Abstract
Smad proteins transduce the transforming growth factor-β (TGF-β) signal at the cell surface into gene regulation in the nucleus. Upon TGF-β treatment, the highly homologous Smad2 and Smad3 are phosphorylated by the TGF-β receptor at the SSXS motif in the C-terminal tail. Here we show that in addition to the C-tail, three (S/T)-P sites in the Smad3 linker region, Ser208, Ser204, and Thr179 are phosphorylated in response to TGF-β. The linker phosphorylation peaks at 1 h after TGF-β treatment, behind the peak of the C-tail phosphorylation. We provide evidence suggesting that the C-tail phosphorylation by the TGF-β receptor is necessary for the TGF-β-induced linker phosphorylation. Although the TGF-β receptor is necessary for the linker phosphorylation, the receptor itself does not phosphorylate these sites. We further show that ERK is not responsible for TGF-β-dependent phosphorylation of these three sites. We show that GSK3 accounts for TGF-β-inducible Ser204 phosphorylation. Flavopiridol, a pan-CDK inhibitor, abolishes TGF-β-induced phosphorylation of Thr179 and Ser208, suggesting that the CDK family is responsible for phosphorylation of Thr179 and Ser208 in response to TGF-β. Mutation of the linker phosphorylation sites to nonphosphorylatable residues increases the ability of Smad3 to activate a TGF-β/Smad-target gene as well as the growth-inhibitory function of Smad3. Thus, these observations suggest that TGF-β-induced phosphorylation of Smad3 linker sites inhibits its antiproliferative activity.
Transforming growth factor-β (TGF-β)2 and related factors regulate a wide variety of biological activities, such as cell proliferation, differentiation, migration, adhesion, apoptosis, angiogenesis, and immune function (1). Accordingly, TGF-β family members play an important role in early embryogenesis as well as in the homeostasis of adult tissues. Abnormalities in TGF-β signaling lead to a number of human diseases, such as cancer and fibrosis (2, 3).
TGF-β signals through two types of serine-threonine kinase receptors (2–8). TGF-β binds directly to the type II receptor, which leads to the recruitment of the type I receptor into the ligand-receptor complex. The type II receptor is constitutively active. It transphosphorylates and activates the type I receptor, which then plays a major role in specifying downstream signaling events (2–8).
Smad proteins transduce the TGF-β family signal at the cell surface into gene regulation in the nucleus (2–8). Smad2 and Smad3 are direct substrates of the TGF-β type I receptor (9–12), whereas Smad1, Smad5, and Smad8 are phosphorylated by bone morphogenetic protein receptor kinase (13, 14). These Smads are termed as receptor-regulated Smads (R-Smads). Upon TGF-β treatment, Smad2 and Smad3 are phosphorylated by the TGF-β type I receptor at the SSXS motif in their C-tails (11, 12), form complexes with Smad4, then together accumulate in the nucleus to regulate transcription of target genes (2–8).
Smads contain conserved N-terminal and C-terminal domains, also designated as MH1 and MH2 domains, respectively. In the middle, there is a proline-rich linker region that is divergent in sequence and length (2–8). The R-Smads linker regions contain demonstrated as well as suspected phosphorylation sites for proline-directed kinases, such as cyclin-dependent kinases, ERK MAP kinases, c-Jun N-terminal kinases, p38 MAP kinases, and GSK3, as well as for other kinases, such as Ca2+-calmodulin-dependent kinase II (15–44). The Smad3 linker region contains four proline-directed kinase phosphorylation sites: Thr179, Ser204, Ser208, and Ser213. Previous studies have shown that the Smad3 linker region can be phosphorylated by different kinases under different conditions. For example, CDK4 and CDK2 phosphorylate Smad3 at the Thr8 in the N-terminal domain and Thr179 and Ser213 in the linker region at the basal state (15). CDK phosphorylation of Smad3 inhibits its transcriptional activity and antiproliferative function (15). In response to EGF treatment, ERK phosphorylates Ser208, Ser204, and Thr179 in the Smad3 linker region and Ser208 is the best ERK site in Smad3 (15, 33). JNK and p38 can phosphorylate the Smad3 linker region in response to hepatocyte growth factor or TGF-β in certain cell types (35, 37, 38). In addition, we have shown that the linker region contains a transcriptional activation domain (45). An independent study has also shown that the linker region is necessary for Smad3 to activate transcription (46).
GSK3 is involved in insulin and Wnt signaling pathways (47). Recent studies indicate that GSK3 also regulates the signaling pathways of the TGF-β family members (39–42). GSK3 is phylogenetically most closely related to the CDKs, such as CDK1 and CDK2 (48). GSK3 refers to two isoforms: GSK3α and GSK3β. The two isoforms are encoded by different genes and share nearly identical sequences in their kinase domains. Outside of the kinase domain, their sequences differ substantially, but currently little is known about isoform-specific functions. Most of the studies were carried out with GSK3β.
