Abstract
Therapeutic antibodies directed against the type 1 insulin-like growth factor receptor (IGF-1R) have recently gained significant momentum in the clinic because of preliminary data generated in human patients with cancer. These antibodies inhibit ligand-mediated activation of IGF-1R and the resulting down-stream signaling cascade. Here we generated a panel of antibodies against IGF-1R and screened them for their ability to block the binding of both IGF-1 and IGF-2 at escalating ligand concentrations (>1 μm) to investigate allosteric versus competitive blocking mechanisms. Four distinct inhibitory classes were found as follows: 1) allosteric IGF-1 blockers, 2) allosteric IGF-2 blockers, 3) allosteric IGF-1 and IGF-2 blockers, and 4) competitive IGF-1 and IGF-2 blockers. The epitopes of representative antibodies from each of these classes were mapped using a purified IGF-1R library containing 64 mutations. Most of these antibodies bound overlapping surfaces on the cysteine-rich repeat and L2 domains. One class of allosteric IGF-1 and IGF-2 blocker was identified that bound a separate epitope on the outer surface of the FnIII-1 domain. Using various biophysical techniques, we show that the dual IGF blockers inhibit ligand binding using a spectrum of mechanisms ranging from highly allosteric to purely competitive. Binding of IGF-1 or the inhibitory antibodies was associated with conformational changes in IGF-1R, linked to the ordering of dynamic or unstructured regions of the receptor. These results suggest IGF-1R uses disorder/order within its polypeptide sequence to regulate its activity. Interestingly, the activity of representative allosteric and competitive inhibitors on H322M tumor cell growth in vitro was reflective of their individual ligand-blocking properties. Many of the antibodies in the clinic likely adopt one of the inhibitory mechanisms described here, and the outcome of future clinical studies may reveal whether a particular inhibitory mechanism leads to optimal clinical efficacy.
The type I insulin-like growth factor receptor (IGF-1R)2 is a large transmembrane receptor tyrosine kinase expressed on most somatic cells. IGF-1R is activated by the binding of its constitutive ligands, IGF-1 and IGF-2 (and at a much lower affinity, insulin). Ligand binding to the IGF-1R extracellular domains leads to activation of its cytoplasmic tyrosine kinase domain, receptor autophosphorylation, and phosphorylation of downstream targets such as insulin receptor substrate-1 (IRS-1), the Src homology and collagen domain protein (Shc), and others (1, 2). Phosphorylation of IRS-1 activates the phosphoinositol kinase 3/AKT cellular growth and survival pathways, and Shc phosphorylation leads to the activation of other signal cascades, including the extracellular signal-regulated kinase(Erk)/mitogen-activated protein kinase (MAPK) cellular growth and proliferation pathways (3).
Human IGF-1R is synthesized as a 1368-amino acid polypeptide whose primary and tertiary structures have been reviewed (4, 5). The N-terminal region (consisting of residues 1-903 of the mature protein sequence) is extracellular and highly glycosylated. C-terminal to the extracellular region are a transmembrane helix (residues 904-928) and a cytoplasmic tyrosine kinase signaling domain (residues 963-1239). The extracellular region can be subdivided into six distinct protein domains as follows: an N-terminal receptor L domain (L1), a cysteine-rich repeat (CRR) domain, a second receptor L domain (L2), and three type III fibronectin domains denoted FnIII-1, FnIII-2, and FnIII-3. FnIII-2 contains a long linker sequence that gets clipped between residues 708 and 710, resulting in two disulfide-linked polypeptides known as the IGF-1R α- and β-chains (5). Similar to the insulin receptor (IR), the IGF-1R extracellular region is responsible for the constitutive dimerization via a large protein interface that includes L1, L2, FnIII-1, and FnIII-2 (6). Alanine scanning studies have shown that residues important for binding IGF-1 and IGF-2 to IGF-1R reside in the L1 domain and the linker region embedded in the FnIII-2 domain (7-9). A few residues in the CRR domain have also been shown to affect IGF-1 binding.
As a growth mediator, IGF-1R has been implicated in various forms of cancer (1, 2). Epidemiological studies have shown that irregular IGF-1/insulin-like growth factor-binding protein levels in human serum predispose individuals to a higher risk for common cancers. Loss of imprinting and chromosomal aberrations leading to increased IGF-2 expression or IGF-1R activity have also been linked to Ewing's sarcoma and peripheral neuroectodermal tumors (10, 11). IGF-1R activity is often a late event in tumorigenesis, promoting growth and survival of cancer cells. Additionally, IGF-1R activity has been linked to the survival of tumor detachment events required for metastasis (2, 12).
The successful development of anti-tumor agents against epidermal growth factor receptor, HER-2, and vascular endothelial growth factor receptor provided compelling evidence that targeting receptor tyrosine kinase family proteins can be both efficacious and tolerated (13). The development of therapeutics against IGF-1R has included small molecule inhibitors of the tyrosine kinase domain and antibodies to the extracellular domain that inhibit receptor signaling by blocking one or both ligands or by inducing receptor down-regulation. The development of small molecule inhibitors of IGF-1R is complicated by the extremely high homology between the kinase domains of IGF-1R and IR (93% identity). Overall, IGF-1R is only 83% identical to IR, which has enabled the development of noncross-reactive anti-IGF-1R antibody inhibitors.
Several companies have published the results of preclinical and clinical studies pertaining to inhibitory anti-IGF-1R antibodies (1, 14). To date, results from human clinical trials have been very encouraging. Overall, the toxicity profile has been manageable, and clinical activity has been observed in certain settings both as a single agent or in combination with standards of care (1, 14). When combined with carboplatin and paclitaxel in a recent phase II study in non-small cell lung cancer (NSCLC), CP,751,871 antibody (Pfizer) led to a significant increase in overall objective response rates, particularly in the squamous cell carcinoma population, over carboplatin and paclitaxel alone. Both CP,751,871 and AMG479 antibodies (Amgen) have shown efficacy in Ewing sarcoma, a tumor class with known links to IGF-1R biology (15-17). The number of antibody therapeutics directed against IGF-1R advancing through clinical trials has grown substantially over the past few years with many reaching advanced stages (phase II/III trials) (1, 14). Although it has been perceived that most of these antibodies exert their activity through similar mechanisms, such as receptor down-regulation, and therefore should have similar attributes, data from current clinical studies suggest that there are subtle differences among these antibodies with regard to both activity and toxicity. This may be attributed to epitope specificity and the mode of action of different antibodies. Thus, studies elucidating the structure-function relationship of inhibitory anti-IGF-1R antibodies may provide some understanding of differences in the clinical activity and safety profiles of the various antibodies currently in the clinic.
In this study, we evaluated the ability of a panel of inhibitory anti-IGF-1R antibodies to block one or both of the IGF ligands. Antibodies were found that inhibit IGF-1 binding only, IGF-2 binding only, and the binding of both IGF-1 and IGF-2. We generated full-length and truncated ectodomain constructs of IGF-1R and a set of 54 variant IGF-1R constructs that include surface mutations at 64 sites primarily in the α-chain (between L1 and FnIII-2). Screening the antibodies against these IGF-1R constructs enabled residue-specific identification of all the antibody epitopes. Very subtle epitope differences are described that result in the ability of each antibody to preferentially block one or both of the ligands. Additionally, we biophysically characterized the activity of select dual IGF-blocking antibodies in detail to probe the mechanistic aspects behind IGF-1R inhibition.
EXPERIMENTAL PROCEDURES
Recombinant Construction of hIGF-1R-(1-903), mIGF-1R-(1-904)-Fc, hIGF-1R-(1-462)-Fc, IGF-1, and IGF-2—The ectodomain of human IGF-1R was constructed from Gen-Bank™ image clones AI633655, BM905286, and BU849760. The human IGF-1R ectodomain sequence encoding the polypeptide signal and residues 1-903 was subcloned into mammalian expression vectors with or without fusion sequences encoding a C-terminal human IgG1-Fc (constant domain fragment containing an IgG1 hinge, CH2, and CH3 domains). The expressed proteins are denoted hIGF-1R-(1-903)-Fc and hIGF-1R-(1-903), respectively. The primers used to subclone the ectodomain protein in the absence of the IgG1-Fc enabled the addition of a C-terminal decahistidine tag. A truncated human IGF-1R construct (hIGF-1R(1-462)-Fc) was subcloned into the expression vector using the parental hIGF-1R-(1-903)-Fc vector as template. A similar truncated version of the IGF-1R ectodomain has been described (9, 18). A comparable murine IGF-1R ectodomain insert was constructed using overlapping oligonucleotide synthesis and subcloned into the same expression vector (mIGF-1R(1-904)-Fc). Generation of stable bulk CHO pools or CHO cell lines expressing the soluble IGF-1R proteins was facilitated using methods described previously (19). The IgG1-Fc-containing fusion proteins were purified from CHO supernatants by passage over a protein A affinity column followed by preparative size exclusion chromatography (SEC) as described previously for other Fc fusion proteins (18). CHO culture supernatants containing the 10-histidine tag IGF-1R construct, hIGF-1R-(1-903), were concentrated by tangential flow filtration (molecular mass cutoff, 10,000 kDa, Millipore). The protein was purified by batch purification using Ni2+-nitrilotriacetic acid-agarose (Qiagen), followed by Q-Sepharose (Fast Flow) and preparative SEC using Superdex 200 (GE Healthcare).
Synthetic genes encoding human IGF-1 and IGF-2 with N-terminal octahistidine tags were synthesized by GeneArt AG and subcloned into pPICZαA (Invitrogen). The recombinant ligands were expressed in Pichia and purified using Ni2+-nitrilotriacetic acid-agarose. The ligands were further purified by reverse phase chromatography using a Waters Delta Prep HPLC and a preparative XBridge C18 column with H2O/acetonitrile (0.1% trifluoroacetic acid) gradients. The polypeptides were lyophilized, resuspended in deionized H2O, and dialyzed (molecular mass cutoff, 1000, Supelco) against PBS before use. Untagged recombinant human IGF-1 was purchased (Chemicon).
