Abstract
The cytoplasmic deacetylase HDAC6 is an important regulator of cellular pathways that include response to stress, protein folding, microtubule stability, and cell migration, thus representing an attractive target for cancer chemotherapy. However, little is known about its upstream regulation. Our previous work has implicated HDAC6 as a new protein target for the farnesyltransferase inhibitors (FTIs), although HDAC6 lacks a farnesylation motif. Here we show that the protein farnesyltransferase (FTase) and HDAC6 are present in a protein complex together with microtubules in vivo and in vitro. FTase binds microtubules directly via its α subunit, and this association requires the C terminus of tubulin. Treatment with an FTI removed FTase, but not HDAC6, from the protein complex, suggesting that the active form of FTase is bound to microtubules. Importantly, the removal of FTase from microtubules abrogated HDAC6 activity, as did a stable knockdown of the α subunit of FTase (FTαKD). Interestingly, the FTαKD cells showed increased sensitivity to the antiproliferative effects of Taxol and the FTI lonafarnib when used either as single agents or in combination as compared with parental cells. Altogether, these data suggest that FTase, via its tubulin-association, is a critical upstream regulator of HDAC6 activity and that FTase expression could help stratify cancer patients that would most benefit from this treatment.
The protein farnesyltransferase (FTase)4 is a prenylation enzyme that recognizes proteins with a COOH terminus CAAX motif and catalyzes the transfer of a 15-carbon farnesyl group from a farnesyl pyrophosphate to the C-terminal cysteine. FTase is an αβ heterodimer in which the α subunit is shared with another prenylation enzyme, the geranylgeranyltransferase I, whereas the β subunit is responsible for substrate specificity (for a review, see Ref. 1). Among the more than 50 proteins known to contain a farnesylation motif, Ras has been the most studied for its prevalence in malignant transformation. Based on evidence that Ras downstream signaling was dependent on Ras association with the plasma membrane through the addition of a farnesyl group, inhibitors of the protein farnesyltransferase (FTIs) were developed for cancer chemotherapy (for a review, see Ref. 2). Unexpectedly, it turned out that K-Ras and N-Ras could be alternatively geranylgeranylated and thus reduced the anticipated clinical activity of FTIs. Moreover, FTIs were shown to retain antitumor properties in cells where K-Ras remained anchored at the plasma membrane, suggesting that other proteins in addition to Ras were affected by FTI treatment, contributing to the overall antitumor activity of this class of drugs. Another problem that has hindered the clinical development of FTIs has been the poor understanding of the proteins that are regulated by FTase, thus, prohibiting the identification of tumor types and/or individual cancer patients more likely to benefit from this type of treatment.
Interestingly, the combination of FTIs with taxanes, among most other classes of cancer chemotherapeutics, was shown to be very effective in preclinical cancer cell models as well as in early clinical trials (3–6). However, the mechanism behind the FTI/taxane synergy remains yet to be elucidated. We have recently shown that the FTI lonafarnib (LNF) inhibited the tubulin deacetylase function of HDAC6, thus increasing tubulin acetylation and stability and facilitating Taxol binding to the stabilized microtubule, which is the preferred substrate for taxanes (7, 8). Although these results implicate HDAC6 as a new protein target for the FTIs, the exact mechanism of FTI-induced HDAC6 inhibition remains to be understood.
HDAC6 is a Class IIb histone deacetylase (HDAC), which, unlike most other histone deacetylases, is mainly cytoplasmic and targets nonhistone substrates, with tubulin being the first identified target protein (9–11). To date, HDAC6 has been shown to regulate the acetylation and activity of several other proteins, such as the protein chaperone hsp90 (12, 13), the actin-binding protein cortactin (14), β-catenin (15), and the peroxiredoxins I and II (16). Apart from its deacetylase activity, HDAC6 also plays an important role in the aggresome formation, through its binding and transport of polyubiquinated misfolded proteins (17), and localizes to stress granules upon exposure of cells to environmental stress (18). Despite the wealth of emerging data on HDAC6 target proteins, only little is known about the upstream proteins/pathways that regulate HDAC6 activity. HDAC6 was shown to be phosphorylated and activated by the Aurora-A kinase in the cilia (19) and to be involved in the transforming growth factor-β-induced epithelial to mesenchymal transition (20). Interestingly, although HDAC6-null mice are viable and do not appear to have any developmental defect, HDAC6 was recently found to be required for Ras-induced oncogenic tumorigenesis (21). Thus, HDAC6 represents an attractive target for cancer chemotherapy, and a better understanding of its cellular regulation is needed for the development of effective targeted HDAC6 therapies.
In the present study, we report that the protein FTase is an important upstream regulator of HDAC6 activity. We identify FTase in a protein complex with microtubules and HDAC6, and we show that removal of FTase from this tripartite complex abrogates HDAC6 activity. FTase does not bind HDAC6 directly but regulates its activity via the binding of each respective protein to microtubules. The binding of FTase to microtubules is direct and mediated by the α subunit of FTase and the C terminus of tubulin. Stable knockdown of FTase-α results in HDAC6 inactivation, phenocopying the pharmacological inhibition of FTase. Additionally, FTase-α knockdown sensitized cells to Taxol or FTI treatment, further suggesting that FTase expression and activity are important determinants of HDAC6 activity and HDAC6 target proteins. These results provide new insight into the cellular regulation of HDAC6 with important clinical implications.
