)-activated rLAP55. Samples (∼200 μg) were loaded on a Superdex™200, 10/300 GL column with 0.2 m Na3PO4, pH 7.0, as the mobile phase. Estimates of molecular size were calculated from the elution of gel filtration standards that included thyroglobulin, IgG, ovalbumin, myoglobin, and vitamin B12. Abs, absorbance.
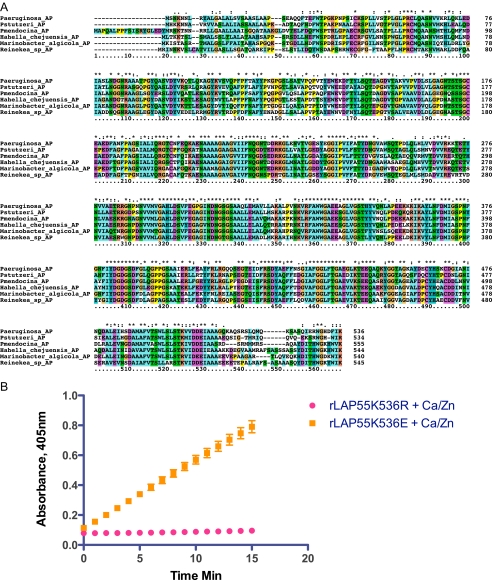
Mechanistic Insight; Inhibition of LAP Activity via the Basic Side Chain of the Last Amino Acid—A comparison of primary PA-LAP sequences reported in Cahan et al. (17) shows high sequence homology between S. griseus LAP (Sg-AP) and PA-LAP around the active site. However, there is no similarity when residues at the C terminus are compared. With the advent of various “biodiscovery projects,” the genomes of bacterial organisms recovered from distinct environments, including marine locations, have been sequenced. Blast searches of the NCBI microbial genome data base revealed the existence of three potential structural genes closely related to PA-LAP that encode putative aminopeptidases; that is, one from Reinekea, one from Hahella and one from Marinobacter species. In each case, the LAP has a C terminus that closely resembles PA-LAP (Fig. 6A). Each has a terminal lysine or arginine preceded by an isoleucine and a phenylalanine. The conservation of a terminal basic amino acid together with its apparent role in maintaining PA-LAP as a proenzyme prompted us to investigate the activity of substitutions for lysine at position 536. In a rapid renaturation assay, the replacement of lysine with arginine resulted in refolding of rLAP55K536R to the proenzyme state Figs. 1B and 6B. However, mutating the terminal basic amino acid to an acidic amino acid, rLAP55K536E, resulted in substantial activity without the need of a processing protease Figs. 1B and 6B. From this we conclude that lysine, by virtue of its basic side chain, likely interacts electrostatically with another portion of the PA-LAP molecule to maintain the proenzyme state. Disruption of this interaction allows for enzyme activity to be manifest (see “Discussion” and Fig. 7 for a model).
FIGURE 6.
A, comparison of primary sequences from closely related enzymes (putative aminopeptidases). Putative leucine aminopeptidases (AP) from Pseudomonas, Hahella, Reinekea, and Marinobacter species were identified by BLAST sequence analysis. These were then compared using ClustalX (Version 2.09). B, aminopeptidase activity of two C-terminal mutants measured after rapid renaturation. Results are reported as the change in absorbance at 405 nm over15 min with Leu-NA as the substrate.
FIGURE 7.
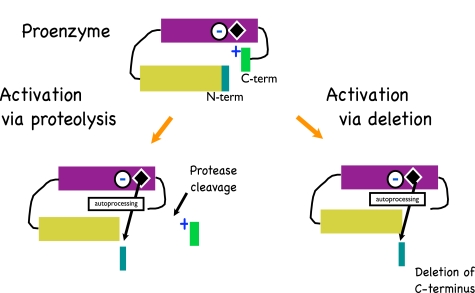
A model for the activation of pro-aminopeptidase is provided. The proenzyme form of Leu-AP is maintained via an interaction between the basic side chain of amino acid 536 with a portion of the enzyme near its active site. Activation occurs when this interaction is disrupted either by protease treatment near the C terminus or by deletion of 1 or more amino acids. The active site then removes the 12 amino acid prosequence at the N terminus and the enzyme is active.
A Model for Activation of PA-LAP—A combination of experimental data from our study from Cahan et al. (17) and the results of a Clustal sequence alignment (Fig. 6A) has allowed us to formulate a model (Fig. 7) for the activation of PA-LAP. We propose that PA-LAP is secreted to the growth media as a proenzyme. Support for this is based on the presence of an intact C terminus when PA-LAP is first secreted (Fig. 3D). Furthermore, we propose that the proenzyme state is maintained via an electrostatic interaction between the C-terminal lysine and some portion of the enzyme domain (Fig. 6B). Activation is initiated via cleavage between residues 512 and 518. A combination of protease cleavage results and deletion analysis confirms that the exact site of cleavage is not overly important. It is noteworthy that when all six LAPs are compared at the primary sequence level (Fig. 6A), amino acids 510-529 (using the PA numbering scheme) are among the least conserved residues. From this we speculate that the ∼510-529 segment may constitute a protease sensitive loop. Cleavage within this segment apparently uncovers the enzyme active site, and this facilitates processing at the N terminus. Although the details are not yet complete, our experimental data suggests that N-terminal processing occurs in cis and occurs between Trp-36 and Thr-37. The combination of C- and N-terminal processing promotes activation and the folding of the enzyme into a more compact size.