Anti-IGF-1R Antibodies—Antibodies were generated against the hIGF-1R-(1-903)-Fc protein by both murine immunization and subsequent hybridoma generation (including BIIB1-3) and in vitro library/phage display (including BIIB4-5) methods (20, 21). BIIB2 protein was produced as a murine monoclonal antibody by culturing its original hybridoma cell line. The BIIB1 and BIIB3 murine heavy chain variable domains (VH) and light chain variable domains (VL) and the BIIB4 and BIIB5 human VH and VL domains were subcloned into heavy and light chain vectors containing either engineered human IgG4 heavy or κ light chain constant domains, respectively, to form full-length chimeric or human antibodies as described (20, 22). The expression plasmids were transfected into CHO-DG44 or CHO-DG44i (a host strain selected to be insulin-independent), and stable bulk CHO pools or CHO cell lines expressing each antibody were produced and cultured as described (19, 20). The antibodies were purified using protein A (protein A-Sepharose FF, GE Healthcare) using PBS as the running buffer and 0.1 m glycine, pH 3.0, as the elution buffer. Eluants were neutralized to pH 8.5 using Tris base. The eluants were further purified by capture on an anion exchange resin (TMAE Fractogel®, EMD Biosciences), followed by elution using a 20 mm Tris buffer, pH 8.5, and a 10 mm to 1 m NaCl gradient. Each antibody was dialyzed into PBS. A commercially available inhibitory anti-IGF-1R antibody (αIR-3 (23)) was purchased from Calbiochem.
BIIB4 and BIIB5 Fab Preparation—BIIB4 and BIIB5 antibodies were digested using immobilized papain resin (Pierce). The papain resin was washed with a solution of 20 mm sodium phosphate, 10 mm EDTA, and 20 mm cysteine (reduced) at pH 7.0. Each antibody was mixed with immobilized papain resin in 500 mm EDTA and 100 mm cysteine, pH 7.0, and digested for 3 h in a 37 °C water bath, followed by mixing on an inverting shaker overnight at room temperature. Completion of the digestion was determined by analytical SEC. The resin was removed from the digested material using a sintered glass funnel filter washed with 20 mm acetate, pH 5.0. The flow-through was collected and diluted 10-fold with 20 mm acetate, pH 5.0. Fabs were purified by S-Sepharose cation exchange chromatography using a linear salt gradient. Fabs were collected after identification using analytical SEC. The Fabs were dialyzed into PBS. A solution containing 1 m Tris, 200 mm iodoacetate, pH 8.5, was diluted 10-fold into the Fab solutions to alkylate free thiols within the hinge region generated during the digestion. The resulting solutions were incubated on an inverting shaker for 20 min at room temperature, and excess iodoacetate was removed by exhaustive dialysis into PBS.
Analytical SEC/Static Light Scattering and Differential Scanning Calorimetry (DSC)—The oligomeric state of each protein sample used in this report was analyzed by SEC/static light scattering. Between 30 and 100 μl protein was injected onto a TSK-gel G3000SWXL, 7.8 mm × 30 cm, 250-Å analytical SEC column (Tosoh Biosciences) equilibrated in 10 mm phosphate, 150 mm NaCl, 0.02% NaN3, pH 6.8, using an Agilent 1100 HPLC system. Light scattering data for material eluting from the SEC column were collected using a mini-DAWN static light scattering detector coupled to an in-line refractive index meter (Wyatt Technologies). UV data were analyzed using HPCHEM (Agilent), and light scattering data were analyzed using ASTRA V (Wyatt Technologies).
DSC scans were performed using an automated capillary DSC (capDSC, MicroCal, LLC). hIGF-1R-(1-903) protein solutions and reference (buffer) solutions were sampled automatically from 96-well plates using the robotic attachment. Prior to each protein scan, two buffer scans were performed to define the base line for subtraction. All 96-well plates containing protein were stored within the instrument at 6 °C. Each sample was diluted to 1 mg/ml in PBS. Scans were performed from 10 to 95 °C at 2 °C/min using the low feedback mode. Scans were analyzed using the Origin software supplied by the manufacturer. Subsequent to the subtraction of reference base-line scans, nonzero protein scan base lines were corrected using a third-order polynomial. The unfolding parameters for hIGF-1R-(1-903) were deconvoluted using the multiple peak fitting routine within the software assuming non-two-state unfolding behavior.
Antibody Cross-blocking Studies—The ability of various anti-IGF-1R antibodies to block BIIB4 or BIIB5 was determined using biotinylated versions of both antibodies and hIGF-1R-(1-903)-Fc. BIIB4 and BIIB5 were biotinylated using the protocols within the EZ-link Sulfo-NHS-LC-Biotin kit (Pierce). Fifty μl of 2 μg/ml hIGF-1R-Fc in PBS were coated per well of a 96-well clear MaxiSorp plate (Nunc) for 2 h at room temperature and no shaking. Plates were washed with PBS and blocked overnight at 2-8 °C using a PBS, 1% BSA solution. Plates were washed and incubated with a 100 μl mixture of biotinylated BIIB4 or BIIB5 (80 ng/ml) and competitor antibody for 1 h at room temperature. Inhibitor antibodies were serially diluted (5-fold dilutions) from 40 μg/ml to 3 ng/ml. A control was also performed by serial dilution of a nonspecific IgG4 isotype control antibody with biotinylated BIIB4 or BIIB5. Plates were washed and shaken for 1 h at room temperature with 100 μl/well streptavidin-HRP (1:4000 dilution into blocking buffer, Southern Biotech). Plates were washed, and 100 μl/well SureBlue Reserve TMB Microwell peroxidase substrate (Kirkegaard & Perry Laboratories) was added to the wells. Detection of the presence of biotinylated BIIB4 or BIIB5 was performed by reading the absorbance at 650 nm every 5 min using a Wallac 1420-041 Multilabel Counter plate reader.
IGF-1 and IGF-2 Blocking Assays—hIGF-1R-Fc was biotinylated using protocols within the EZ-Link Sulfo-NHS-LC-Biotin kit (Pierce). Biotinylated hIGF-1R-Fc at 5 μg/ml was added to the wells of SigmaScreen streptavidin-coated 96-well plates (Sigma) at 100 μl/well and incubated overnight at 2-8 °C. The plates were then washed four times with 200 μl/well PBST. IGF-1 (8× histidine-tagged) was added at 320 nm in PBST with 1.0 mg/ml BSA. To test the ability of the antibodies to compete with IGF-1 for hIGF-1R-(1-903)-Fc binding, serial dilutions of anti-IGF-1R antibodies (generally starting between 1 and 10 μm) were made into the IGF-1 solutions. IGF-2 binding was assessed in a similar manner. IGF-2 (8× histidine-tagged) was added at 640 nm in PBST with 1.0 mg/ml BSA. The dilutions were added to the plates in duplicate at 100 μl/well, and the plates were incubated at room temperature for 1 h. The plates were then washed four times with 200 μl/well PBST. An HRP-conjugated anti-His tag antibody (Penta-His HRP conjugate, Qiagen) was diluted 1:1000 in PBST and added to plates at 100 μl/well, and the plates were incubated at room temperature for 1 h. The plates were then washed four times with 200 μl/well PBST. SureBlue Reserve TMB Microwell peroxidase substrate (Kirkegaard & Perry Laboratories) was added to plates at 100 μl/well followed by 1% phosphoric acid at 100 μl/well once the desired reaction was observed. The absorbance of each well was determined at 450 nm.
Kinetic Analysis of the Binding of IGFs and Antibodies to hIGF-1R-(1-903)—All surface plasmon resonance (SPR) experiments were performed on a Biacore 3000 set to 25 °C using HBS-EP running buffer (Biacore). For the kinetic analyses, a biotin-labeled anti-His tag antibody (biotin-Penta-His, Qiagen) was immobilized to saturation on a Biacore SA chip surface by injection at 500 nm in HBS-EP buffer. hIGF-1R-(1-903) was captured on the biotin-Penta-His surface by injecting 20 μl of 40 nm protein at 2 μl/min. Serially diluted concentration series (starting at 64 nm) of antibodies, antibody Fabs, or IGF-1 were injected over the hIGF-1R-(1-903) surface at 20 μl/min, followed minimally by a 20-min dissociation period where buffer was run over the sensorchip surface. Each injection was regenerated using 3× 10-μl injections of 10 mm acetate, pH 4.0, at 20 μl/min. Each curve was double referenced using the following: 1) data obtained from a streptavidin surface devoid of hIGF-1R-(1-903), and 2) data from a primary injection of hIGF-1R-(1-903), followed by a secondary injection of HBS-EP buffer. The sensorgrams for each construct were fit to the 1:1 Langmuir binding model within BiaEval 3.0 (Biacore). Binding of IGF-1 to IGF-1R was also performed in the presence of BIIB5. In these studies, 400 nm BIIB5 was added to the mobile phase, the hIGF-1R-(1-903) solution, and the IGF-1 solutions used to measure the ligand/receptor kinetics.
Equilibrium IGF-1 Binding to IGF-1R in the Presence and Absence of Inhibitory Antibodies Using SPR—hIGF-1R-(1-903)-Fc was immobilized (∼15,000 RUs) to a CM5 chip surface using standard amine chemistry protocols provided by Biacore. At this high immobilization level, flowing IGF-1 over the IGF-1R(1-903)-Fc surface led to mass transfer limited linear binding curves whose initial velocity of binding, Vi (RU/s), depended linearly on the concentration of the IGF-1 solution flowed over the chip surface. Binding constants between hIGF-1R-(1-903) and IGF-1 could be determined by flowing mixtures of hIGF-1R-(1-903) and IGF-1 over the sensorchip surface containing sIGF-1R-Fc. The hIGF-1R(10-903)-Fc sensorchip surface measures the concentration of unbound IGF-1 in each hIGF-1R-(1-903)/IGF-1 mixture. The equilibrium dissociation constant, KD, and binding stoichiometry, n, between IGF-1 and hIGF-1R-(1-903) are determined by the concentration of unbound IGF-1 using Equation 1,

where Vi is initial rate of binding; m is slope of the IGF-1 concentration-dependent standard curve; [IGF-1]f is unbound IGF-1 concentration = Vi/m; [IGF-1]t is total IGF-1 concentration; and [R]t is the total hIGF-1R-(1-903) concentration (24).
IGF-1 binding to precomplexed mixtures of hIGF-1R-(1-903) and inhibitory mAbs was performed in an identical fashion. The mAbs and hIGF-1R-(1-903) were equimolar (240 nm) in these experiments. Analytical SEC was used to ensure that the mixtures were precisely equimolar. Additionally, the hIGF-1R-(1-903)/mAb mixtures were passed over the hIGF-1R-(1-903)-Fc sensorchip in the absence of IGF-1 to ensure no residual mAb binding to the surface occurred that might complicate the results.