We show in this report that TGF-β treatment induces rapid phosphorylation of Ser208, Ser204, and Thr179 in the Smad3 linker region. Although phosphorylation of Smad3 at the C-tail SSXS motif by the TGF-β type I receptor is necessary for the linker phosphorylation, the receptor itself does not phosphorylate the linker region. We also show that ERK is not responsible for phosphorylation of the linker region in the presence of TGF-β. We further show that the TGF-β-induced phosphorylation of Ser204 is dependent on GSK3. Flavopiridol, a chemical inhibitor that potently inhibits all members of the CDK family and also inhibits GSK3 (Refs. 49–56 and references therein), abolished TGF-β-induced phosphorylation of Thr179 and Ser208, suggesting that CDK family members are responsible for their phosphorylation. Mutation of Ser208 into nonphosphorylatable alanine abolished Ser204 phosphorylation in response to TGF-β, suggesting that Ser208 serves as the priming site for Ser204 phosphorylation. We further show that mutation of the linker phosphorylation sites increases the ability of Smad3 to activate a TGF-β/Smad target gene and to inhibit cell proliferation. Taken together, our observations indicate that TGF-β-induced phosphorylation of Smad3 linker sites inhibits Smad3 activity.
EXPERIMENTAL PROCEDURES
Cell Lines—The Mv1Lu mink lung epithelial cells, the HaCaT human keratinocytes, the HepG2 human hepatocellular carcinoma cells, and the 293 human kidney epithelial cells were maintained as previously described (15, 33). The L17 cell line is a derivative of the Mv1Lu cell line. The immortalized GSK3β–/– mouse embryonic fibroblasts (MEF) and the control wild type MEF were generously provided by Dr. James R. Woodgett (Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Canada) (57). Smad3+/– mice were generously provided by Dr. Xiao-Fan Wang (Duke University, Durham, NC) (58). Smad3+/– mice were mated with each other and Smad3–/– primary MEF were isolated as previously described (15, 58).
Smad3 Phosphopeptide Antibodies and Other Antibodies—The Smad3 phosphopeptide antibodies against Ser208, Ser204, Thr179, and Ser213 were affinity purified against the phosphopeptide antigen, and cross-absorbed against the unphosphorylated peptide of the same sequence. The specificities of these phosphate-specific antibodies have previously been demonstrated by immunoblotting, immunoprecipitation, phosphatase treatment, and confirming that the recognized band is Smad3 by comparing wild type versus Smad3-deficient cells (supplemental Fig. 4 in Ref. 15). In brief, each of the phosphopeptide antibodies recognizes only the wild type Smad3 but not the corresponding mutant Smad3 by immunoblotting; each of the phosphopeptide antibodies can recognize overexpressed wild type Smad3 but not the corresponding mutant form in an immunoprecipitation assay; treatment of the phosphorylated Smad3 with the λ phosphatase leads to the disappearance of the phosphorylated band; the band recognized by each of the phosphopeptide antibodies is Smad3, as none of these antibodies can detect a band that comigrates with Smad3 using cell extracts from Smad3 knock-out mouse embryonic fibroblasts (supplemental Fig. 4 in Ref. 15). The Smad3 (C-tail) phosphopeptide antibody was generously provided by Dr. Edward B. Leof (Mayo Clinic Cancer Center, Rochester, MN). The antibody against Smad3 was from Zymed Laboratories Inc. The ERK antibody was from Cell Signaling Solutions. The antibodies against phospho-ERK (pERK), CDK2, and CDK4 were purchased from Santa Cruz Biotechnology, Inc. The antibodies against phospho-JNK (pJNK) and phospho-p38 (pp38) were from Cell Signaling Technology. The HA antibody was from Roche Applied Science.
Ligands and Chemical Inhibitors—For treatment of Mv1Lu or HaCaT cells with TGF-β, 300 or 200 pm TGF-β was used unless otherwise indicated. The incubation time was 1 h unless otherwise indicated. For treatment with EGF, Mv1Lu cells were treated with EGF (50 ng/ml) for 15 min, at which time the phosphorylation of the three sites were maximally induced (33). For treatment with MEK1 inhibitors PD98059 or U0126, Mv1Lu cells were pretreated with PD98059 (final concentration of 50 μm) or U0126 (final concentration of 3 μm) for 1 h prior to the addition of EGF or TGF-β. For treatment with LiCl, Mv1Lu cells were incubated with LiCl (final concentration of 120 mm) for 1 h prior to the addition of TGF-β. For treatment with flavopiridol, Mv1Lu cells were incubated with flavopiridol (final concentration of 250 nm) for 8 h prior to the addition of TGF-β.
Immunoblotting—Immunoblotting was performed essentially as previously described (15, 33). In brief, cells were lysed in the TNE buffer (10 mm Tris-HCl, pH 7.8, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40) in the presence of protease and phosphatase inhibitors, and 30 μg of cell lysates were analyzed by immunoblotting. Ser(P)208, Ser(P)204, and Thr(P)179 antibodies were used at 0.15 μg/ml, and Ser(P)213 antibody was used at 0.6 μg/ml. The concentrations of other antibodies were used according to the manufacturers' instructions.
In Vitro Kinase Assay—TβRII and TβRI-HA or TβRI (KR)-HA were cotransfected into 293 cells. 40 h post-transfection, cells were treated with TGF-β for 1 h. Cells were harvested and lysed in TNE buffer in the presence of protease and phosphatase inhibitors. The cell lysates were then immunoprecipitated with HA antibody. The immunoprecipitates were used in a nonradioactive kinase reaction containing 20 mm HEPES, pH 7.4, 10 mm MgCl2, 0.5 mm ATP, and 0.5 μm GST-Smad3 in 30 μl at 30 °C for 30 min, 1 h, 2 h, or 3 h with similar results. Recombinant GSK3β was purchased from New England Biolab, and the in vitro kinase assay was carried out in a 30-μl reaction containing 20 mm Tris, pH 7.5, 10 mm MgCl2, 5 mm dithiothreitol, 0.5 mm ATP, 0.5 μm GST-Smad3, and 100 ng of GSK3β at 30 °C for 1 h. CDK2 and CDK4 were immunoprecipitated from endogenous Mv1Lu cells, and the kinase reactions were carried out in 30 μl containing 50 mm HEPES, pH 7.4, 15 mm MgCl2, 1 mm EGTA, 0.1% Tween 20, 1 mm dithiothreitol, 0.5 mm ATP, and 1 μm GST-Smad3 at 30 °C for 1 h. The kinase reactions were terminated by addition of SDS protein gel sample buffer. The reaction products were then analyzed by immunoblotting using Smad3 phosphopeptide antibodies.