EXPERIMENTAL PROCEDURES
Materials, Cell Lines, and Plasmids—A549 and HEK293 cell lines were obtained from the ATCC. MCF7:GFP-tubulin cells were a gift from Mary Ann-Jordan (University of California, Santa Barbara, CA) (22). All cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. Paclitaxel, vinblastine, colchicine, and trichostatin A (TSA) were purchased from Sigma. LNF, tipifarnib, and FTI-277 were from Schering-Plough, Johnson & Johnson, and Calbiochem, respectively. FLAG-tagged HDAC6 (WT and mutant) and HA-tagged FTase-β (WT and Y361L mutant) expression plasmids were described previously (23, 24). Antibodies against HDJ-2 were from Labvision NeoMarkers (Fremont, CA), and HDAC6 and β-tubulin were from Cell Signaling. Mouse monoclonal antibodies against FTase-α and FTase-β, H-Ras, and HIF-1α were from BD Biosciences. Rabbit polyclonal antibodies against FTase-α and FTase-β were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies against acetylated α-tubulin, HA, glutathione S-transferase (GST), and FLAG were from Sigma. Rabbit polyclonal antibody against MBP was from New England Biolabs.
In Vitro Tubulin Deacetylation Assay—The tubulin deacetylation assay was carried out essentially as described (9). Briefly, microtubule-associated protein (MAP)-enriched tubulin (cytoskeleton) was preformed into microtubules in the absence of paclitaxel or glycerol by incubation for 30 min at 35 °C. Preformed microtubules were incubated at 37 °C for 2 h with either FLAG-HDAC6 immunoprecipitates or affinity-purified His-HDAC6. Samples were then placed on ice for 15 min. The supernatant was collected by centrifugation and analyzed by immunoblotting with antibodies against acetylated α-tubulin and total α-tubulin, respectively.
Immunoprecipitation and Immunoblotting—Cells were lysed in a buffer containing 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, and 0.5% Nonidet P-40 with the protease inhibitor mixture (Roche Applied Science). The whole cell lysate (500 μg) was incubated with preformed protein A/G-Sepharose bead-antibody mixtures or anti-FLAG M2-agarose beads (Sigma) at 4 °C for 2 h. Samples were washed four times with the lysis buffer, resolved by SDS-PAGE, and immunoblotted as described previously (25). 20 μg of total cell lysate was loaded as input control.
In Vitro Protein Binding Assays—For the microtubule-dependent in vitro interaction of HDAC6 and FTase, purified FTase (containing both GST-FTβ and MBP-FTα) was bound through its GST tag and is thus referred to as GST-FT. To assess the role of each FTase subunit in this interaction, GST, GST-tagged FTase-β (GST-FTβ), MBP, and MBP-tagged FTase-α (MBP-FTα) were expressed in E. coli and purified using glutathione-agarose (for the GST tag) or amylose-agarose (for the MBP tag) beads, respectively, as described (23). His-tagged HDAC6 was produced using the baculoviral system and affinity-purified as described (10). MAP-free tubulin (cytoskeleton) was incubated with 20 μm paclitaxel at 35 °C for 30 min to preform microtubules. For the in vitro binding assay, GST alone and GST-FTβ immobilized on glutathione-agarose beads or MBP alone and MBP-FTα immobilized on amylose beads were incubated at 35 °C for 2 h with either preformed microtubules or nonpolymerized MAP-free tubulin (cytoskeleton) in the presence or absence of purified His-HDAC6. The final concentration for each protein was 5 μm. The pellets (beads) were collected by centrifugation, washed three times with PBS buffer containing 300 mm NaCl, and then subjected to SDS-PAGE and immunoblot. For a subset of experiments, the C terminus of α- and β-tubulin in preformed microtubules was removed by extensive digestion with subtilisin as described (26). The total protein bound to the beads (B) and one-fifth of the total unbound supernatant (U) were loaded on an SDS-polyacrylamide gel.
Farnesylation Assay—Farnesylation assays were performed in a buffer containing 50 mm Tris-HCl (pH 7.4), 10 mm MgCl2, 5 μm ZnCl2, and 5 mm dithiothreitol, as described previously (23). The substrates used were GST-fused with short peptides containing C-terminal CAAX motifs, and the prenyl substrates used were [3H]farnesyl pyrophosphate (22.5 Ci/mmol; 1 Ci = 37 GBq). The concentration of the wild type and Y361L mutant GST-FTβ was optimized and was ∼100 nm for each assay.
Stable Knockdown of FTase-α Using shRNA Lentiviral Vectors—Knockdown of human FTase-α (accession number NM_002027) in A549 cells was achieved using shRNA lentiviral vectors following instructions from the manufacturer (OpenBiosystems). Briefly, HEK293 cells were transfected with VSV-G, packaging (both plasmids from Addgene), and FTase-α shRNA plasmids to produce shRNA lentiviral particles. Five different FTase-α shRNAs were tested (TRCN34584, TRCN34585, TRCN34586, TRCN34587, and TRCN34588). Viral supernatants from single shRNA transfections or a pool of all five shRNAs transfected together were used to transduce A549 cells, and selection of FTase-α KD cells was achieved using puromycin. Only lentiviral constructs derived from pooled shRNAs and single shRNAs TRCN34584 and TRCN34585 yielded puromycin-resistant A549 clones. FTase-α knockdown was verified by reverse transcription-PCR using primers described elsewhere (27) or real time quantitative reverse transcription-PCR, as described in Fig. S2A.
Drug Sensitivity Assay—Cytotoxicity assays using the protein-staining sulforhodamine B method were performed in 96-well plates, as described previously (28).