Generation of IGF-1R Constructs to Support Epitope Mapping—A 54-member IGF-1R mutant library was constructed by mutagenizing the wild-type hIGF-1R-(1-903)-Fc pV90 plasmid using the Stratagene site-directed mutagenesis kit following the manufacturer's protocols. Incorporation of each mutant (or double mutant) within the pV90 vector was confirmed by DNA sequencing. Plasmid production was performed by transformation into DH5α (Invitrogen), culturing 100 ml of the transformed Escherichia coli at 37 °C overnight, and isolation of plasmid using the Qiagen Endotoxin-Free MaxiPrep kit. Two hundred μg of each mutant plasmid was transiently transfected into 100 ml of HEK293 cells at 2 × 106 cells/ml using the Polyfect transfection kit (Qiagen) for soluble protein secretion into the media. Cells were cultured at 37 °C in a CO2 incubator in DMEM (Irvine Scientific) with 10% fetal bovine serum (FBS) containing low IgG (Invitrogen) that had been further depleted of bovine IgG by passage over a 20-ml protein A column. After 7 days, supernatants containing each IGF-1R-Fc mutant were collected by centrifugation at 1200 rpm and filtered through a 0.2-μm filter. Each mutant was affinity-purified by standard protein A affinity chromatography methods (18). Mutant proteins were concentrated to ∼300 μl using VivaSpin 6 Mr cutoff 30,000 centrifugal concentrators (Sartorius).
Potential folding or expression issues for each of the hIGF-1R-(1-903)-Fc mutant proteins were monitored by IGF-1R Western blot detection. Samples were run on 4-20% Tris-glycine gels (Invitrogen) using Xcell SureLock mini cell following standard manufacturer's protocols. Samples were transferred to nitrocellulose using the iBlot Dry blotting system and transfer stacks (Invitrogen) following the standard manufacturer's protocol. Membranes were blocked overnight at 4 °C in 25 ml of PBST with 5 mg/ml nonfat dry milk. After blocking, membranes were washed once with 25 ml of PBST for 5 min at room temperature. Membranes were incubated with a primary anti-IGF-1Rβ antibody (Santa Cruz Biotechnology) at 1:100 in 10 ml of PBST for 1 h at room temperature. The membranes were washed three times in 25 ml of PBST for 5 min. For detection, membranes were incubated with a secondary HRP-conjugated goat anti-rabbit IgG-Fc antibody (US Biological) at a 1:1000 dilution in 10 ml of PBST for 1 h at room temperature. Membranes were washed three times in 25 ml of PBST for 5 min followed by 1 wash in 25 ml of PBST for 20 min. Protein bands were detected using the ECL Western blotting analysis system (GE Healthcare) following the manufacturer's protocol.
Epitope Mapping—The ability of BIIB4 and BIIB5 to bind hIGF-1R-(1-903)-Fc, mIGF-1R(1-904)-Fc, and hIGF-1R(1-462)-Fc was determined by SPR. Experiments were performed on a Biacore 3000 set to 25 °C using HBS-EP as the running buffer. An anti-human IgG-Fc antibody (2C11, Biogenesis) was injected at 40 μg/ml over a CM5 sensorchip surface in 10 mm acetate, pH 5.0, and immobilized to ∼8000 RU using standard amine chemistry protocols (Biacore). hIGF-1R-(1-903)-Fc, mIGF-1R(1-904)-Fc, or hIGF-1R(1-462)-Fc was captured on the 2C11 chip surface by injecting 40 μl of 20 nm protein at 3 μl/min. Following capture of the receptor, 40-μl injections of BIIB4 or BIIB5 Fabs (serial dilutions from 25 to 0.39 nm) were performed in running buffer at 30 μl/min. Dissociation of the Fabs was measured for ∼30 min. Regeneration of the chip surface was achieved using 3× 10-μl injections of 10 mm glycine, pH 2.0, at 60 μl/min. Each curve was double referenced using the following: 1) data obtained from a CM5 chip surface devoid of IGF-1R-Fc, and 2) data from a primary injection of each IGF-1R-Fc construct followed by a secondary injection of HBS-EP buffer. The concentration-dependent binding of the Fabs was fit to the 1:1 Langmuir binding model within BiaEval 3.0.
Binding of the 54 hIGF-1R-(1-903)-Fc mutant constructs to all five BIIB antibodies and αIR-3 was assessed by SPR. The BIIB4 and BIIB5 surfaces were used to capture the wild-type hIGF-1R-(1-903)-Fc and mIGF-1R(1-903)-Fc controls and the set of 54 mutant hIGF-1R-(1-903) constructs. Both hIGF-1R-Fc and mIGF-1R-Fc proteins oligomerize because of the incorporation of two separate homodimeric regions (IGF-1R ectodomain and IgG1-Fc). The ability of mIGF-1R-Fc to bind the antibody surfaces was the result of the high apparent avidity of the oligomeric protein. The mutant IGF-1R-Fc constructs were captured onto the BIIB4 and BIIB5 surfaces by injection of 60 μl of the affinity-purified, concentrated material at 1 μl/min. After completion of the receptor-Fc loading step, flow rates were elevated to 5 μl/min. In each case, >100 RU of each receptor construct was assessed for binding to the sensorchip surfaces prior to subsequent addition of BIIB1-5 or αIR-3 for screening purposes. The binding of BIIB1-4 and αIR-3 to each IGF-1R-Fc construct was assessed using receptor bound to the noncompetitive BIIB5 surface. The binding of BIIB5 to each IGF-1R-Fc construct was assessed using receptor bound to the noncompetitive BIIB4 surface. BIIB1-4 and αIR-3 (50 μl injections) were screened at 30, 10, and 3 nm concentrations. BIIB5 was screened at 10, 3, and 1 nm. Dissociation was measured for 7 min after the antibody injections were complete, after which the flow rate was elevated to 30 μl/min and the chip surfaces were regenerated by 2× 10-μl injections of 0.1 m glycine, pH 2.0.
Isothermal Titration Calorimetry (ITC)—BIIB3 (75 μm), BIIB4 (67 μm), BIIB5 (75 μm), IGF-1 (60 μm), and hIGF-1R-(1-903) (5 μm) were dialyzed against PBS, pH 7.2, in the same container. These protein solutions were adjusted to the concentrations listed above by dilution using the PBS dialysate. ITC experiments were performed on an ITC200 microcalorimeter (MicroCal, LLC). For each binding reaction, the reaction cell (containing roughly 250 μl) was filled with 5 μm hIGF-1R-(1-903). The instrument syringe (40 μl) was filled with antibody or ligand and titrated into the reaction cell using 1.5- or 2.0-μl injections. A 4-min equilibration period was used between all injections with an initial delay of 60 s. To investigate the effect of inhibitory antibodies on IGF-1R/IGF-1 binding, antibodies were titrated to a 2:1 saturation of their receptor-binding sites, followed by a second set of injections with IGF-1. Numerical integration of the data was performed using the ITC data analysis software supplied by MicroCal (Origin). ΔHA°(T) values (where ΔHA°(T) is the molar enthalpy of association) were calculated based on the difference between the average heat liberated/absorbed during the binding phase of the injections and the average heat of dilution found once the receptor, IGF-1R, was saturated with antibody or ligand.
Circular Dichroism—CD spectra were taken on a Chirascanplus spectrophotometer (Applied Photophysics Ltd.). Far UV CD scans (260-195 nm) were performed at 1.4 μm hIGF-1R-(1-903), 5.4 μm IGF-1, 3.5 μm antibody Fab, or combinations of hIGF-1R-(1-903) and IGF-1 or Fab at the same protein concentrations. The signal averaging time was 1 s/λ, and each scan was repeated two times for signal averaging. The step sizes were 0.5 nm and, bandwidth was 1 nm. Temperature ramps monitored by far-UV CD (250-195 nm) were performed at 0.3 mg/ml for both hIGF-1R-(1-903) (1.4 μm) and IGF-1 (37.5 μm). All far-UV temperature ramp spectra were taken using a 0.5-mm cell and a bandwidth between 0.7 and 1.0-nm bandwidth. Temperature ramps of hIGF-1R-(1-903) monitored by near UV CD (320-250 nm) were performed using 0.5 mg/ml protein in a 1-cm UV cell and a 2-nm bandwidth. Temperature ramp rates for the far and near UV temperature-dependent studies were 1 °C/min. Signal averaging times were 0.9 and 0.65 s/λ for the far and near UV temperature ramps, respectively, using a 1.0-nm step size. This enabled the collection of 1 scan/°C without interrupting the scan rate. PBS (Irvine Scientific) was used as the buffer for all samples. Temperature-dependent protein unfolding parameters were analyzed using the Global 3 module of the dynamic multimode spectroscopy software of the manufacturer.
Immunoprecipitation of IGF-1R·IGF-1 Complexes Using BIIB5—Resuspended protein A/G beads (300 μl, Pierce) were washed with PBS and mixed with 1.0 mg of BIIB5 in a 1.5-ml Eppendorf tube on a rotary shaker for 2 h at room temperature. In a separate tube, 100 μl containing 12 μg of hIGF-1R-(1-903) and 460 ng of human IGF-1 were mixed (1:1 molar ratio, 500 nm protein) for 1 h at room temperature. Protein A/G beads with bound BIIB5 were washed with PBS and incubated with the hIGF-1R-(1-903)/IGF-1 mixture for 30 min at room temperature. The beads with bound protein were then washed extensively with PBS followed by elution of bound protein with 300 μl of 0.1 m glycine, pH 3.0. For the negative control, addition of 12 μg of human IGF-1R(1-903) in the absence of IGF-1 was also immunoprecipitated. The protein A/G bead eluants were denatured in Tris-glycine SDS sample buffer (Novex®) and boiled before application to a 4-20% Tris-glycine SDS-polyacrylamide gel. The molecular weight marker was Mark 12™ (Invitrogen). Protein bands were transferred to nitrocellulose membranes using the iBlot ™ system. IGF-1 and IGF-1R were detected with an anti-human IGF-1 primary antibody (rabbit anti-human IGF-1 biotin) and a mouse anti-human IGF-1R α-chain primary antibody (IGF-1Rα, 1H7), respectively. Secondary detection was performed with HRP-labeled streptavidin (Southern Biotech) and HRP-labeled goat anti-mouse IgG (US Biological).