Transfection and Reporter Gene Assay—HepG2 cells were transfected and analyzed for luciferase activities as previously described (45). In brief, HepG2 cells were seeded overnight in 60-mm dishes. Cells were transfected by DEAE-dextran (125 μg/ml) for 3 h. Cells were treated with TGF-β for 24 h and luciferase activities were then analyzed. The luciferase activities were normalized by the cotransfected Renilla luciferase control pRL-TK (Promega). The results represent the mean ± S.D. of four independent transfection experiments.
Retroviral Infection, Northern Blot Analysis, and [3H]Thymidine Incorporation Assay—Smad3 and each of the phosphorylation mutants were cloned into the pLZRSΔ-IRES-GFP retroviral vector, and the resulting retroviral plasmids were transfected into ecotropic phoenix packaging cells to produce retroviruses as previously described (15). Smad3–/– primary MEF (passage 3) were infected with the various retroviruses. Total RNA were isolated from the infected cells, and analyzed by Northern blot analysis as previously described (15). For the [3H]thymidine incorporation assay, the infected Smad3–/– MEF were seeded in 6-well plates for 24 h, then treated with or without 500 pm TGF-β for 24 h. During the last 4 h, 5 μCi of [3H]thymidine was added to the culture, and [3H]thymidine incorporation was analyzed as previously described (15). The results indicate the average and standard deviation of four independent experiments.
RESULTS
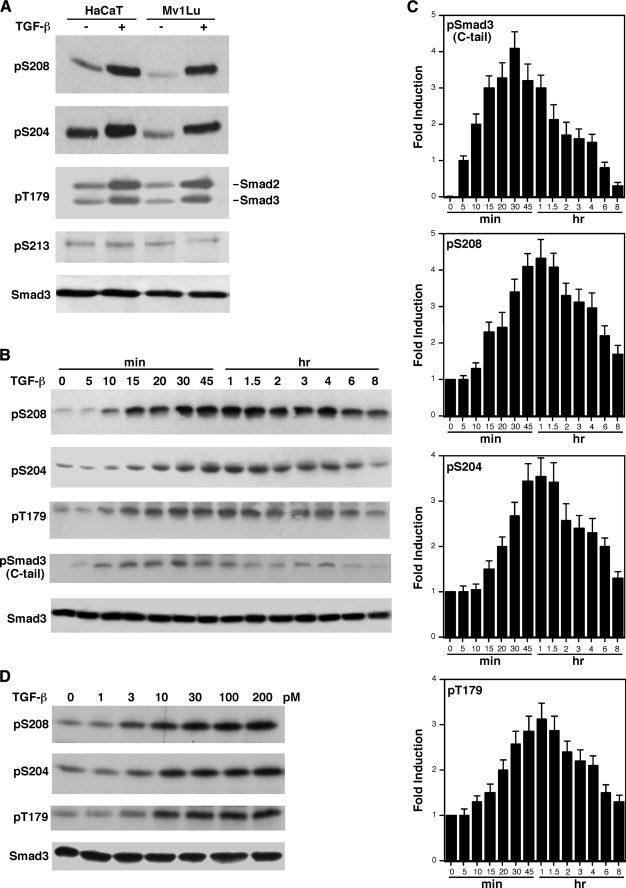
TGF-β Induces Rapid Phosphorylation of the Smad3 Linker Sites—The Smad3 linker region contains four (S/T)-P phosphorylation sites: Thr179, Ser204, Ser208, and Ser213. We have generated phosphopeptide antibodies against each of the four phosphorylation sites in Smad3 (15). The specificities of these phosphopeptide antibodies were previously demonstrated by immunoblotting, immunoprecipitation, phosphatase treatment, and confirming that the recognized band is Smad3 by comparing wild type versus Smad3-deficient cells (supplemental Fig. 4 in Ref. 15). The availability of these phosphate-specific antibodies allowed us to analyze whether the Smad3 linker phosphorylation is regulated by TGF-β treatment. Mv1Lu mink lung epithelial cells and HaCaT human keratinocytes are highly responsive to TGF-β. Mv1Lu and HaCaT cells were treated with TGF-β for 1 h. Cells were harvested and cell lysates were analyzed by immunoblotting with the Smad3 phosphopeptide antibodies. As shown in Fig. 1A, endogenous Smad3 phosphorylation levels at three linker sites Ser208, Ser204, and Thr179 were induced in response to TGF-β in both Mv1Lu and HaCaT cells. We could not detect induction of endogenous Ser213 phosphorylation in response to TGF-β in either cell types (Fig. 1A). The phosphopeptide antibodies against Ser(P)204, Ser(P)208, and Ser(P)213 recognize only the phosphorylated forms of Smad3 (Fig. 1A). The phosphopeptide antibody against Thr(P)179 in Smad3 also recognizes the analogous position in Smad2, which is also phosphorylated in response to TGF-β (Fig. 1A). The subsequent studies were focused on the Smad3 linker phosphorylation.