RESULTS
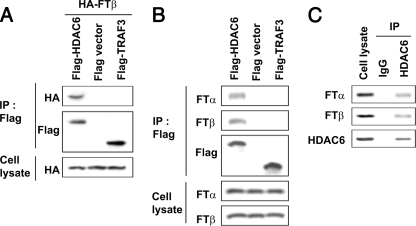
HDAC6 Interacts with FTase—We have previously shown that FTIs increased tubulin acetylation and microtubule stability by inactivating the tubulin deacetylase HDAC6. Although this finding provided a molecular explanation for the reported synergy between FTIs and taxanes, known to preferentially bind stable microtubules, it did not explain how an inhibitor of protein farnesylation could affect HDAC6 and tubulin, given that neither protein contains a CAAX farnesylation motif. To explore the functional relationship between FTase and HDAC6, we first examined whether the two proteins interact with each other. To do so, we co-transfected plasmids encoding either HA-FTase-β or FLAG-HDAC6 constructs in HEK293 cells, immunoprecipitated HDAC6 with an anti-FLAG antibody, and assessed for the presence of FTase using an anti-HA antibody (Fig. 1A). HA-FTase-β was only immunoprecipitated by FLAG-HDAC6 but not by a control vector or an irrelevant FLAG-tagged protein like Traf3, a protein involved in the tumor necrosis factor receptor signaling. Furthermore, exogenously expressed HDAC6 was able to co-precipitate endogenous FTase in the A549 lung cancer cell model previously used in our studies of the FTI/Taxane synergy (7) (Fig. 1B). Finally, the FTase-HDAC6 interaction was also observed with the endogenous proteins in A549 cells (Fig. 1C). Collectively, these results show that there is a physical association between HDAC6 and FTase in vivo.
FIGURE 1.
HDAC6 interacts with FTase. A, exogenous HDAC6 interacts with exogenous FTase. HEK293 cells were transfected with different FLAG-tagged plasmids (empty vector, WT HDAC6, or TRAF3) and HA-FTβ plasmid. Cell lysates (500 μg) were immunoprecipitated (IP) with an anti-FLAG antibody and immunoblotted (IB) as indicated. B, exogenous HDAC6 interacts with endogenous FTase-α and FTase-β. A549 cells were transfected with different FLAG-tagged constructs (HDAC6, vector, or TRAF3), immunoprecipitated as in A, and immunoblotted for endogenous FTase subunits, as indicated. C, endogenous HDAC6 interacts with endogenous FTase. A549 cells were immunoprecipitated with anti-HDAC6 or IgG control and immunoblotted as indicated.
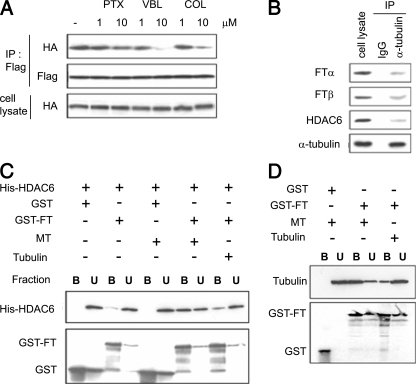
Microtubules Are Required for the FTase-HDAC6 Interaction—Our data revealed that all classes of FTIs when combined with Taxol synergistically enhance tubulin acetylation (Fig. S1), thereby implicating microtubule function in their mechanism of synergy. To assess the role of the microtubule cytoskeleton in the newly identified HDAC6-FTase complex (Fig. 1) we treated HEK-293 cells co-transfected with HA-FTase-β and FLAG-HDAC6 with either Taxol (microtubule stabilizer) or the tubulin-depolymerizing drugs vinblastine and colchicine. The interaction between HDAC6 and FTase was then evaluated by co-immunoprecipitation. Treatment with vinblastine or colchicine (Fig. 2A) inhibited the HDAC6-FTase interaction only at concentrations (10 μm) that induced robust microtubule depolymerization (data not shown). Treatment with paclitaxel, on the other hand, did not affect the HDAC6-FTase protein complex, despite the ability of the drug to polymerize cellular microtubules (data not shown). Taken together, these results suggest that the presence of microtubule polymers in cells is required for the physical association between these two proteins. To examine whether tubulin is part of the HDAC6-FTase complex, we immunoprecipitated endogenous tubulin from A549 cells and observed that the two FTase subunits, α and β, together with HDAC6 were present in the tubulin immunoprecipitates (Fig. 2B).
FIGURE 2.
Microtubules are required for the HDAC6-FTase interaction. A, effects of microtubule inhibitors on the HDAC6-FTase interaction. HEK293 cells were transfected as in Fig. 1A, treated with 10 μm Taxol (PTX), vinblastine (VBL), or colchicine (COL) for 6 h at 37°C. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibodies and immunoblotted as indicated. B, endogenous α-tubulin interacts with endogenous FTase and HDAC6. A549 cells were immunoprecipitated with anti-α-tubulin or IgG and immunoblotted as indicated. C, role of microtubules and soluble tubulin dimers on the HDAC6-FTase interaction in vitro. Purified His-HDAC6 was incubated with purified GST or GST-FTase immobilized on glutathione-agarose beads in the presence of preformed Taxol-stabilized, MAP-free microtubules (MT), or MAP-free nonpolymerized tubulin (Tubulin). The final concentration for each protein was 5 μm. Samples were centrifuged, and proteins bound to the GST-FTase beads (B) or unbound fractions (U) were detected by immunoblotting. D, FTase interacts directly with microtubules in vitro. MAP-free Taxol-stabilized preformed microtubules or nonpolymerized soluble tubulin were incubated with purified GST or GST-FTase immobilized on glutathione-agarose beads. Samples were centrifuged, and proteins bound to the beads or present in the unbound fraction were detected by immunoblotting.
Based on these results, we determined and assessed whether the interaction between HDAC6 and FTase is direct or mediated by microtubules. An in vitro GST pull-down assay was performed in which purified GST-FTase immobilized on glutathione-agarose beads was incubated together with purified His-HDAC6 in the presence of purified microtubule polymers (MT) or tubulin dimers (Tubulin) (Fig. 2C). As seen in lane 3, HDAC6 was hardly detectable in the FTase pull-down in the absence of exogenous tubulin, indicating that HDAC6 is unable to interact directly with FTase in vitro. The addition of preformed MAP-free microtubule polymers (lane 7) allowed the binding of His-HDAC6 to GST-FTase. When soluble tubulin (lane 9) was added to the reaction, some binding of His-HDAC6 was observed, albeit to a lesser extent. These results clearly show that microtubule polymers, even in the absence of MAPs, are necessary and sufficient for the HDAC6-FTase interaction.