IGF-1R Down-regulation by IGF-1R Antibodies—H322M cells of human NSCLC origin were seeded into 12-well culture plates and grown in RPMI 1640 medium containing 10% FBS (Irvine Scientific) overnight. The next day, cells were treated with 15 μg/ml of an in-house nonspecific isotype control antibody, BIIB4, or BIIB5 for 1, 4, or 24 h. Cellular proteins were extracted in a cell lysis buffer (Meso Scale Discovery). Protein concentrations in the cell lysates were measured using the BCA protein assay kit (Pierce), and equal amounts of total protein were separated by electrophoresis on a NUPAGE 4-12% BisTris gel and transferred to a nitrocellulose membrane (0.45 μm pore). The blot was probed with anti-total-IGF-1R (Cell Signaling Technology) and then with a secondary antibody conjugate, anti-rabbit-IgG-HRP (Jackson ImmunoResearch). Blots were developed with the Supersignal Western Substrate kit (Pierce), and the chemiluminescence image was captured using Versa-Doc 5000 imaging system (Bio-Rad).
Inhibition of Tumor Cell Growth by IGF-1R Antibodies—NSCLC H322M cells were seeded at 8000 cells per well in 96-well clear-bottom TC-treated plates (Wallac) and grown in RPMI 1640 medium (ATCC) containing 2% FBS overnight. Cells were then switched to serum-free medium (SFM) and incubated in medium supplemented with 1, 10, 100, or 500 nm of the nonspecific IgG4 control antibody, BIIB4, or BIIB5 for 1 h at 37 °C followed by stimulation with 200 ng/ml of IGF-1 and IGF-2 (R & D Systems). After 3 days in culture, cell viability was determined by measuring ATP levels with the Cell Titer Glo reagent (Promega). The percentage of cell growth inhibition was calculated according to the formula [1-(Ab-SFM)/(IGF-SFM)] × 100%, with IGF-only and SFM-only normalized to 0 and 100%, respectively.
RESULTS
Ligand Blocking Properties of Anti-IGF-1R Abs—A panel of antibodies derived from both in vitro panning of a phage display library (21) and traditional hybridoma methods against hIGF-1R-(1-903)-Fc was evaluated for the ability to block both IGF-1 and IGF-2 from binding hIGF-1R-(1-903)-Fc in an ELISA format. Four separate classes of inhibitory antibodies were discovered (Table 1 and Fig. 1, A and B). One class included two nonoverlapping epitopes on IGF-1R (described below). Here we describe results of studies with individual representatives from each of these five antibody classes. Four of the five antibody classes do not appear to compete directly with the IGF ligands for binding to IGF-1R and were initially categorized as heterotropic allosteric inhibitors (or modulators) that bind to secondary (nonligand-interacting) sites on the receptor. These apparent allosteric inhibitors likely induce a conformational change in IGF-1R that reduces ligand binding affinity. Experiments to elucidate the difference between the allosteric inhibitors and the single competitive inhibitor will be described throughout the “Results.”
TABLE 1.
IGF-1 and IGF-2 blocking characteristics of select BIIB anti-IGF-1R antibodies and αIR-3
| Antibody | IC50, 320 nm IGF-1 blockade | % IGF-1 binding at antibody saturation | IC50, 640 nm IGF-2 blockade | % IGF-2 binding at antibody saturation | ||
|---|---|---|---|---|---|---|
| Allosteric IGF-1 blocker | ||||||
| BIIB1 | 0.4 ± 0.1 | 38 ± 4 | Did not block | 50 ± 4 | ||
| αIR-3 | 4.9 ± 0.5 | 23 ± 2 | Did not block | 60 ± 3 | ||
| Allosteric IGF-2 blocker | ||||||
| BIIB2 | Did not block | 80 ± 8 | 1.7 ± 1.0 | 34 ± 11 | ||
| Allosteric IGF-1 and IGF-2 blocker | ||||||
| BIIB5 | 1.3 ± 0.1 | 25 ± 4 | 1.9 ± 0.3 | 16 ± 5 | ||
| BIIB3 | 2.9 ± 0.9 | 24 ± 6 | 1.0 ± 0.5 | 22 ± 4 | ||
| Competitive IGF-1 and IGF-2 blocker | ||||||
| BIIB4 | 13 ± 5 | 1 ± 1 | 7.9 ± 3.2 | 1 ± 1 | ||
FIGURE 1.
Ligand-blocking properties of inhibitory anti-IGF-1R antibodies in competitive IGF-1 and IGF-2 ELISAs. IGF-1-(A) and IGF-2 (B)-blocking behavior of representative antibodies of the following four categories is shown: allosteric IGF-1-only blocker; allosteric IGF-2-only blocker; allosteric IGF-1 and IGF-2 blocker; and competitive IGF-1 and IGF-2 blocker. Dependence of IGF-1 concentration on the potency and activity of BIIB5 (allosteric inhibitor) (C) and BIIB4 (competitive inhibitor) (D) is shown.
The first class of inhibitory antibodies, which includes the commercially available αIR-3 murine monoclonal antibody and an in-house representative denoted BIIB1, inhibited IGF-1 binding to IGF-1R but did not inhibit IGF-2 binding. Allostery was assumed because these antibodies did not bring IGF-1 binding to base line but instead led to a partial inhibition even at saturating antibody concentrations. A second class of inhibitory antibody, represented by an antibody denoted BIIB2, inhibited IGF-2 binding to IGF-1R but had no effect on IGF-1 binding. BIIB2 also did not bring IGF-2 binding to base line. A third class, including antibodies BIIB3 and BIIB5, inhibited both IGF-1 and IGF-2, again without bringing IGF binding to base line. The last class (represented by BIIB4) completely inhibited IGF-1 and IGF-2 at high antibody concentrations, suggesting a competitive blocking mode.
To investigate the mechanism of inhibition, the competitive ELISA was performed at a number of different IGF-1 and IGF-2 concentrations across the linear range of the IGF-1 and IGF-2 ELISA binding curves. Increasing the ligand concentration in the presence of an allosteric inhibitor (see Fig. 1C for BIIB5 inhibitory curves) led to a greater percentage of IGF-1 or IGF-2 bound to the receptor with virtually no change in the antibody IC50 (where IC50 is the half-maximal inhibitory concentration of a substance). The IC50 of BIIB4, the only apparent competitive blocker, was attenuated by increasing ligand concentrations, consistent with a competitive mechanism of inhibition (Fig. 1D).
Each of these antibodies was tested for its ability to block labeled BIIB4 and BIIB5 binding to IGF-1R using both competitive ELISA and SPR-based methods. These two antibodies were chosen because BIIB4 appeared to be the only purely competitive inhibitor, whereas BIIB5 was the most clearly allosteric inhibitor (described below). Results of the cross-blocking studies indicated that the antibodies could be grouped into two separate epitope categories (Table 1). BIIB1, BIIB2, BIIB3, BIIB4, and αIR-3 all competed with labeled BIIB4, but not labeled BIIB5, for binding to IGF-1R. Inhibition was not always complete, suggesting nonidentical epitopes for BIIB1, BIIB2, BIIB3, BIIB4, and αIR-3, as might be expected based on the different ligand-blocking behaviors of the antibodies. Other antibodies from the original panel were found that blocked labeled BIIB5 from binding IGF-1R, but they all displayed a similar ligand-blocking behavior and had near identical epitopes as BIIB5 (see below); therefore, only the activity profile of BIIB5 was investigated further. These data provide clear evidence that at least two structurally separate regions of IGF-1R can be targeted for inhibiting ligand binding to IGF-1R.
Epitope Mapping of Inhibitor Antibodies—To investigate in more detail the epitopes recognized by the five different BIIB antibodies and αIR-3, we used SPR to measure the binding of the antibodies to three separate IGF-1R constructs as follows: 1) hIGF-1R-(1-903)-Fc; 2) mIGF-1R(1-904)-Fc, a mouse ectodomain fusion protein; and 3) hIGF-1R(1-462)-Fc, a human construct containing the three N-terminal three IGF-1R domains (L1-CRR-L2). For BIIB4 and BIIB5, Fab protein was available and used for screening because the Fab proteins yielded single-exponential association and dissociation kinetics. Comparative binding was first investigated using hIGF-1R-(1-903)-Fc and mIGF-1R(1-904)-Fc. All the antibodies demonstrated significantly diminished binding to mIGF-1R(1-904)-Fc compared with what was observed with hIGF-1R-(1-903)-Fc (apparent KD > 10-fold weaker; supplemental Fig. 1, A and B, and Fig. 2, A and B give examples of BIIB5 and BIIB4 Fabs binding to hIGF-1R-(1-903)-Fc and mIGF-1R(1-904)-Fc, respectively). BIIB5 was the only antibody demonstrating decreased binding to the truncated receptor construct, hIGF-1R(1-462)-Fc, with an affinity >100-fold weaker than what was observed for hIGF-1R-(1-903). The bindings of the BIIB5 and BIIB4 Fabs to hIGF-1R(1-462) are shown in supplemental Fig. 1C and Fig. 2C, respectively. These results indicate that the epitopes recognized by BIIB1, BIIB2, BIIB3, BIIB4, and αIR-3 are fully contained within the first three domains of IGF-1R, whereas the epitope group recognized by BIIB5 at least partially overlaps with an area in the FnIII domains.
Within the α-chain, human and mouse IGF-1R only differ from one another at 32 amino acid positions. Because none of the inhibitory antibodies bound mouse IGF-1R with an affinity similar to human IGF-1R, we generated a library of hIGF-1R-(1-903)-Fc constructs with single or double mutations corresponding to mouse receptor amino acids to screen against the antibodies in an effort to identify their epitopes. Using the available crystal structure of the first three domains of IGF-1R (9) and the IR ectodomain crystal structure (6), we determined that only 24 of the amino acid differences between human and murine IGF-1R are on the surface of the protein (>30% exposed to solvent) and capable of facilitating interactions with an antibody. We produced 19 single- and double-mutant constructs that encompass all 24 surface residues and screened them against BIIB4 and BIIB5 (Table 2 and supplemental Fig. 1 and Fig. 2 for screening examples with BIIB5 and BIIB4, respectively). Double mutants were made if two adjacent nonidentical residues between mouse and human existed in the IGF-1R primary sequence. The binding of BIIB4 to IGF-1R was decreased by two separate mutations, one in the CRR domain and one in the L2 domain (S257F and E303G), whereas BIIB5 binding to IGF-1R was completely ablated by a single mutation in the FnIII-1 domain (H464E).