FIGURE 1.
TGF-β induces rapid phosphorylation of Smad3 linker sites. A, phosphorylation of Ser208, Ser204, and Thr179 in the Smad3 linker region is significantly increased by TGF-β. Mv1Lu and HaCaT cells were treated with 300 pm TGF-β for 1 h. Phosphorylation of Ser208, Ser204, Thr179, and Ser213 sites were detected by immunoblots using specific phosphopeptide antibodies. B, time course of TGF-β-inducible phosphorylation of Smad3. Mv1Lu cells were treated with 300 pm TGF-β for the indicated periods of time. Phosphorylation of Ser208, Ser204, and Thr179 as well as the C-tail were analyzed by specific phosphopeptide antibodies. Smad3 protein levels were also examined by immunoblot as a control. C, the average of four independent time course experiments from B. The error bars indicate the standard deviation. D, TGF-β dose curve for Ser208, Ser204, and Thr179 phosphorylation. Mv1Lu cells were treated for 1 h with the indicated concentrations of TGF-β. Phosphorylation of Ser208, Ser204, and Thr179 and Smad3 expression levels were analyzed by immunoblots.
The Smad3 C-tail Phosphorylation Precedes the Linker Phosphorylation—We analyzed the time courses of TGF-β-induced phosphorylation of Smad3 at the Ser208, Ser204, and Thr179 sites. For comparison, we also analyzed the Smad3 C-tail phosphorylation at the SSXS motif. Fig. 1B shows a representative time course for each of the phosphorylated sites in an experiment. Fig. 1C shows the average of four independent experiments. As shown in Fig. 1, B and C, the increases of phosphorylation at the Ser208, Ser204, and Thr179 sites can be detected at ∼10–15 min and peaks at 1 h after TGF-β treatment. The C-tail phosphorylation can be detected at 5 min and peaks at 30 min after TGF-β treatment. Thus, the phosphorylation of C-tail precedes the phosphorylation of the linker sites.
Low Doses of TGF-β Can Induce the Smad3 Linker Phosphorylation—To determine whether the Smad3 linker phosphorylation can be induced by physiological concentrations of TGF-β, we performed a TGF-β dose curve experiment. Mv1Lu cells were treated with different concentrations of TGF-β for 1 h. The phosphorylation levels of Ser208, Ser204, and Thr179 were then analyzed by immunoblots. As shown in Fig. 1D, as low as 10 pm TGF-β was sufficient for almost maximal induction of phosphorylation at the Ser208, Ser204, and Thr179 sites. Thus, these sites are fully expected to be phosphorylated at physiological concentrations of TGF-β in living organisms.
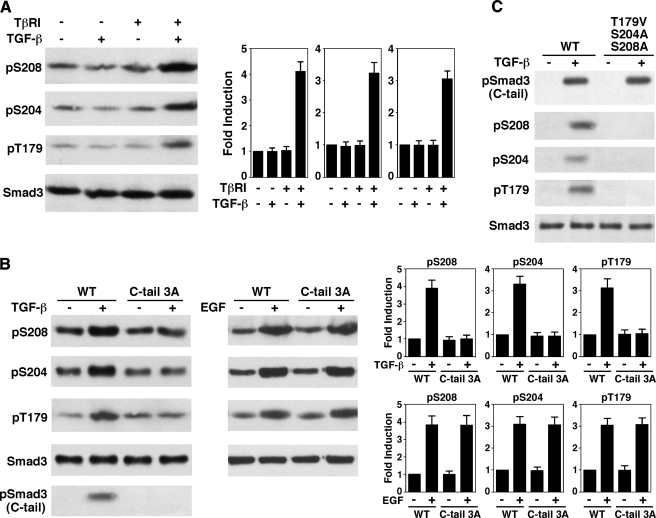
The Smad3 C-tail Phosphorylation May Be Necessary for the TGF-β-induced Linker Phosphorylation—Because TGF-β treatment leads to the induction of phosphorylation at the Ser208, Ser204, and Thr179 in the Smad3 linker region, we asked whether TβRI is required for the induction. As shown in Fig. 2A, TGF-β cannot induce Smad3 linker phosphorylation in L17 cells, a TβRI-deficient cell line derived from the parental Mv1Lu cell line. TGF-β-dependent linker phosphorylation was restored when TβRI was introduced into L17 cells. This result indicates that the linker phosphorylation is mediated by signaling events through the TGF-β receptors.
FIGURE 2.
The C-tail phosphorylation by the TGF-β receptor is necessary for TGF-β-induced phosphorylation of the Smad3 linker sites. A, TβRI is required for TGF-β-induced Smad3 linker phosphorylation. Mv1Lu-derived TβRI-deficient L17 cells were transfected with empty vector or TβRI. Cells were then treated with or without 300 pm TGF-β for 1 h. Phosphorylation of Ser208, Ser204, and Thr179 were analyzed by immunoblots. A representative experiment is shown. The averages of four independent experiments were plotted. The error bars indicate standard deviation. B, the Smad3 C-tail phosphorylation is necessary for TGF-β-induced linker phosphorylation. L17 cells were cotransfected with TβRI along with either wild type (WT) Smad3 or the C-tail 3A mutant. Cells were then treated with TGF-β for 1 h or with EGF for 15 min for maximal induction. Phosphorylation of Ser208, Ser204, and Thr179 were then analyzed by immunoblots. The bar graphs represent the averages of four independent experiments. The error bars indicate standard deviation. C, mutation of the Smad3 linker phosphorylation sites has essentially no effect on C-tail phosphorylation. L17 cells were cotransfected with TβRI along with either the wild type Smad3 or the linker phosphorylation mutant T179V/S204A/S208A. Cells were then treated with TGF-β for 1 h. Phosphorylation of the C-tail was examined by immunoblot.