DISCUSSION
Many proteases are expressed initially as proenzymes, and it is not unusual for them to auto-activate. Here we report on an unusual mode of activation for a secreted aminopeptidase. Cleavage at the enzyme C terminus leads to autoprocessing at the N terminus and the removal of 12 amino acids. When the cleavage step was recapitulated by deleting C-terminal residues, an identical autoprocessing event was noted at the N terminus. This suggests that the C terminus is key to keeping the enzyme in a proenzyme state. When this was investigated in detail, it was determined that the basic side chain contributed by the last amino acid is responsible for maintaining the proenzyme state. Furthermore, the necessary order of events for activation was established by experiments showing that deletions at the C terminus but not the N terminus were necessary to produce an active enzyme.
The unusual mode of activation centers on the “removal” of 12 amino acids from the N terminus of PA-LAP. This step is mediated by the active site of PA-LAP, and herein lies a dilemma. If the autoprocessing event is an “en bloc” endoproteolytic cleavage, as it appears to be, this would be quite unusual. At the time of submission, the authors are unaware that such a cleavage step has been described previously for an exoprotease. The fact that autoprocessing appears to be exclusively an intramolecular event also seems to be an important feature of this process. Another explanation for N-terminal processing is the sequential exoproteolytic removal of each of the 12 amino acids. PA-LAP has high activity against, Leu, and Met-NA substrates but none against glutamic acid (there are two glutamates at the N terminus; Fig. 1A) and very little against alanine substrates (data not shown). Because PA-LAP does not have broad substrate specificity and no known activity against many of the residues in the prosequence, this pathway seems less likely. A further argument against exoproteolytic autoprocessing centers on our N-terminal sequencing results. After performing many sequencing reactions, we never encountered intermediates with partial removal of the N-terminal prosequence.
A blast search of microbial genomes revealed the close relationship of six putative LAP gene products, three from pseudomonad family members and three from unrelated organisms (Fig. 6A). Of potential interest is the fact that each of the non-pseudomonads is annotated as a “marine” organism. PA-LAP and the five putative LAP genes were aligned using a ClustalX algorithm. When the N-terminal and C-terminal prosequences were considered, several features stood out. At the N terminus, tryptophan at position 36 was invariant (all residue numbers are directly keyed to the primary sequence of PA-LAP presented in Fig. 1A). In each case Trp-36 was preceded by an aromatic amino acid, either phenylalanine or tyrosine, and followed by either threonine or serine. This conservation of amino acids at the putative site of autoprocessing was not seen for the other residues of the prosequence, supporting its potential role as a recognition and cleavage site. When the C terminus was considered, there was a highly conserved seven amino sequence at the end of the molecule. Tryptophan 530 followed by glycine 531 was invariant. Residue 534 was always an aromatic, amino acid 535 was either valine or isoleucine, and 536 was always a basic amino acid, usually lysine. Between residue 510 and 529 there was a high degree of sequence variability. We speculate that residues 510-529 may represent a “bait” region for cleavage by proteases of varying specificity. The mass spectrometry data indicate distinct cleavage sites for elastase, trypsin, and chymotrypsin in this bait region. Once cleavage occurs it is not clear how the enzyme is activated. Although our data do not address this directly, they are consistent with the proenzyme being maintained by an electrostatic interaction between the basic residue at 536 and a negatively charged residue or residues such that access to the active site is blocked. Cleavage in the bait region could potentially allow this interaction to dissociate irreversibly and thereby “unblock” access to the active site. Activation is presumably followed immediately by autopressing at the N terminus. Future experiments will focus on whether removal of the N-terminal prosequence is required for LAP activity against substrates such as Leu-NA.
P. aeruginosa infects cystic fibrosis patients chronically and is most noteworthy for growing in the airways of affected individuals but can also be isolated from the intestinal tract. We chose to evaluate activation via proteases expressed by epithelia that might interact with the secreted products of P. aeruginosa. Of these, trypsin, chymotrypsin, and Pseudomonas elastase were each capable of activating rLAP. It is noteworthy that both PA-LAP and elastase are positively regulated by the Pseudomonas quorum sensing system and, thus, likely to be expressed at the same time in the growth cycle. Neutrophil infiltrates are common in the lungs of cystic fibrosis patients (20). We, therefore, tested human neutrophil elastase as a potential activating enzyme and found little evidence that it could cleave or activate rLAP. Aminopeptidases are functionally important because of their potential to provide nutrient support, degrade host defense proteins, and assist in the processing and maturation of other exoproducts. Recent work from the Kuehn and Bauman (21) indicates the PA-LAP associates with vesicles released from P. aeruginosa, and this fact may be important in the characterization of PA-LAP as a virulence factor.