TABLE 2.
The effects of IGF-1R mutation on the binding of inhibitory antibodies
a No effect (NE), measured KD within 2.5-fold of WT hIGF-1R-Fc; weak (W), measured KD between 2.5- and 10-fold higher than WT; medium (M), measured KD between 10- and 100-fold higher than WT; strong (S), binding to antibody was ablated by mutation; and ND, not determined. DNE means did not express or expressed poorly because of intolerance of the protein fold for the mutation. M*, measured >10-fold increase in affinity upon mutation. W*, measured >5-fold increase in affinity upon mutation.
Based on these results, we generated an additional 20 mutations in the CRR/L2 domains and 21 mutations in the FnIII-1 domain surrounding those residue positions where mutation led to decreases in BIIB4 or BIIB5 affinity for IGF-1R. BIIB1, BIIB2, BIIB3, BIIB4, and αIR-3 were screened against the IGF-1R constructs containing CRR/L2 mutations, and BIIB5 was screened against the IGF-1R constructs containing FnIII-1 mutations. The data from these screens enabled residue-specific epitope mapping of all five BIIB antibodies and αIR-3 (Table 2). The epitope of BIIB5 covered a patch on the surface of the FnIII-1 domain opposite the ligand-binding region (Fig. 2 shows the epitope mapped onto the IR ectodomain structure (6)). The epitopes of BIIB1, BIIB2, BIIB3, BIIB4, and αIR-3 all spanned the CRR and L2 domains (Fig. 2 shows the epitope of BIIB4 mapped to the ectodomain structure of IR (6)).
FIGURE 2.
Epitope mapping data for BIIB4 and BIIB5 mapped onto the surface of the x-ray crystal structure of the extracellular domains of IR (6) based on homologous positions determined using a sequence alignment of IR and IGF-1R.
Whereas the epitopes of BIIB1, BIIB2, BIIB3, BIIB4, and αIR-3 overlap with one another as evidenced by their cross-blocking behavior and the screening results, differences were clearly observed that differentiate the unique ligand-blocking properties of each antibody (Table 2 and Fig. 3). BIIB4, the competitive ligand blocker, intrudes into the ligand-binding pocket (7, 8) to a greater extent than any of the other antibodies in the same epitope group. BIIB3, which is the only other antibody of this epitope group that blocks both ligands, also intrudes into the ligand-binding pocket, but to a less significant extent. The epitopes of BIIB1, BIIB2, and αIR-3, which block only IGF-1 or IGF-2, are offset from the ligand-binding pocket.
FIGURE 3.
Epitope mapping data for BIIB1, BIIB2, BIIB3, BIIB4, and αIR-3 onto the surface of the x-ray crystal structure of the N-terminal L1-CRR-L2 domains of IGF-1R (9). Shown for reference in the last panel are the positions of alanine mutations that had an effect on IGF-1 binding to IGF-1R as published previously (7).
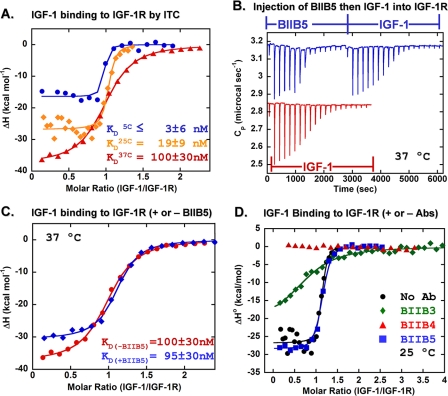
Characterization of the Mechanism of IGF/IGF-1R Blockade by BIIB3, BIIB4, and BIIB5—BIIB3-5 were shown to block both IGF-1 and IGF-2 from binding IGF-1R. Although it is known that antibody binding can lead to IGF-1R down-regulation potentially contributing to efficacy regardless of the ligand-blocking behavior (1, 25), evidence from other receptor tyrosine kinases indicates that in some tumors dysregulation of the receptor internalization machinery can lead to constant receptor levels available on the cell surface for continued signaling through ligand binding (26, 27). In such cases, antibodies with intrinsic IGF-1- and IGF-2-blocking properties may provide an advantage over antibodies dependent on receptor down-regulation for activity. Therefore, we focused our studies on characterizing the mechanism of ligand blocking for the dual ligand inhibitors to gain insights into their specific activities. To facilitate detailed biophysical analyses, we used hIGF-1R-(1-903), a construct lacking a fusion partner. The protein was purely dimeric (∼250 kDa), as determined by SEC/static light scattering, and stable for months at 2-8 °C. hIGF-1R-(1-903) demonstrated a single apparent unfolding transition with a thermal midpoint of unfolding (TM) of 61 °C as determined by DSC (supplemental Fig. 3). The protein reduced to two polypeptides of predicted molecular weight for the α- and β-chains when analyzed by SDS-PAGE.
ITC with BIIB3, BIIB4, and BIIB5—The ability of BIIB3, BIIB4, and BIIB5 to alter the ligand-binding behavior of IGF-1R was investigated by ITC. Initially, the binding of IGF-1 to hIGF-1R-(1-903) was studied to provide a base line for the activity of the receptor/ligand pair. Similar to what has been described previously (28), we found that IGF-1 binds to the hIGF-1R-(1-903) dimer with a 1:1 stoichiometry, suggesting that when one IGF molecule binds it leads to a conformational change in the receptor that occludes a possible second high affinity binding site. The affinity of IGF-1 for IGF-1R was strongly temperature-dependent, becoming weaker as the temperature was elevated from 5 to 37 °C (Table 3 and Fig. 4A). Neither IGF-1 nor hIGF-1R-(1-903) demonstrated structural changes between 5 and 50 °C based on temperature-dependent near and far-UV CD studies (Fig. 5, A-C). The comparatively large entropy penalty for IGF-1 binding at 37 °C does suggest ordering of one or both of the reaction components upon complex formation. Comparison of the CD spectra of the complex compared with the sum of the spectra of the individual protein components indicates an ordering of β-sheet structure upon binding (Fig. 6A). The ordered β-structure can likely be linked to the receptor as the ligands are known to be helical (29).
TABLE 3.
ITC-derived temperature dependence of IGF-1 and antibody binding to IGF-1R
| Temperature | ΔHo | -TΔSo | ΔGo | Kd | Stoichiometry of binding (n) |
|---|---|---|---|---|---|
| °C | kcal/mol | kcal/mol | kcal/mol | nm | |
| IGF-1 | |||||
| 5 | −16.4 ± 0.5 | 5.6 | −10.8 | 3.3 ± 6.0 | 0.93 |
| 25 | −26.8 ± 0.6 | 16.2 | −10.6 | 19 ± 9 | 1.00 |
| 37a | −33.4 ± 0.5 | 23.6 | −9.8 | 130 ± 60 | 1.02 |
| BIIB5 | |||||
| 5b | −11.3 ± 0.8 | 0.89 | |||
| 15b | −14.4 ± 0.6 | 0.98 | |||
| 25c | −20.1 ± 2.5 | 7.8d | −12.3d | 1 ± 0.2d | 0.98 |
| 37b | −29.2 ± 0.4 | 0.97 | |||
| BIIB4 | |||||
| 5b | −6.7 ± 1.0 | 0.90 | |||
| 25 | −26.6 ± 0.6 | 15.1d | −11.5d | 4 ± 0.5d | 0.91 |
| 37b | −39.5 ± 1.2 | 0.70 | |||
| BIIB3 | |||||
| 25c | −14.1 ± 0.6 | 0.84 |
Data are average of three experiments.
Tight binding resulted in too few points in the transition region to measure Kd.
Data are average of two experiments.
Data were determined using both SPR and ITC experiments.
FIGURE 4.
ITC results for the binding of IGF-1 to IGF-1R in the absence and presence of saturating levels of inhibitory antibodies. A, temperature dependence of IGF-1 binding to hIGF-1R-(1-903) as follows: 5 °C (•), 25 °C (♦), and 37 °C (▴). B, raw ITC data at 37 °C for the dual titration of BIIB5 followed by IGF-1. C, transformed ITC curves for IGF-1 binding to hIGF-1R-(1-903) in the absence (•) and presence (♦) of BIIB5 at 37 °C. D, transformed ITC curves for the binding of IGF-1 to hIGF-1R(-1903) in the absence and presence of saturating levels (∼2-fold excess) of BIIB3, BIIB4, and BIIB5 at 25 °C.
FIGURE 5.
A, temperature-dependent and far-UV; B, near UV CD spectra of hIGF-1R-(1-903); C, far-UV CD spectra of IGF-1.
FIGURE 6.
CD studies demonstrate that both the ligand and the BIIB4 and BIIB5 Fabs induce structural changes in IGF-1R. Far UV CD spectra of apo-hIGF-1R-(1-903) and hIGF-1R-(1-903) in the presence of the following: A, IGF-1; B, BIIB4 Fab; C, BIIB5 Fab; and D, a nonspecific antibody Fab.
The temperature dependence of antibody binding to IGF-1R was also investigated prior to studies with ternary antibody·ligand·receptor complexes. At temperatures between 5 and 37 °C, the affinities of the antibodies for IGF-1R were sufficiently strong to preclude the determination of affinity constants (Table 3). BIIB4 and BIIB5 exhibited particularly strong enthalpies of binding. The affinity of the Fab forms of both BIIB5 (1 nm) and BIIB4 (4 nm) for hIGF-1R-(1-903) was determined by kinetic SPR and identical to what was observed using hIGF-1R-(1-903)-Fc during the epitope mapping studies (supplemental Figs. 1 and 2). With knowledge of both the affinity and enthalpy of binding, the entropy contribution to the interaction of hIGF-1R-(1-903) to both BIIB4 and BIIB5 was estimated (Table 3). A strong entropy penalty exists for both BIIB4 and BIIB5 binding to IGF-1R, particularly at 37 °C, suggestive of conformational ordering within the receptor, the antibodies, or both. Differences in the far-UV CD spectra between (i) the sum of the spectra of uncomplexed hIGF-1R-(1-903) and BIIB4 or BIIB5 and (ii) the spectra of the antibody·receptor complexes indicate ordering upon the binding of both antibodies to IGF-1R (Fig. 6, B and C). A control experiment combining hIGF-1R-(1-903) with a nonspecific antibody Fab is shown in Fig. 6D for comparison. A weakening of the absolute value of the CD band at 217 nm is often associated with ordered β-sheet structures in the absence of denaturants; the opposite is evident in the temperature-induced unfolding of hIGF-1R-(1-903) (Fig. 5A) and is a common feature of thermally induced unfolding profiles of antibody domains, which form loosely associated structures with presumably more overall β-sheet content in their denatured states than in their native proteins.