We next compared the TGF-β-induced linker phosphorylation of wild type Smad3 with its C-tail phosphorylation mutant, Smad3 C-tail 3A. The Smad3 C-tail 3A mutant was generated by mutation of the three serine residues in the SSXS motif at the C-tail into alanines, thus abolishing receptor-mediated phosphorylation. As shown in Fig. 2B, phosphorylation of the linker sites was induced by TGF-β in wild type Smad3, whereas the induction was abolished in the C-tail 3A mutant. We have previously shown that Ser208, Ser204, and Thr179 are phosphorylated by ERK in response to EGF (33). To determine whether the inability of the C-tail 3A mutant to be phosphorylated in response to TGF-β was due to disruption of Smad3 structure, we asked whether the C-tail 3A mutant can be phosphorylated in the three linker sites in response to EGF. As shown in Fig. 2B, the C-tail 3A mutant can be phosphorylated at the three linker sites in response to EGF to the same extent as the wild type Smad3. Thus, these observations suggest that phosphorylation of the Smad3 C-tail is necessary for TGF-β-induced phosphorylation of the linker sites.
On the other hand, mutation of Ser208, Ser204, and Thr179 in Smad3 did not affect the C-tail phosphorylation (Fig. 2C). The control blots confirmed that Smad3 was not phosphorylated at the Ser208, Ser204, and Thr179 sites (Fig. 2C). These results suggest that the linker phosphorylation may not have a feedback role on the C-tail phosphorylation.
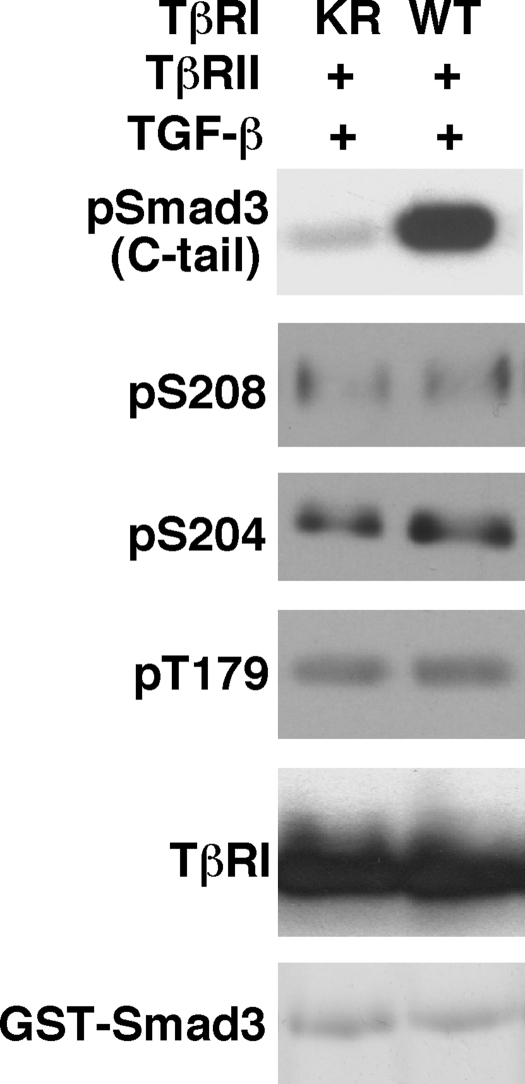
The TGF-β Type I Receptor Is Not the Kinase for Phosphorylation of the Linker Region—We next analyzed whether the TGF-β type I receptor phosphorylates the linker sites. 293 cells were cotransfected by TβRII and HA-tagged wild type TβRI or kinase-deficient TβRI (KR). Cells were then treated with TGF-β, and cell lysates were immunoprecipitated by the HA antibody. The immunoprecipitates were subjected to a nonradioactive in vitro kinase assay using GST-Smad3 as a substrate. The reaction products were then analyzed by immunoblot with Smad3 phosphopeptide antibodies. In addition, the reaction products were analyzed by immunoblot with HA antibody to verify that the TβRI (wild type) and TβRI (KR) were present at very similar levels. As shown in Fig. 3, wild type TβRI markedly phosphorylated the C-tail of GST-Smad3. In contrast, wild type TβRI was incapable of phosphorylating the linker sites of GST-Smad3. The background bands, which were detected after a long exposure time, were essentially at the same levels for TβRI (wild type) versus the kinase-deficient TβRI (KR). Immunoblot analysis confirmed that TβRI (wild type) and TβRI (KR) were present at the same levels (Fig. 3). GST-Smad3 was also present at the same levels as confirmed by Coomassie Blue staining (Fig. 3). Thus, TβRI does not phosphorylate the Smad3 linker sites.
FIGURE 3.