Future experiments aimed at understanding the molecular details of activation will be focused on obtaining structural information from growing crystals of both the pro- and active forms of LAP and comparing these structures. And information related to virulence is being pursued via the association of PA-LAP with vesicles that interact with mammalian cells. Also, sera from cystic fibrosis patients are being analyzed for evidence that increases in anti-PA-LAP titers coincide with high density growth in affected individuals.
Acknowledgments
We thank Susan Gottesman and Peter FitzGerald for advice in preparing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health, Intramural Research Program, NCI, Center for Cancer Research.
Footnotes
The abbreviations used are: QS, quorum sensing; LAP, leucine aminopeptidase; PA-LAP, P. aeruginosa LAP; rLAP, recombinant LAP; NA, nitroanilide; DTT, dithiothreitol; TOF, time of flight.
References
- 1.Prince, A. S. (2002) N. Engl. J. Med. 347 1110-1111 [DOI] [PubMed] [Google Scholar]
- 2.Dulon, S., Leduc, D., Cottrell, G. S., D'Alayer, J., Hansen, K. K., Bunnett, N. W., Hollenberg, M. D., Pidard, D., and Chignard, M. (2005) Am. J. Respir. Cell Mol. Biol. 32 411-419 [DOI] [PubMed] [Google Scholar]
- 3.Smith, L., Rose, B., Tingpej, P., Zhu, H., Conibear, T., Manos, J., Bye, P., Elkins, M., Willcox, M., Bell, S., Wainwright, C., and Harbour, C. (2006) J. Med. Microbiol. 55 1641-1644 [DOI] [PubMed] [Google Scholar]
- 4.Vasil, M. L., Krieg, D. P., Kuhns, J. S., Ogle, J. W., Shortridge, V. D., Ostroff, R. M., and Vasil, A. I. (1990) Infect. Immun. 58 4020-4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wieland, C. W., Siegmund, B., Senaldi, G., Vasil, M. L., Dinarello, C. A., and Fantuzzi, G. (2002) Infect. Immun. 70 1352-1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woods, D. E., Cryz, S. J., Friedman, R. L., and Iglewski, B. H. (1982) Infect. Immun. 36 1223-1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuster, M., Lostroh, C. P., Ogi, T., and Greenberg, E. P. (2003) J. Bacteriol. 185 2066-2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner, V. E., Bushnell, D., Passador, L., Brooks, A. I., and Iglewski, B. H. (2003) J. Bacteriol. 185 2080-2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, P., de Groot, A., Bitter, W., and Tommassen, J. (1998) J. Bacteriol. 180 3467-3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner, V. E., Gillis, R. J., and Iglewski, B. H. (2004) Vaccine 22 Suppl. 1, 15-20 [DOI] [PubMed] [Google Scholar]
- 11.Matsui, M., Fowler, J. H., and Walling, L. L. (2006) Biol. Chem. 387 1535-1544 [DOI] [PubMed] [Google Scholar]
- 12.Gonzales, T., and Robert-Baudouy, J. (1996) FEMS Microbiol. Rev. 18 319-344 [DOI] [PubMed] [Google Scholar]
- 13.Carl-McGrath, S., Lendeckel, U., Ebert, M., and Rocken, C. (2006) Histol. Histopathol. 21 1339-1353 [DOI] [PubMed] [Google Scholar]
- 14.Endo, H., Takenaga, K., Kanno, T., Satoh, H., and Mori, S. (2002) J. Biol. Chem. 277 26396-26402 [DOI] [PubMed] [Google Scholar]
- 15.Tokuhara, T., Hattori, N., Ishida, H., Hirai, T., Higashiyama, M., Kodama, K., and Miyake, M. (2006) Clin. Cancer Res. 12 3971-3978 [DOI] [PubMed] [Google Scholar]
- 16.Nouwens, A. S., Beatson, S. A., Whitchurch, C. B., Walsh, B. J., Schweizer, H. P., Mattick, J. S., and Cordwell, S. J. (2003) Microbiology 149 1311-1322 [DOI] [PubMed] [Google Scholar]
- 17.Cahan, R., Axelrad, I., Safrin, M., Ohman, D. E., and Kessler, E. (2001) J. Biol. Chem. 276 43645-43652 [DOI] [PubMed] [Google Scholar]
- 18.Kramer, M. D., Binninger, L., Schirrmacher, V., Moll, H., Prester, M., Nerz, G., and Simon, M. M. (1986) J. Immunol. 136 4644-4651 [PubMed] [Google Scholar]
- 19.Taylor, A. (1993) FASEB J. 7 290-298 [DOI] [PubMed] [Google Scholar]
- 20.Smith, R. S., Fedyk, E. R., Springer, T. A., Mukaida, N., Iglewski, B. H., and Phipps, R. P. (2001) J. Immunol. 167 366-374 [DOI] [PubMed] [Google Scholar]
- 21.Bauman, S. J., and Kuehn, M. J. (2006) Microbes Infect. 8 2400-2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biotechniques Fuqua, W. C. (1992) 12 223-225 [PubMed]