Finally, we examined the effect of titrating IGF-1 into solutions containing pre-formed complexes with BIIB3, BIIB4, or BIIB5. Fig. 4B gives an example of the raw data from a dual titration, first with BIIB5 followed by subsequent titration of IGF-1 into a solution containing hIGF-1R-(1-903). BIIB5, which binds the FnIII-1 domain, did not have a measurable effect on IGF-1 binding to hIGF-1R-(1-903) under the ITC conditions, although it was shown to be an allosteric IGF-1 and IGF-2 blocker in the competitive ELISA (Table 4 and Fig. 4, B-D). BIIB3 led to an apparent 50-fold decrease in the affinity of IGF-1 for hIGF-1R-(1-903) (Table 4 and Fig. 4D). Finally, BIIB4 appeared to completely abrogate IGF-1 binding to the receptor under the ITC conditions consistent with its classification as a competitive inhibitor (Table 4 and Fig. 4D).
TABLE 4.
IGF-1/IGF-1R binding parameters determined at 25 °C in the absence and presence of saturated levels of inhibitory antibodies
| IGF-1R antibody complex |
ITC results
|
Equilibrium SPR results | |||||
|---|---|---|---|---|---|---|---|
| ΔH° () | - T ΔS° | Δ G° | Kd | na | Kd | na | |
| kcal/mol | kcal/mol | kcal/mol | nm | nm | |||
| IGF-1R | − 26.8 ± 0.6 | 16.2 | − 10.6 | 19 ± 9 | 1.00 | 11 ± 2 | 0.66 |
| IGF-1R·BIIB5b | − 28.4 ± 2.0 | 17.9 | − 10.5 | 20 ± 4 | 1.09 | 150 ± 15 | 0.65 |
| IGF-1R·BIIB3b | − 21.7 ± 1.8 | 13.5 | − 8.2 | 1000 ± 100 | 0.80 | 300 ± 40 | 0.5 |
| IGF-1R·BIIB4c | -d | - | - | - | - | - | - |
IGF-1·hIGF-1R-(1-903) binding stoichiometry.
Apparent allosteric inhibitor.
Apparent competitive inhibitor.
No IGF-1 binding was observed.
Equilibrium SPR Studies of the Ligand-blocking Properties of BIIB3, BIIB4, and BIIB5—We next investigated the ability of BIIB3, BIIB4, and BIIB5 to block IGF-1 binding to hIGF-1R-(1-903) using equilibrium SPR performed under mass transfer-limited conditions (24). To detect IGF-1 binding, the ligand was injected over a surface containing high levels of immobilized hIGF-1R-(1-903)-Fc, yielding an IGF-1 binding standard curve where the IGF-1 signal was linearly dependent on the concentration of IGF-1 (Fig. 7A). To measure the affinity of IGF-1 for hIGF-1R-(1-903), we added free receptor in solution at either 24, 64, or 240 nm (Fig. 7A). The standard curve was offset by the concentration of hIGF-1R-(1-903), and the shape of the curve, prior to resuming a linear curve parallel to the standard curve, provided a measure of bound versus free IGF-1 in solution, which was used to determine the IGF-1 affinity for the receptor. At 64 and 240 nm hIGF-1R-(1-903), sufficient curvature was observed to measure the equilibrium dissociation constant (KD) of IGF-1 for hIGF-1R-(1-903) at 25 °C (Table 4). The KD, 11 nm, was within a factor of 2 of what was obtained by ITC (KD = 19 nm).
FIGURE 7.
SPR-based equilibrium IGF-1/IGF-1R binding studies in the absence and presence of BIIB3, BIIB4, and BIIB5. A, free IGF-1 concentration (nm) was measured by SPR in the absence (▴) and presence of 24 nm (▪), 64 nm (♦), and 240 nm (•) hIGF-1R-(1-903). B, free-IGF-1 concentration (nm) was measured by SPR in the presence of 240 nm hIGF-1R and the absence (▪) or presence of 240 nm BIIB5 (•), BIIB3 (▴), or BIIB4 (♦). Theoretical curves for the equilibrium concentration of free-IGF-1 in the presence of 240 nm hIGF-1R-(1-903) over a 6 nm to 20 μm range of theoretical KD values between the ligand and the receptor are shown.
Next, we evaluated the effect of adding an equimolar (240 nm) amount of BIIB3, BIIB4, or BIIB5 in solution with the hIGF-1R-(1-903) used in the equilibrium-binding experiment. Theoretical IGF-1 binding curves with the KD as a variable are shown in Fig. 7B. The IGF-1 binding curve in the absence of antibody, as measured above, was between the 6 and 24 nm theoretical curves. The IGF-1 binding curve in the presence of BIIB5 was shifted to between the theoretical KD = 96 and 384 nm curves (Fig. 7B). The IGF-1 binding curve in the presence of BIIB3 was shifted closer to the KD = 384 nm theoretical curve (Fig. 7B). Consistent with BIIB4 being a competitive inhibitor, the IGF-1 curve in the presence of BIIB4 was similar to the standard curve with no measurable IGF-1 binding to hIGF-1R-(1-903) in solution (Fig. 7B). Binding parameters derived by fitting the curves of IGF-1 binding to hIGF-1R-(1-903) in the absence and presence of each antibody are provided in Table 4.
Further Investigation of the Allosteric Mechanism of BIIB5—The competition ELISA, epitope mapping, ITC, and equilibrium SPR binding studies are all consistent with the role of BIIB5 as an allosteric inhibitor. However, unlike BIIB3 and BIIB4, the measured extent of IGF-1 blockade across methods was not consistent. Therefore, additional studies were performed to investigate the allosteric mechanism behind BIIB5 inhibition of IGF-1 binding to IGF-1R. First, hIGF-1R-(1-903) was immunoprecipitated by BIIB5 in the presence and absence of IGF-1. These experiments were performed at 500 nm hIGF-1R-(1-903) and IGF-1 concentrations, above the IGF-1/hIGF-1R-(1-903) KD value measured using ITC or equilibrium SPR in the presence of BIIB5. Immunoprecipitation of hIGF-1R-(1-903) by BIIB5 in the presence of IGF-1 yielded near stoichiometric levels of IGF-1 bound to the receptor (Fig. 8A), demonstrating that ternary engagement of hIGF-1R-(1-903) occurs, consistent with an allosteric mechanism for BIIB5. Next, we tracked the EC50 value (where EC50 is the half-maximal effective concentration) of IGF-1 binding to hIGF-1R-(1-903) at a saturating (400 nm) concentration of BIIB5 in the competitive ELISA. The EC50 value was reduced ∼50-fold in the presence of BIIB5 (Fig. 8B). Finally, kinetic IGF-1/hIGF-1R-(1-903) binding experiments were performed in the absence and presence of 500 nm BIIB5 using SPR. In the absence of BIIB5, kinetic analysis of IGF-1 binding to hIGF-1R-(1-903) resulted in single exponential association and dissociation kinetics that fit well to a 1:1 model yielding a KD = 20 nm (Fig. 8C). In the presence of BIIB5, a decrease in the Rmax (i.e. maximum achievable signal) was observed (Fig. 8D). The curves did not fit well to a 1:1 binding model in the presence of BIIB5; however, the best fits suggest the KD is reduced ∼3.5-fold in the presence of BIIB5. These experiments confirm the role of BIIB5 as an allosteric inhibitor; however, the relative strength of BIIB5 inhibition of IGF-1 binding to IGF-1R remains unclear.
FIGURE 8.
Multiple effects of BIIB5 on IGF-1 binding to IGF-1R. A, co-immunoprecipitation of IGF-1·IGF-1R complexes from a 1:1 mixture of 500 nm IGF-1 and hIGF-1R-(1-903) by BIIB5. Lanes 1-4 provide a titration of purified IGF-1 for quantitation. The remaining lanes are blots of 50 ng of purified hIGF-1R-(1-903) (lane 5), 50 ng of purified BIIB5 (lane 6), and co-immunoprecipitation of a hIGF-1R-(1-903)·IGF-1 complex by BIIB5 (lane 7), and unsuccessful immunoprecipitation of IGF-1 by BIIB5 in the absence of hIGF-1R-(1-903) (lane 8). B, direct IGF-1 binding ELISA in the absence (•) and presence of 400 nm BIIB5 (▪). C and D, kinetic SPR curves of IGF-1 binding to hIGF-1R-(1-903) at 25 °C in the absence (C) and presence (D) of 500 nm BIIB5. Concentrations of IGF-1 in the SPR studies were 1:1 serial dilutions from 64 to 0.125 nm.
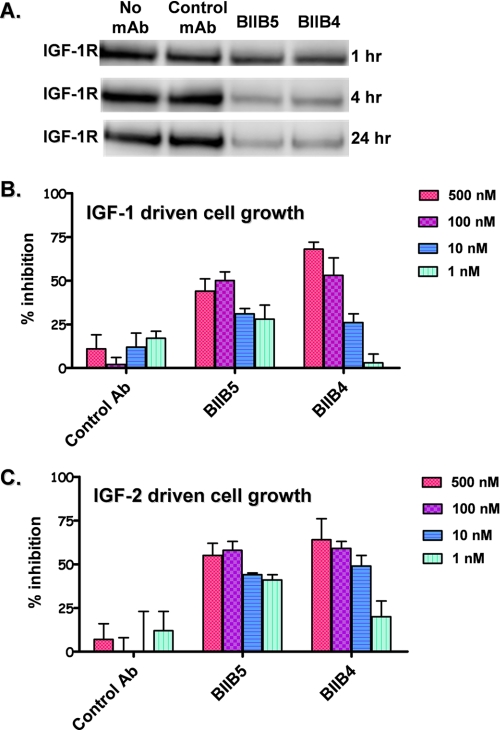
IGF-1R Down-regulation and in Vitro Tumor Cell Growth Inhibition by BIIB4 and BIIB5—Many studies have shown that antibody binding can lead to down-regulation of IGF-1R, potentially contributing to observed cellular signaling and cell growth responses (30-34). Based on the biochemical and biophysical studies, we investigated the ability of both BIIB4 (competitive inhibitor) and BIIB5 (allosteric inhibitor) to degrade IGF-1R over time when incubated with H322M cells (derived from NSCLC cells). The data showed that incubation with both BIIB4 and BIIB5 led to IGF-1R degradation over a period of hours (Fig. 9A). No significant differences were observed in the apparent kinetics or overall ability of the two antibodies to degrade IGF-1R.