TβRI does not phosphorylate the Smad3 linker sites. 293 cells were cotransfected with TβRII and HA-tagged wild type TβRI or a kinase-deficient mutant TβRI (KR). Following TGF-β treatment, cells were harvested and cell lysates were immunoprecipitated by HA antibody. The immunoprecipitates were subjected to a nonradioactive in vitro kinase assay using GST-Smad3 as a substrate. The reactions were carried out at 30 °C for 30 min. Similar results were obtained when the reactions were carried out for 1, 2, or 3 h (data not shown). Phosphorylation at the Smad3 C-tail, Ser208, Ser204, and Thr179 were examined by immunoblots with specific phosphopeptide antibodies. The wild type TβRI and the mutant TβRI (KR) expression levels were analyzed by immunoblot with the HA antibody. Equal GST-Smad3 levels in the kinase reactions were confirmed by Coomassie Blue staining.
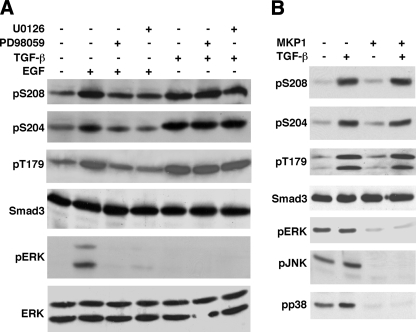
ERK Is Not Responsible for Phosphorylating the Linker in Response to TGF-β—As described above, Ser208, Ser204, and Thr179 are phosphorylated by ERK in response to EGF (15, 33) (Fig. 2B). We therefore asked whether ERK is the kinase that phosphorylates the same three sites in the presence of TGF-β. We performed a TGF-β time course to monitor the changes in ERK activity, which was analyzed by a phosphopeptide antibody that detects only the activated ERK, pERK. TGF-β induces a very rapid, transient, and modest activation of ERK in Mv1Lu cells. Upon TGF-β treatment, ERK activity is increased within 2-fold ∼5 min after TGF-β treatment. The ERK activity then rapidly declines to basal levels within minutes (data not shown). Our observations are consistent with a previous study on the time course of ERK activity in the presence of TGF-β in Mv1Lu cells (59). When TGF-β induces maximal induction of phosphorylation at Ser208, Ser204, and Thr179 sites at 1 h, ERK activity is essentially at the basal level (Fig. 4A, compare lane 1 with lane 5 for the pERK panel). Moreover, pretreatment of cells with MEK1 inhibitors PD98059 or U0126 for 1 h prior to addition of TGF-β has little effect on the TGF-β-induced phosphorylation of the three sites (Fig. 4A).
FIGURE 4.
ERK is not responsible for phosphorylation of the Smad3 linker sites in response to TGF-β. A, ERK is not the kinase that phosphorylates the Smad3 linker sites in the presence of TGF-β. Mv1Lu cells were pretreated with or without 3 μm U0126, 50 μm PD98059, or the dimethyl sulfoxide vehicle control for 1 h. Cells were then treated with 300 pm TGF-β for 1 h or 50 ng/ml EGF for 15 min. Phosphorylation of Ser208, Ser204, and Thr179 were analyzed by immunoblots with specific phosphopeptide antibodies. ERK activities were analyzed by the phospho-ERK (pERK) levels in the immunoblot. Smad3 and ERK expression levels were also analyzed by immunoblots as controls. B, MKP1 effectively inactivates ERK, JNK, and p38, but has little effect on the TGF-β-induced linker phosphorylation. L17 cells were transfected with TβRI in the absence or presence of the cotransfected MKP1, a MAP kinase phosphatase. Cells were treated with TGF-β for 1 h. Phosphorylation of Ser208, Ser204, and Thr179 were analyzed by immunoblots. ERK, JNK, and p38 activities were analyzed by pERK, pJNK, and pp38 levels, respectively. Smad3 expression levels were also examined as a control.
As positive controls, we have also prepared plates in parallel for treatment with EGF with or without pretreatment with a MEK1 inhibitor for 1 h. As shown in Fig. 4A, EGF treatment activated ERK (compare lane 1 with lane 2 for the pERK panel), which was inhibited by pretreatment with MEK1 inhibitors. EGF induced the phosphorylation at Ser208, Ser204, and Thr179, and the phosphorylation was inhibited by pretreatment with MEK1 inhibitors (Fig. 4A). Taken together, these results indicate that EGF activates ERK, which phosphorylates Ser208, Ser204, and Thr179. The TGF-β-induced phosphorylation of the same three sites, however, does not result from phosphorylation by ERK.
To further confirm that ERK is not responsible for the TGF-β-induced phosphorylation of the three sites, we transfected L17 cells with MKP1 (MAP kinase phosphatase 1), a specific MAP kinase superfamily phosphatase that inactivates ERK, JNK, and p38 by dephosphorylating them (60). The TGF-β type I receptor was also cotransfected. Cells were treated with or without TGF-β for 1 h, and then analyzed for Smad3 linker phosphorylation and ERK, JNK, and p38 activities by pERK, pJNK, and pp38 levels, respectively. As shown in Fig. 4B, MKP1 effectively dephosphorylated and thus inactivated ERK, JNK, and p38. TGF-β-induced linker phosphorylation, however, was not affected by the phosphatase. This result complements our conclusions above based on chemical inhibitors.