HDAC6 has been previously shown to bind microtubules directly (9–11). Here we show that HDAC6, in the absence of microtubule protein, does not bind FTase directly, suggesting that FTase itself probably interacts with microtubules. To investigate whether FTase binds tubulin or microtubules in vitro, we incubated purified GST-FTase with either preformed MAP-free microtubule polymers or MAP-free tubulin dimers. As seen in Fig. 2D, microtubules were specifically pulled down by GST-FTase but not by GST alone (lanes 3 and 1, respectively). By contrast, only a small amount of soluble tubulin was associated with GST-FTase (lane 5). These results thus indicated that FTase binds tubulin directly and interacts preferentially with microtubule polymers.
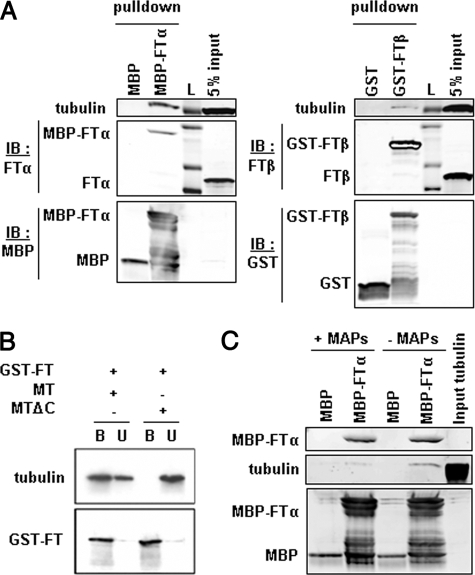
FTase Is a Direct Binding Partner for Microtubules via Its α Subunit—FTase is formed by the association of two subunits, α and β, both of which are required for the catalytic activity of the enzyme. In the experiments described thus far, we used both FTase subunits; therefore, we set out to determine which subunit mediates the association with microtubules. We performed pull-down experiments using either purified MBP-FTα or purified GST-FTβ in the presence of cell lysates from NIH3T3 or A549 cells (data not shown). As seen in Fig. 3A, tubulin was more readily pulled down by MBP-FTα than by GST-FTβ, suggesting that the FTase-α subunit mediates binding of FTase to microtubules. Although the GST or MBP tag used here could affect binding to tubulin, in a way that could impair the GST-FTβ binding, our previous results (Fig. 2) showing efficient binding of GST-tagged FTase-α and -β suggest that this is not the case.
FIGURE 3.
FTase interacts directly with microtubules via its α subunit. A, FTase binding to tubulin is mediated by the FTase-α subunit. Purified MBP-tagged FTα (MBP-FTα) or MBP alone immobilized on amylase beads and GST-tagged FTase-β (GST-FTβ) or GST alone immobilized on glutathione beads were incubated with cell lysates. Bound protein fractions were resolved by SDS-PAGE and immunoblotted (IB) for the presence of FTase subunits and tubulin. No tubulin was detected in control MBP or GST beads. L, protein ladder. B, deletion of the C terminus of tubulin abolishes the interaction between FTase and microtubules. The C terminus of α- and β-tubulin in preformed microtubules was removed by proteolysis with subtilisin (MTΔC). The in vitro interaction between FTase and intact (MT) or cleaved microtubules was evaluated as in A. C, MAPs interfere with the binding of tubulin to FTase. MBP control or MBP-FTα beads were incubated with preformed MAP-containing or MAP-free microtubule protein.
Importantly, subtilisin digestion of tubulin, which removes the C-terminal tails of α-and β-tubulin, completely abolished the interaction of FTase with microtubules (Fig. 3B), showing that the C-terminal tail of tubulin mediates the FTase binding. Intriguingly, this region of tubulin is also the binding site for known MAPs, such as MAP2. We then wondered whether the presence of MAPs would affect the binding of FTase to microtubules. To test this hypothesis, we performed a pull-down assay using MBP-FTase-α incubated with either purified preformed MAP-containing or MAP-free microtubules (Fig. 3C). Our results show that the presence of MAPs inhibited the binding of tubulin to MBP-FTase-α, suggesting that FTase might compete with MAPs for microtubule binding.
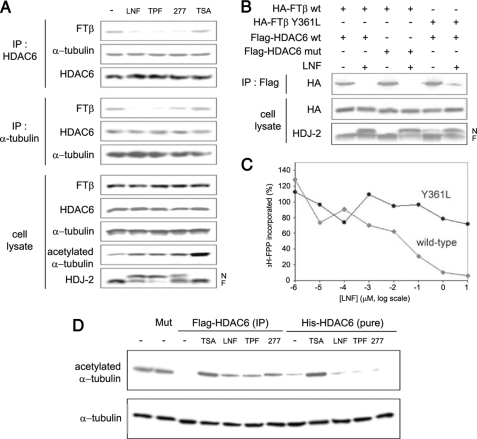
Effects of FTIs on the HDAC6-Microtubule-FTase Protein Complex and Activity—We next sought to investigate the effect of FTI treatment on the HDAC6-FTase-microtubule protein complex by co-immunoprecipitation assays (Fig. 4A). We treated A549 cells with different classes of FTIs, using concentrations that effectively inhibited the enzymatic activity of FTase, as evidenced by the inhibition of HDJ-2 farnesylation, and observed that drug treatment inhibited the HDAC6-FTase as well as the FTase-microtubule interactions, whereas it had no effect on the HDAC6-tubulin association. Notably, FTIs also inhibited the HDAC6 tubulin deacetylase function (increased tubulin acetylation), in agreement with our previously published data (7). On the other hand, pharmacological inhibition of HDAC6 by the pan-histone deacetylase inhibitor TSA had no effect on the formation of the triprotein complex nor on FTase activity (lack of inhibition of HDJ-2 farnesylation), whereas it did inhibit the tubulin deacetylase activity of HDAC6, as expected. These results suggest that FTase, via its microtubule association, is involved in the regulation of HDAC6 activity and that FTI-mediated inhibition of HDAC6 may result from the disruption of the FTase-HDAC6 protein complex.