FIGURE 9.
A, antibody (Ab)-induced degradation of IGF-1R in H322M cells. B, inhibition of IGF-1-induced H322M cell growth by BIIB4 and BIIB5. C, inhibition of IGF-2-induced H322M cell growth by BIIB4 and BIIB5.
Next, we examined the relative ability of BIIB4 and BIIB5 to inhibit H322M cell growth using a cell viability assay. Both BIIB4 and BIIB5 exhibited dose-dependent inhibition of IGF-1- or IGF-2-stimulated cell growth (Fig. 9, B and C). The two antibodies did, however, demonstrate different dose titration behavior in both the IGF-1- and IGF-2-stimulated cell growth assays. BIIB5, the allosteric inhibitor, showed potent activity at low antibody concentrations (1 and 10 nm), whereas its activity reached a saturating plateau at high concentrations (100 and 500 nm). In contrast, BIIB4, the competitive inhibitor, showed less activity at low antibody concentrations, but its activity steadily increased with increasing antibody concentration, reaching a higher maximal activity than BIIB5. Interestingly, the dose responses of BIIB4 and BIIB5 in the cell-based assay reflected the ligand-blocking properties determined biochemically and biophysically. Because BIIB4 and BIIB5 both degrade IGF-1R at a similar rate, the observed difference in cell growth was likely because of their distinct epitope and ligand-blocking properties.
DISCUSSION
Here we link several classes of inhibitory anti-IGF-1R antibodies with highly specific epitopes on the surface of IGF-1R. Previous biochemical studies have shown that both agonistic and inhibitory epitopes exist within the first four ectodomains of both IR and IGF-1R (35-40). The first and most well characterized inhibitory antibody to IGF-1R, αIR-3, was developed 25 years ago in mice (40). This antibody was shown to specifically block IGF-1 from binding IGF-1R using an apparent allosteric mechanism, while having only a weak effect on the binding of IGF-2 (23). Our panel contains antibodies with both discriminatory and nondiscriminatory blocking activity toward IGF-1 and IGF-2. Many of the IGF-1R residues involved in IGF-1 and IGF-2 binding have been mapped using alanine-scanning mutagenesis (7, 8). In these studies, a subset of residues in the CRR domain (residues 240-242 and 251) appeared to be important for IGF-1 binding but not IGF-2 binding (8, 9). Based on the homology between IGF-1 and IGF-2 (67% identity), it can be surmised that antibodies that specifically block IGF-1 must have subtle epitope differences from those that specifically block IGF-2.
We found that each antibody group (differentiated by their ability to block (i) IGF-1-only, (ii) IGF-2-only, or both IGF-1 and IGF-2 (iii) allosterically, or (iv) competitively) that recognizes the CRR/L2 region has unique epitope characteristics (Table 2). The antibodies that specifically blocked IGF-1, αIR-3 and BIIB1, were specifically attenuated from binding IGF-1R by mutation at residues 251 and 255, whereas BIIB2, which only blocked IGF-2, was unaffected by these mutations. BIIB2 was specifically attenuated by an IGF-1R mutation at residue 250, whereas αIR-3 and BIIB1 were unaffected by this mutation. BIIB3 and BIIB4, which blocked both IGF-1 and IGF-2, were uniquely attenuated by mutations at residues 259 and 260, and the determined epitopes appear to position BIIB3 and BIIB4 more in-line with the ligand-binding pocket than the IGF-1-only or IGF-2-only blockers. Together, these data suggest very small residue subsets within each epitope differentiate the ligand-blocking behavior of the inhibitory antibodies that recognize the CRR/L2 region.
It is apparent from the competitive ELISA studies that IGF-1 and IGF-2 can engage IGF-1R while saturated with αIR-3, BIIB1, BIIB2, or BIIB3, although the association is much weaker with a reduced affinity. The fact that these antibody epitopes weakly overlap with residues shown to affect IGF binding suggests that the ligand inhibition properties of these antibodies may not be excluded to a “second site” as indicated by a purely allosteric compound like BIIB5. Therefore, the mechanism behind ligand blockade for αIR-3, BIIB1, BIIB2, and BIIB3 may be that by binding IGF-1R, the antibodies strip away residue interactions weakly associated with ligand binding, thus changing the ligand affinity for IGF-1R without completely displacing the ligand. This may happen in concert with an inhibitory conformational change in the receptor.
BIIB5 represents the inhibitory antibody class we found that recognized an epitope well removed from the ligand-binding region. Soos et al. (35) demonstrated using chimeric IGF-1R/IR constructs that an inhibitory epitope does exist in the FnIII-1 domain. They also showed that a similar inhibitory epitope exists within the IR (36). Mapping our epitope on the recent structure of the IR extracellular domain, crystallized in the presence of this inhibitory anti-IR 83-14 antibody (6), demonstrates that the BIIB5 epitope partially overlaps the inhibitory epitope found on the IR. Based on the distance of the epitope from the ligand-binding pocket and our biophysical studies, it is obvious why BIIB5 is an allosteric inhibitor. We do show that the binding of BIIB5 leads to conformational changes in IGF-1R that must be inimical to ligand binding. The structural aspects of the conformational change induced by BIIB5, however, remain unclear.
Our ITC and CD studies demonstrate that binding of IGF-1, BIIB4, and BIIB5 to IGF-1R induces conformational changes in the receptor, including the ordering of receptor polypeptide regions. BIIB4 exhibits a much larger ΔCP° of binding (-990 cal mol-1 °C-1, where ΔCP° is molar heat capacity of association) compared with IGF-1 or BIIB5 (-530 and -560 cal mol-1 °C-1, respectively). Based on the epitope mapping studies, the binding of BIIB4 to IGF-1R can be attenuated by residues across a wide area of IGF-1R spanning both the CRR and L2 domains. This area appears too large to be entirely contained within a single antibody epitope. Many mutations in both the CRR and L2 domains only mildly affect IGF-1R binding of BIIB4 and other antibodies within this epitope class. It may be that some of these IGF-1R mutations do not directly interrupt antibody interactions with the receptor, but instead lead to a change in the dynamics or structure of the CRR region, provided the CRR domain does not form a cooperatively folded structure. The CRR domain does not have a hydrophobic core and like many repeat domains may not be expected to unfold cooperatively (41). It is plausible that antibodies like BIIB4, which bind the CRR region, lead to increased structure formation within the CRR domain, the L2 domain, or even the FnIII-2 loop region that is accompanied by the burial of additional surface area, resulting in the large ΔCP°. Consistent with conformational changes in the receptor upon antibody or ligand binding, increases in ordered structure were observed using CD spectroscopy when BIIB4, BIIB5, or IGF-1 were mixed with IGF-1R. Unfolded and partially folded polypeptide regions of proteins are increasingly being implicated in biological activities such as the regulation of binding affinity or activity (42). It seems increasingly likely based on our studies and previous studies (28) that receptor ordering upon ligand or antibody binding may be an important aspect of IGF-1R activity, leading to both active and inactive forms of the receptor.
The existence of a wide variety of IGF-1R antibody inhibitors, including those that inhibit allosterically or competitively or those that inhibit one or both of the ligands, poses questions as to the relative efficacy each class of antibody may have in the clinic. How important will the mechanism of inhibition be for achieving clinical results? Certainly, the results described here suggest that the activity profiles of anti-IGF-1R inhibitors are broad and occur through multiple mechanisms of action. An additional complication is that some inhibitory antibodies lead to IGF-1R internalization and down-regulation. It is currently unknown whether receptor down-regulation or receptor inhibition/ligand blocking will dominate in the therapeutic setting. The data we show with the H322M cell line suggest that the specific mechanism of ligand blocking can in certain circumstances be reflected in the activity profile of the inhibitory antibody. However, therapeutic activity may depend on tumor phenotype, the status of internalization machinery, and the availability of ligand, either in the serum or in the tumor microenvironment. It is known that the antibodies currently in clinical trials display a spectrum of ligand-blocking and receptor down-regulation behaviors (25). It will be interesting to see whether specific ligand-blocking properties of these therapeutic antibodies lead to any differentiation in the efficacy of these molecules in the clinic.
Supplementary Material
Acknowledgments
We thank Dyax Corp. (Cambridge, MA) for the antibody discovery efforts behind the identification of BIIB4 and BIIB5. We thank Dr. Marcel Maeder and John Brazel for developing the models for the global fit routines of the CD spectra and the software for the analysis, respectively, and Dr. Fred Taylor and Renee Shapiro for their initial efforts on the project. We also thank Dr. Kristin Demarest for critical reading of the manuscript.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-3.
Footnotes
The abbreviations used are: IGF-1R, type I insulin-like growth factor receptor; CHO, Chinese hamster ovary; CRR, cysteine-rich region; DSC, differential scanning calorimetry; Fab, antibody antigen-binding fragment; Fc, IgG constant domain fragment; HPLC, high pressure liquid chromatography; ITC, isothermal titration calorimetry; PBS, phosphate-buffered saline; RU, resonance unit; SEC, size exclusion chromatography; SPR, surface plasmon resonance; mAb, monoclonal antibody; NSCLC, non-small cell lung cancer; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; ELISA, enzyme-linked immunosorbent assay; HRP, horseradish peroxidase; IR, insulin receptor.