Although it is not the major points for Fig. 4B, it is worth pointing out why pERK, pJNK, and pp38 levels showed only modest or no induction by TGF-β when no MKP1 was cotransfected in lane 2 of Fig. 4B. These are all because TGF-β activates ERK, JNK, and p38 in a time-dependent manner. As described above, the activation of ERK by TGF-β is rapid and transient. The pERK level peaks at ∼5 min after TGF-β addition and rapidly declines, and is at the basal level after incubation for TGF-β for 1 h (59).3 The analysis of the Ser208, Ser204, and Thr179 phosphorylations was carried out after a 1-h TGF-β treatment. This is why the pERK level did not show induction by TGF-β in Fig. 4B. Similarly, TGF-β activates JNK rapidly. The pJNK level increases ∼5 min after TGF-β addition and peaks at ∼10 min (34, 35).3 pJNK level then quickly declines, and is only slightly higher than the basal level after treatment with TGF-β for 1 h (34, 35).3 This is why the pJNK level shows only a slight increase in response to TGF-β in Fig. 4B. For the p38, TGF-β activated by p38 can be detected 30 min after addition of TGF-β, and the pp38 level peaks at ∼2 h (35, 61).3 This is why the pp38 level shows only a modest increase in response to TGF-β in Fig. 4B.
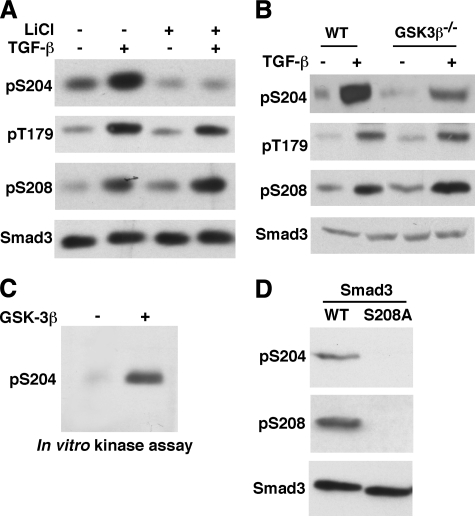
GSK3 Is Responsible for Ser204 Phosphorylation in Response to TGF-β—We employed a number of chemical inhibitors as one approach to identify the responsible kinases. Lithium ions inhibit a number of enzymes, but the only known kinase target is GSK3 (62). We pretreated Mv1Lu cells with LiCl or NaCl as a control before addition of TGF-β. As shown in Fig. 5A, among the three sites, only Ser204 phosphorylation was abolished by LiCl (Fig. 5A), suggesting that Ser204 is phosphorylated by GSK3.
FIGURE 5.
GSK3 is the kinase for TGF-β-induced phosphorylation of Ser204. A, LiCl selectively inhibits Ser204 phosphorylation. Mv1Lu cells were pretreated with 120 mm NaCl (control) or LiCl for 1 h before addition of TGF-β. The Smad3 linker phosphorylation was analyzed by phosphospecific antibodies. B, TGF-β-induced phosphorylation of Ser204 is selectively inhibited in GSK3β–/– MEF. GSK3β–/– MEF and the wild type (WT) littermate control MEF were treated with TGF-β for 1 h. Phosphorylation levels of the three linker sites in Smad3 were examined. C, GSK3β can phosphorylate the Ser204 site in vitro. 100 ng of recombinant GSK3β was used to phosphorylate 0.5 μm GST-Smad3 in a nonradioactive reaction. The reaction products were analyzed for Ser204 phosphorylation by immunoblotting with the Ser(P)204 antibody. D, Ser208 is the priming site for Ser204 phosphorylation. 293 cells were transfected with wild type Smad3 or the S208A mutant, together with the activated TβRI. Cell lysates were analyzed for Ser204 phosphorylation by the phosphospecific antibody.
GSK3 consists of GSK3α and GSK3β. Most GSK3 studies are focused on GSK3β. The GSK3β knock-out mouse has been generated (57). We analyzed Smad3 linker phosphorylation in GSK3β knock-out MEF and the wild type littermate control MEF. As shown in Fig. 5B, TGF-β-induced phosphorylation of Ser204 was strongly reduced in GSK3β knock-out MEF, consistent with LiCl data. To provide further evidence that GSK3 is the kinase for the Ser204 site, we show that recombinant GSK3β can phosphorylate the Ser204 site in an in vitro kinase assay (Fig. 5C).
GSK3 often requires priming phosphorylation of a substrate by other kinases (47). The priming site is located at the p + 4 position of a GSK3 site. Interestingly, the positioning of Ser208 and Ser204 exactly fits the p + 4 rule, because Ser208 is four amino acids downstream of Ser204. We therefore asked whether Ser208 phosphorylation is required for Ser204 phosphorylation. As shown in Fig. 5D, when Ser208 is mutated into alanine (S208A), the Ser204 site is no longer phosphorylated. This supports the notion that Ser208 is the priming site for Ser204 phosphorylation by GSK3.