FIGURE 4.
Effects of FTIs on the HDAC6-microtubule-FTase complex and activity. A, FTIs abrogate the FTase-HDAC6 interaction and the FTase-microtubule (MT) interaction but not the HDAC6-microtubule interaction. A549 cells were treated with 10 μm FTIs or 0.5 μm TSA for 16 h at 37 °C. Cell lysates were immunoprecipitated (IP) with anti-HDAC6 or anti-α-tubulin antibodies and immunoblotted as indicated. Tubulin acetylation and HDJ-2 farnesylation were examined to show the activity of TSA and FTIs, respectively. N, nonfarnesylated HDJ-2; F, farnesylated HDJ-2. B, interactions between WT and Y361L mutant FTase with WT and H216A/H611A mutant HDAC6 and the effect of LNF on these interactions. HEK293 cells were transfected with various plasmid combinations, treated or untreated with 10 μm LNF for 16 h at 37 °C, immunoprecipitated, and immunoblotted as indicated. C, LNF inhibits the activity of wild type but not the Y361L mutant FTβ. D, effects of FTIs on the tubulin deacetylase activity of HDAC6 in vitro. Preformed MAP-enriched microtubules that are heavily acetylated (lane 1) were incubated with FLAG-HDAC6 immunoprecipitates (IP) from A549 cells transfected with FLAG-HDAC6 or incubated with affinity-purified His-HDAC6 in the presence of various compounds for 2 h at 37°C. HDAC6 activity was evaluated by the decrease of acetylated tubulin examined by SDS-PAGE and immunoblotting. FLAG-HDAC6 (H216A/H611A) mutant IP (Mut) was included as a control. FTIs were used at 10 μm and TSA at 0.5 μm.
To examine whether the enzymatic activity of FTase or HDAC6 is required for their association, we examined the interactions of exogenously expressed HA-tagged WT or Y361L mutant FTase-β with FLAG-tagged WT or H216A/H611A catalytically inactive double mutant HDAC6 by co-immunoprecipitation (Fig. 4B). Our results show that the catalytic activity of HDAC6 is not required for the formation of the FTase-HDAC6 complex nor for its susceptibility to disruption by FTI (LNF) treatment. To assess the requirement for the FTase enzymatic activity, we used the Y361L FTase-β mutant, previously shown to retain 70% of FTase activity and to be resistant to various FTIs, including the tricyclic SCH44342 and SCH56582 as well as the peptidomimetic BMS193269 and B1088 (23). Here we show that the Y361L mutant FTase-β is also resistant to the effects of lonafarnib, as evidenced by the inability of LNF to decrease [3H]farnesyl diphosphate at concentrations that significantly inhibited the activity of WT FTase (Fig. 4C).
Our co-immunoprecipitation experiment showed that the Y361L FTase-β mutant protein retained its ability to associate with WT HDAC6 and remained resistant to the effects of LNF treatment, which was unable to inhibit the formation of the tripartite protein complex, consistent with its impaired ability to inhibit the mutant FTase in vitro (Fig. 4C). Taken together, our results show that the enzymatic activity of FTase is required for the enzyme's physical association with HDAC6, an interaction that, in turn, appears to regulate HDAC6 function (Fig. 4A).
So far, our data show that FTase activity is essential for the regulation of HDAC6 function, although HDAC6 is not a direct binding partner for FTase. To rule out the possibility that FTIs act directly on HDAC6, independently of FTase or other cellular proteins, we assessed the tubulin deacetylase function of HDAC6 in vitro by co-incubating purified MAP-enriched bovine brain tubulin with either purified His-HDAC6 protein or HDAC6 cellular immunoprecipitates in the presence of different classes of FTIs. The levels of tubulin acetylation, assessed by immunoblotting, were used as a read-out for HDAC6 activity (Fig. 4D). As expected, FLAG-HDAC6 immunoprecipitates caused a complete deacetylation of bovine brain tubulin (lane 3), which is normally heavily acetylated (lane 1). The catalytically inactive H216A/H611A mutant HDAC6 (Mut) was used as a negative control (lane 2). Pharmacologic inhibition of HDAC6 by the addition of TSA restored acetyl-tubulin levels, as expected (lane 4). The FTIs also increased tubulin acetylation (lanes 5–7), indicating that FTIs can inhibit HDAC6 tubulin deacetylase activity. Similarly, the addition of the affinity-purified His-HDAC6 protein resulted in complete tubulin deacetylation, which was readily reversed by TSA (lanes 8 and 9). However, the addition of FTIs had no effect on the activity of His-HDAC6 in this cell-free system (lanes 10–12), where HDAC6, tubulin, and FTIs were the only components present in the reaction. This last result demonstrates that FTIs do not act directly on HDAC6 and further suggests that FTase and perhaps other proteins present in the FLAG-HDAC6 immunocomplex are involved in the FTI-mediated inhibition of HDAC6 activity.