References
- 1.Chitnis, M. M., Yuen, J. S. P., Protheroe, A. S., Pollak, M., and Macaulay, V. M. (2008) Clin. Cancer Res. 14 6364-6370 [DOI] [PubMed] [Google Scholar]
- 2.Pollak, M. N., Schernhammer, E. S., and Hankinson, S. E. (2004) Nat. Rev. Cancer 4 505-518 [DOI] [PubMed] [Google Scholar]
- 3.Adams, T. E., McKern, N. M., and Ward, C. W. (2004) Growth Factors 22 89-95 [DOI] [PubMed] [Google Scholar]
- 4.Lawrence, M. C., McKern, N. M., and Ward, C. W. (2007) Curr. Opin. Struct. Biol. 17 699-705 [DOI] [PubMed] [Google Scholar]
- 5.Adams, T. E., Epa, V. C., Garrett, T. P., and Ward, C. W. (2000) Cell. Mol. Life Sci. 57 1050-1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKern, N. M., Lawrence, M. C., Streltsov, V. A., Lou, M.-Z., Adams, T. E., Lovrecz, G. O., Elleman, T. C., Richards, K. M., Bentley, J. D., Pilling, P., Hoyne, P. A., Cartledge, K. A., Pham, T. M., Lewis, J. L., Sankovich, S. E., Stoichevska, V., Da Silva, E., Robinson, C. P., Frenkel, M. J., Sparrow, L. G., Fernley, R. T., Epa, V. C., and Ward, C. W. (2006) Nature 443 218-221 [DOI] [PubMed] [Google Scholar]
- 7.Whittaker, J., Groth, A. V., Mynarcik, D. C., Pluzek, L., Gadsboll, V. L., and Whittaker, L. J. (2001) J. Biol. Chem. 276 43980-43986 [DOI] [PubMed] [Google Scholar]
- 8.Sorensen, H., Whittaker, L., Hinrichsen, J., Groth, A., and Whittaker, J. (2004) FEBS Lett. 565 19-22 [DOI] [PubMed] [Google Scholar]
- 9.McKern, N. M., Lou, M., Frenkel, M. J., Verkuylen, A., Bentley, J. D., Lovrecz, G. O., Ivancic, N., Elleman, T. C., Garrett, T. P. J., Cosgrove, L. J., and Ward, C. W. (1997) Protein Sci. 6 2663-2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan, S., Shapiro, D. N., and Helman, L. J. (1995) Oncogene 11 2503-2507 [PubMed] [Google Scholar]
- 11.Karnieli, E., Werner, H., Rauscher, F. J., III, Benjamin, L. E., and LeRoith, D. (1996) J. Biol. Chem. 271 19304-19309 [DOI] [PubMed] [Google Scholar]
- 12.Baserga, R., Peruzzi, F., and Reiss, K. (2003) Int. J. Cancer 107 873-877 [DOI] [PubMed] [Google Scholar]
- 13.Press, M. F., and Lenz, H. J. (2007) Drugs 67 2045-2075 [DOI] [PubMed] [Google Scholar]
- 14.Rodon, J., DeSantos, V., Ferry, R. J., and Kurzrock, R. (2008) Mol. Cancer Ther. 7 2575-2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toretsky, J. A., Kalebic, T., Blakesley, V., LeRoith, D., and Helman, L. J. (1997) J. Biol. Chem. 272 30822-30827 [DOI] [PubMed] [Google Scholar]
- 16.Leavey, P. J., and Collier, A. B. (2008) Expert Rev. Anticancer Ther. 8 617-624 [DOI] [PubMed] [Google Scholar]
- 17.Yee, D., Favoni, R. E., Lebovic, G. S., Lombana, F., Powell, D. R., Reynolds, C. P., and Rosen, N. (1990) J. Clin. Investig. 86 1806-1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demarest, S. J., Hopp, J., Chung, J., Hathaway, K., Mertsching, E., Cao, X., George, J., Miatkowski, K., LaBarre, M. J., Shields, M., and Kehry, M. R. (2006) J. Biol. Chem. 281 30755-30767 [DOI] [PubMed] [Google Scholar]
- 19.Brezinsky, S. C. G., Chiang, G. G., Szilvasi, A., Mohan, S., Shapiro, R. I., MacLean, A., Sisk, W., and Thill, G. (2003) J. Immunol. Methods 277 141-155 [DOI] [PubMed] [Google Scholar]
- 20.Hariharan, K., Graff, C., Glaser, S., Garber, E., Reyes, C. L., and Demarest, S. (October 18, 2007) U. S. Patent 20070243194
- 21.Hoet, R. M., Cohen, E. H., Kent, R. B., Rookey, K., Schoonbroodt, S., Hogan, S., Rem, L., Frans, N., Daukandt, M., Pieters, H., van Hegelsom, R., Neer, N. C., Nastri, H. G., Rondon, I. J., Leeds, J. A., Hufton, S. E., Huang, L., Kashin, I., Devlin, M., Kuang, G., Steukers, M., Viswanathan, M., Nixon, A. E., Sexton, D. J., Hoogenboom, H. R., and Ladner, R. C. (2005) Nat. Biotechnol. 23 344-348 [DOI] [PubMed] [Google Scholar]
- 22.Galanis, M., Irving, R. A., and Hudson, P. J. (2001) Current Protocols in Immunology, Unit 17.1, John Wiley & Sons, Inc., New York [DOI] [PubMed]
- 23.Jacobs, S., Cook, S., Svoboda, M. E., and Van Wyk, J. J. (1986) Endocrinology 118 223-226 [DOI] [PubMed] [Google Scholar]
- 24.Day, E. S., Cachero, T. G., Qian, F., Sun, Y., Wen, D., Pelletier, M., Hsu, Y. M., and Whitty, A. (2005) Biochemistry 44 1919-1931 [DOI] [PubMed] [Google Scholar]
- 25.Pollak, M. (2008) Curr. Opin. Pharmacol. 8 384-392 [DOI] [PubMed] [Google Scholar]
- 26.Bache, K. G., Slagsvold, T., and Stenmark, H. (2004) EMBO J. 23 2707-2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roepstorff, K., Grøvdal, L., Grandal, M., Lerdrup, M., and van Deurs, B. (2008) Histochem. Cell Biol. 129 563-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jannson, M., Hallen, D., Koho, H., Andersson, G., Berghard, L., Heidrich, J., Nyberg, E., Uhlen, M., Kordel, J., and Nilsson, B. (1997) J. Biol. Chem. 272 8189-8197 [DOI] [PubMed] [Google Scholar]
- 29.Brzozowski, A. M., Dodson, E. J., Dodson, G. G., Murshudov, G. N., Verma, C., Turkenburg, J. P., de Bree, F. M., and Dauter, Z. (2002) Biochemistry 41 9389-9397 [DOI] [PubMed] [Google Scholar]
- 30.Burtrum, D., Zhu, Z., Lu, D., Marie, D., Anderson, M., Prewett, M., Pereira, D. S., Bassi, R., Abdullah, R., Hooper, A. T., Koo, H., Jimenez, X., Johnson, D., Apblett, R., Kussie, P., Bohlen, P., Witte, L., Hicklin, D. J., and Ludwig, D. L. (2003) Cancer Res. 63 8912-8921 [PubMed] [Google Scholar]
- 31.Shang, Y., Mao, Y., Batson, J., Scales, S. J., Phillips, G., Yang, J., Tang, Z., Modrusan, Z., Tan, C., Liang, W.-C., Tsai, S. P., Vanderbilt, A., Kozuka, K., Hoeflich, K., Tien, J., Ross, S., Li, C., Lee, S. H., Song, A., Wu, Y., Stephan, J.-P., Ashkenazi, A., and Zha, J. (2008) Mol. Cancer Ther. 7 2599-2608 [DOI] [PubMed] [Google Scholar]
- 32.Cohen, B. D., Baker, D. A., Soderstrom, C., Tkalcevic, G., Rossi, A. M., Miller, P. E., Tengowski, M. W., Wang, F., Gualberto, A., Beebe, J. S., and Moyer, J. D. (2005) Clin. Cancer Res. 11 2063-2073 [DOI] [PubMed] [Google Scholar]
- 33.Goetsch, L., Gonzalez, A., Leger, O., Beck, A., Pauwels, P. J., Haeuw, J. F., and Corvaia, N. (2005) Int. J. Cancer 113 316-328 [DOI] [PubMed] [Google Scholar]
- 34.Wang, Y., Hailey, J., Williams, D., Wang, Y., Lipari, P., Malkowski, M., Wang, X., Xie, L., Li, G., Saha, D., Ling, W. L. W., Cannon-Carlson, S., Greenberg, R., Ramos, R. A., Shields, R., Presta, L., Brams, P., Bishop, W. R., and Pachter, J. A. (2005) Mol. Cancer Ther. 4 1214-1221 [DOI] [PubMed] [Google Scholar]
- 35.Soos, M. A., Field, C. E., Lammers, R., Ullrich, A., Zhang, B., Roth, R. A., Andersen, A. S., Kjeldsen, T., and Siddle, K. (1992) J. Biol. Chem. 267 12955-12963 [PubMed] [Google Scholar]
- 36.Soos, M. A., Siddle, K., Baron, M. D., Heward, J. M., Luzio, J. P., Bellatin, J., and Lennox, E. S. (1986) Biochem. J. 235 199-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth, R. A., Maddux, B., Wong, K. Y., Styne, D. M., Van Vliet, G., Humbel, R. E., and Goldfine, I. D. (1983) Endocrinology 112 1865-1867 [DOI] [PubMed] [Google Scholar]
- 38.Roth, R. A., Cassell, D. J., Wong, K. Y., Maddux, B. A., and Goldfine, I. D. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 7312-7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kull, F. C. J., Jacobs, S., Su, Y.-F., and Cuatrecasas, P. (1982) Biochem. Biophys. Res. Commun. 106 1019-1026 [DOI] [PubMed] [Google Scholar]
- 40.Kull, F. C. J., Jacobs, S., Su, Y.-F., Svoboda, M. E., Van Wyk, J. J., and Cuatrecasas, P. (1983) J. Biol. Chem. 258 6561-6566 [PubMed] [Google Scholar]
- 41.Main, E. R., Lowe, A. R., Mochrie, S. G., Jackson, S. E., and Regan, L. (2005) Curr. Opin. Struct. Biol. 15 464-471 [DOI] [PubMed] [Google Scholar]
- 42.Wright, P. E., and Dyson, H. J. (1999) J. Mol. Biol. 293 321-331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.