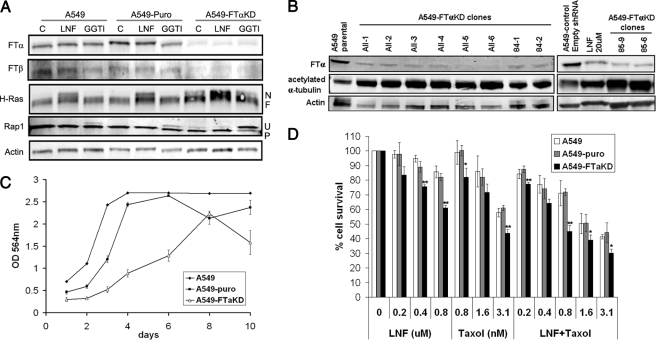
Knockdown of the α Subunit of FTase Results in Inhibition of HDAC6 Activity and Sensitizes Cells to Taxol—To further assess the role of FTase in the regulation of HDAC6 activity, we stably knocked down the tubulin-interacting α subunit of FTase in A549 cells (FTαKD). Quantitative reverse transcription-PCR studies confirmed a successful knockdown of FTase-α with only 20% of mRNA remaining when compared with parental or control shRNA A549 cells (Fig. S2A). Western blot analysis of cell lysates prepared from several clones derived from two single FTase-α shRNA target sequences (namely 84-1 and 84-2 for shRNA TRCN34584 and 85-6 and 85-9 for shRNA TRCN34585) and from a pool of five different FTase-α shRNAs (clones All-1 to -6) all show efficient decrease of FTase-α proteins (Fig. 5, A and B). We also observed reduced protein levels of the FTase-β subunit, in concordance with earlier studies showing that the α and β subunits of FTase are co-translationally associated and that expression of one subunit without the other destabilizes this subunit (29, 30). Inhibition of basal FTase activity in the FTαKD cells was detected by the appearance of the more slowly migrating unfarnesylated form of H-Ras, which, in the shRNA control cells (A549-puro), was apparent only after FTI treatment. Note that FTαKD cells remained sensitive to FTI treatment, as observed with further H-Ras inhibition, which can be explained in part due to the remaining 20% of FTase-α proteins.
FIGURE 5.
Effect of FTase-αKD on HDAC6 activity. A, Western blotting of cell lysates from parental A549, control (puro) and FTase-α (FTαKD) shRNA cell lines. Inhibition of FTase and GGTase activity is shown by the appearance of a nonfarnesylated (N) band for H-Ras or a nonprenylated (U) band for Rap1, respectively. Note the appearance of the nonfarnesylated and nonprenylated bands in untreated FTαKD cells, indicating basal level inhibition of protein farnesylation and geranylgeranylation, as opposed to their appearance in treated-only parental and control cells. B, effect of FTase-αKD on the tubulin deacetylase activity of HDAC6. Note the increase in basal levels of tubulin acetylation concomitant with decrease of FTase-α proteins in several of the FTαKD clones obtained form pooled shRNAs (clones All) and from single shRNAs (84-1 and 84-2 from shRNA TRCN34584 and 85-6 and 85-9 from shRNA TRCN34585) compared with the A549 parental or puro control cells. C, knockdown of FTase-α in cells results in slower cell growth evaluated by sulforhodamine B staining and OD measurement at 564 nm. D, increased sensitivity of A549-FTαKD cells to Taxol, FTI, and their combination. Cells were treated with different concentrations of LNF, Taxol, or their combination for 72 h. Cell cytotoxicity was then evaluated by sulforhodamine B staining, and cell survival was determined as the percentage of cells remaining after treatment. Student's t test was performed, and results are labeled as p < 0.05 (*) and p < 0.005 (**).
The effect of FTase-α knockdown on HDAC6 activity was then analyzed by looking at the acetylation status of the HDAC6 substrate tubulin. We observed a robust increase in the basal acetyl-tubulin levels in the A549-FTαKD cells, mimicking or even exceeding the FTI-induced increase in tubulin acetylation previously seen with A549 cells (Fig. S1). To ensure that this result is not clonal but truly reflective of FTase biology, we tested several stable FTαKD clones and found that all of them had increased acetyl-tubulin levels as compared with the A549-puro control cells (C1) (Fig. 5B). Thus, the FTαKD in A549 cells appears to phenocopy the effects of FTI treatment regarding the regulation of HDAC6 activity.
Given the synergistic interaction between FTIs and taxanes reported previously, we set out to investigate whether FTα knockdown would translate into enhanced cell sensitivity to taxane treatment. Therefore, we determined the sensitivity of parental and FTαKD cells to treatment with Taxol, LNF, or their combination in a 72-h cytotoxicity assay. Interestingly, the FTαKD cells were more sensitive to the effects of Taxol alone, LNF treatment alone, and even more so to the FTI/Taxol combination, as compared with A549 parental and A549-puro cells (Fig. 5D). The profound cell sensitization is even more striking, considering that the FTαKD cells have a slower doubling time compared with A549 and A549-puro cells (Fig. 5C), a condition that should render the cells resistant to Taxol, known to target preferentially actively dividing cells. Altogether, the results obtained with the FTαKD cells confirm that FTase is an upstream regulator of HDAC6 activity.
DISCUSSION
Originally described as a histone deacetylase (24), HDAC6 was later found to deacetylate several non-histone cytoplasmic substrates, such as tubulin, hsp90, cortactin, β-catenin, and the peroxiredoxins I and II (9–11, 13–16). Deacetylation of hsp90 was further shown to play a role in the folding of the glucocorticoid receptor and HIF-1α (hypoxia-inducible factor 1), both of which represent additional indirect targets of HDAC6 (13, 31–33). Interestingly, HDAC6 knock-out mice are viable without any apparent developmental defects (34); however, HDAC6-null mice are more resistant to transformation and tumor growth than control mice, suggesting a role for HDAC6 in tumor development (21). Taken together, these results make HDAC6 a prime target for cancer chemotherapy. However, despite the wealth of new emerging data on cellular targets and pathways regulated by HDAC6, very little is known about the upstream factors that modulate HDAC6 activity per se (19, 20).
In this study, we identify one such upstream regulator, namely the protein farnesyltransferase. We show that HDAC6 is in a protein complex with tubulin and FTase in vitro and in cells (Figs. 1 and 2). We also show that treatment with an FTI, an anticancer agent in clinical development, physically removes FTase from the tripartite protein complex and inhibits HDAC6 activity (Fig. 4). Additional support for a functional relationship between HDAC6 and FTase came from a COMPARE analysis that we performed using the NCI panel of 60 cancer cell lines (35), which showed that FTase expression was inversely correlated with acetylated-tubulin protein levels (Fig. S3). Furthermore, stable cellular knockdown of FTase-α resulted in a robust increase in basal levels of tubulin acetylation (Fig. 5B), phenocopying the pharmacological inhibition of FTase, as we have previously shown (7, 8) (Fig. S1). In addition to tubulin, the FTαKD cells exhibited impaired HDAC6-mediated HIF-1 α protein degradation (Fig. S2C), further supporting the compromised activity of HDAC6 in the cellular context of FTαse knockdown. Altogether, these results emphasize the importance of FTase in the regulation of HDAC6 activity.
Intriguingly, since HDAC6 does not possess a CAAX farnesylation motif, it does not belong to the family of “classic” FTase target proteins. Our own results showing that the in vitro tubulin deacetylation activity of HDAC6 immunoprecipitates, but not of purified His-HDAC6 protein, is inhibited upon FTI treatment (Fig. 4D) further suggest that HDAC6 is not a direct substrate of FTase but that additional proteins, potentially farnesylated, are present in the immunocomplex and could mediate the FTase-dependent regulation of HDAC6. Conversely, we have no evidence for a feedback regulation of FTase by HDAC6, since neither the catalytic mutant HDAC6 nor its pharmacological inhibitor TSA disrupted the FTase-HDAC6-tubulin complex or affected protein farnesylation (Fig. 4A).
Our data clearly show that FTase regulates HDAC6 through direct binding to C terminus of microtubule polymers, a known MAP-binding site (Figs. 2 and 3). MAPs, through their microtubule binding, regulate endogenous microtubule dynamics and affect the stability of the polymer. Our results showing that FTase competes with MAPs for microtubule binding together with the fact that neither tubulin nor MAPs possesses a farnesylation motif suggest a role for FTase in the regulation of microtubule dynamics. Such a role for FTase would be compatible with our previous data showing that the FTI lonafarnib suppresses microtubule dynamics in cells (7), whereas lonafarnib alone, in vitro, does not bind tubulin; nor does it affect microtubule polymer mass (36). In addition, our data showing that the active form of the enzyme (both FTase-α and -β subunits) binds tubulin lend further support to the role of FTase in the regulation of HDAC6 and the chemomechanics of the microtubule cytoskeleton.
In that light, one would expect that either pharmacologic inhibition or knockdown of FTase would have an impact on the activity of drugs that target microtubules. We and others have extensively demonstrated the synergistic interaction of FTIs with the microtubule-stabilizing taxanes both in cells and in vivo. Our data herein also show that FTαKD sensitizes cancer cells to the antiproliferative effects of Taxol (Fig. 5C), further implicating FTase expression and the downstream inhibition of HDAC6 activity in taxane sensitivity. These results raise the exciting possibility of enhanced synergy between an HDAC6 inhibitor and the already synergistic FTI/taxane combination. Finally, examination of the integrity of the FTase-HDAC6 axis in clinical samples could help stratify patients more likely to respond to taxanes alone or combined with an FTI.
Collectively, our results identify a novel signaling pathway in which HDAC6 is a downstream target of FTase with microtubules providing the dynamic scaffold for their interaction. Given the widespread use of microtubule inhibitors in clinical oncology and the clinical development of both FTase and HDAC6 inhibitors, our results have important clinical implications and should assist in the rational development of better anticancer therapies.
Supplementary Material
Acknowledgments
We thank Geri Kreitzer for critical reading of the manuscript and for fruitful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA100202 and CA114335 (to P. G.), 1F32CA134148-01 (to C. C. V.), and CA041996 (to F. T.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: FTase, farnesyltransferase; FTα and FTβ, FTase-α and -β, respectively; FTI, farnesyltransferase inhibitor; LNF, lonafarnib; TSA, trichostatin A; WT, wild type; HA, hemagglutinin; GST, glutathione S-transferase; shRNA, short hairpin RNA; KD, knock-down; MAP, microtubule-associated protein; MBP, maltose-binding protein.
References
- 1.Lane, K. T., and Beese, L. S. (2006) J. Lipid Res. 47 681–699 [DOI] [PubMed] [Google Scholar]
- 2.Basso, A. D., Kirschmeier, P., and Bishop, W. R. (2006) J. Lipid Res. 47 15–31 [DOI] [PubMed] [Google Scholar]
- 3.Adjei, A. A. (2003) Lung Cancer 41 Suppl. 1, S55–S62 [DOI] [PubMed] [Google Scholar]
- 4.Kim, E. S., Kies, M. S., Fossella, F. V., Glisson, B. S., Zaknoen, S., Statkevich, P., Munden, R. F., Summey, C., Pisters, K. M., Papadimitrakopoulou, V., Tighiouart, M., Rogatko, A., and Khuri, F. R. (2005) Cancer 104 561–569 [DOI] [PubMed] [Google Scholar]
- 5.Moasser, M. M., Sepp-Lorenzino, L., Kohl, N. E., Oliff, A., Balog, A., Su, D. S., Danishefsky, S. J., and Rosen, N. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 1369–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun, J., Blaskovich, M. A., Knowles, D., Qian, Y., Ohkanda, J., Bailey, R. D., Hamilton, A. D., and Sebti, S. M. (1999) Cancer Res. 59 4919–4926 [PubMed] [Google Scholar]
- 7.Marcus, A. I., Zhou, J., O'Brate, A., Hamel, E., Wong, J., Nivens, M., El-Naggar, A., Yao, T. P., Khuri, F. R., and Giannakakou, P. (2005) Cancer Res. 65 3883–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcus, A. I., O'Brate, A. M., Buey, R. M., Zhou, J., Thomas, S., Khuri, F. R., Andreu, J. M., Diaz, F., and Giannakakou, P. (2006) Cancer Res. 66 8838–8846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubbert, C., Guardiola, A., Shao, R., Kawaguchi, Y., Ito, A., Nixon, A., Yoshida, M., Wang, X. F., and Yao, T. P. (2002) Nature 417 455–458 [DOI] [PubMed] [Google Scholar]
- 10.Matsuyama, A., Shimazu, T., Sumida, Y., Saito, A., Yoshimatsu, Y., Seigneurin-Berny, D., Osada, H., Komatsu, Y., Nishino, N., Khochbin, S., Horinouchi, S., and Yoshida, M. (2002) EMBO J. 21 6820–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, Y., Li, N., Caron, C., Matthias, G., Hess, D., Khochbin, S., and Matthias, P. (2003) EMBO J. 22 1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bali, P., Pranpat, M., Bradner, J., Balasis, M., Fiskus, W., Guo, F., Rocha, K., Kumaraswamy, S., Boyapalle, S., Atadja, P., Seto, E., and Bhalla, K. (2005) J. Biol. Chem. 280 26729–26734 [DOI] [PubMed] [Google Scholar]
- 13.Kovacs, J. J., Murphy, P. J., Gaillard, S., Zhao, X., Wu, J. T., Nicchitta, C. V., Yoshida, M., Toft, D. O., Pratt, W. B., and Yao, T. P. (2005) Mol. Cell 18 601–607 [DOI] [PubMed] [Google Scholar]
- 14.Zhang, X., Yuan, Z., Zhang, Y., Yong, S., Salas-Burgos, A., Koomen, J., Olashaw, N., Parsons, J. T., Yang, X. J., Dent, S. R., Yao, T. P., Lane, W. S., and Seto, E. (2007) Mol. Cell 27 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Y., Zhang, X., Polakiewicz, R. D., Yao, T. P., and Comb, M. J. (2008) J. Biol. Chem. 283 12686–12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parmigiani, R. B., Xu, W. S., Venta-Perez, G., Erdjument-Bromage, H., Yaneva, M., Tempst, P., and Marks, P. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 9633–9638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaguchi, Y., Kovacs, J. J., McLaurin, A., Vance, J. M., Ito, A., and Yao, T. P. (2003) Cell 115 727–738 [DOI] [PubMed] [Google Scholar]
- 18.Kwon, S., Zhang, Y., and Matthias, P. (2007) Genes Dev. 21 3381–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pugacheva, E. N., Jablonski, S. A., Hartman, T. R., Henske, E. P., and Golemis, E. A. (2007) Cell 129 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan, B., Yao, T. P., Nguyen, H. T., Zhuo, Y., Levy, D. R., Klingsberg, R. C., Tao, H., Palmer, M. L., Holder, K. N., and Lasky, J. A. (2008) J. Biol. Chem. 283 21065–21073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, Y. S., Lim, K. H., Guo, X., Kawaguchi, Y., Gao, Y., Barrientos, T., Ordentlich, P., Wang, X. F., Counter, C. M., and Yao, T. P. (2008) Cancer Res. 68 7561–7569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamath, K., and Jordan, M. A. (2003) Cancer Res. 63 6026–6031 [PubMed] [Google Scholar]
- 23.Del Villar, K., Urano, J., Guo, L., and Tamanoi, F. (1999) J. Biol. Chem. 274 27010–27017 [DOI] [PubMed] [Google Scholar]
- 24.Grozinger, C. M., Hassig, C. A., and Schreiber, S. L. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, J., Gupta, K., Yao, J., Ye, K., Panda, D., Giannakakou, P., and Joshi, H. C. (2002) J. Biol. Chem. 277 39777–39785 [DOI] [PubMed] [Google Scholar]
- 26.Westermann, S., Avila-Sakar, A., Wang, H. W., Niederstrasser, H., Wong, J., Drubin, D. G., Nogales, E., and Barnes, G. (2005) Mol. Cell 17 277–290 [DOI] [PubMed] [Google Scholar]
- 27.Khan, S. G., Dummer, R., Siddiqui, J., Bickers, D. R., Agarwal, R., and Mukhtar, H. (1996) Biochem. Biophys. Res. Commun. 220 795–801 [DOI] [PubMed] [Google Scholar]
- 28.Giannakakou, P., Sackett, D. L., Kang, Y. K., Zhan, Z., Buters, J. T., Fojo, T., and Poruchynsky, M. S. (1997) J. Biol. Chem. 272 17118–17125 [DOI] [PubMed] [Google Scholar]
- 29.Chen, W. J., Andres, D. A., Goldstein, J. L., and Brown, M. S. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 11368–11372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, W. J., Andres, D. A., Goldstein, J. L., Russell, D. W., and Brown, M. S. (1991) Cell 66 327–334 [DOI] [PubMed] [Google Scholar]
- 31.Qian, D. Z., Kachhap, S. K., Collis, S. J., Verheul, H. M., Carducci, M. A., Atadja, P., and Pili, R. (2006) Cancer Res. 66 8814–8821 [DOI] [PubMed] [Google Scholar]
- 32.Kong, X., Lin, Z., Liang, D., Fath, D., Sang, N., and Caro, J. (2006) Mol. Cell Biol. 26 2019–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy, P. J., Morishima, Y., Kovacs, J. J., Yao, T. P., and Pratt, W. B. (2005) J. Biol. Chem. 280 33792–33799 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., Kwon, S., Yamaguchi, T., Cubizolles, F., Rousseaux, S., Kneissel, M., Cao, C., Li, N., Cheng, H. L., Chua, K., Lombard, D., Mizeracki, A., Matthias, G., Alt, F. W., Khochbin, S., and Matthias, P. (2008) Mol. Cell Biol. 28 1688–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoemaker, R. H. (2006) Nat. Rev. Cancer 6 813–823 [DOI] [PubMed] [Google Scholar]
- 36.Buey, R. M., Barasoain, I., Jackson, E., Meyer, A., Giannakakou, P., Paterson, I., Mooberry, S., Andreu, J. M., and Diaz, J. F. (2005) Chem. Biol. 12 1269–1279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.