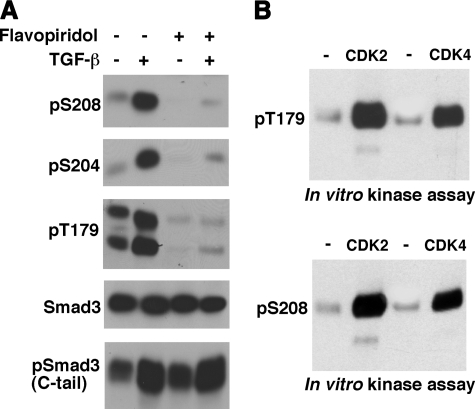
CDK Family Members Are Responsible for TGF-β-induced Phosphorylation of Thr179 and Ser208—A number of chemical inhibitors we examined have little or no effect on Thr179 or Ser208 phosphorylation, but flavopiridol is an exception. Flavopiridol potently inhibits all members of the CDK family (49–55). It also inhibits GSK3 (55, 56), as GSK3 is phylogenetically most closely related to the CDKs (48). As shown in Fig. 6A, treatment of Mv1Lu cells with flavopiridol strongly inhibited TGF-β-dependent phosphorylation of the three sites in the Smad3 linker. As expected, both CDK2 and CDK4 activities were potently inhibited by flavopiridol treatment (data not shown). As a control, the C-tail phosphorylation was little affected (Fig. 6A).
FIGURE 6.
The CDK family members mediate the TGF-β-induced linker phosphorylation. A, flavopiridol, a pan-CDK and GSK3 inhibitor, inhibits TGF-β-induced linker phosphorylation. Mv1Lu cells were treated with or without 250 nm flavopiridol for 8 h. Cells were then incubated with TGF-β for 1 h. Phosphorylation of Smad3 in the linker region was analyzed. The Smad3 C-tail phosphorylation was also analyzed as a control. B, both CDK2 and CDK4 can phosphorylate Thr179 and Ser208 in vitro. CDK2 or CDK4 immunoprecipitated from 240 μg of Mv1Lu cell lysates was used to phosphorylate 1 μm GST-Smad3 in a nonradioactive reaction. IgG was also used to immunoprecipitate the lysates and the immunoprecipitates were used in the control kinase reaction (in the – lane). The reaction products were analyzed by immunoblotting with Thr(P)179 or Ser(P)208 antibodies.
The Thr179 and Ser208 sites can be robustly phosphorylated by both CDK2 and CDK4 in an in vitro kinase assay (Fig. 6B), consistent with our previous studies (15). Ser204 can also be phosphorylated by both CDK2 and CDK4 in the in vitro kinase assay, but the potency is ∼5–10-fold less than that of Thr179 or Ser208 (Ref. 15 and data not shown). Taken together, these observations suggest that CDK family members are responsible for TGF-β-induced phosphorylation of Thr179 and Ser208.
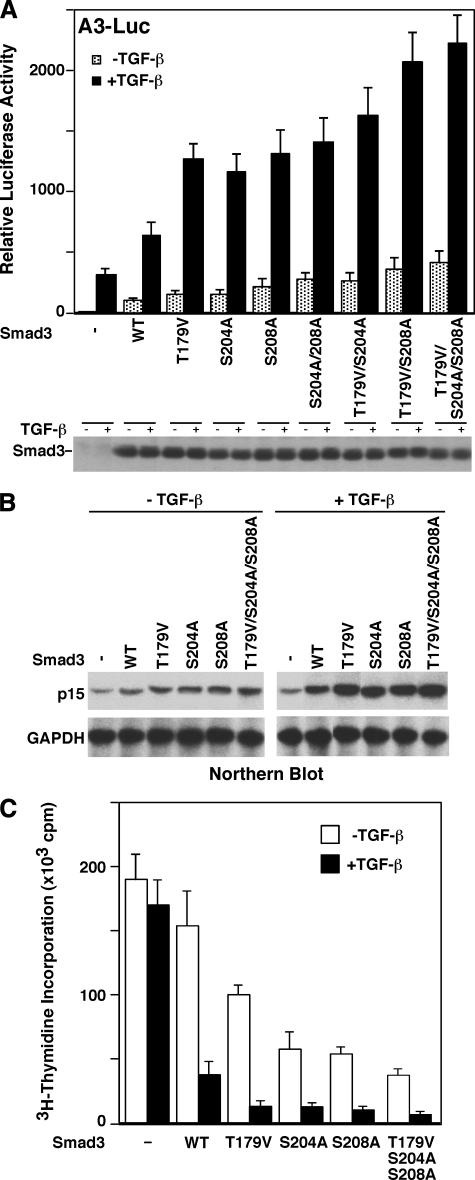
Mutation of the Linker Phosphorylation Sites Increases the Ability of Smad3 to Activate a TGF-β/Smad Reporter Gene—To determine whether linker phosphorylation affects the ability of Smad3 to regulate a TGF-β/Smad responsive gene, we mutated the phosphorylation sites individually or in combination, and then analyzed their ability to activate the A3-Luc reporter gene, which is one of the best characterized TGF-β/Smad reporter genes. The A3 reporter gene contains DNA binding sites for Smads and FoxH1/FAST-1, a winged-helix transcription factor. Smads and FoxH1/FAST-1 together activate the A3 reporter gene (63, 64). As shown in Fig. 7A, the linker phosphorylation mutants have increased capacity to activate the A3-Luc reporter gene. The double mutant T179V/S208A has an activity similar to the triple mutant T179V/S204A/S208A, supporting the notion that Ser208 is a priming site for Ser204. In addition, this suggests different functions of Ser204/Ser208 phosphorylation and Thr179 phosphorylation. Immunoblot analysis indicated that the mutants and the wild type Smad3 were expressed at similar levels.
FIGURE 7.
Mutation of Smad3 linker phosphorylation sites increases its ability to activate a Smad target gene and its antiproliferative function. A, mutation of the Smad3 linker phosphorylation sites enhances the capacity of Smad3 to activate a Smad reporter gene. HepG2 cells were cotransfected with the A3-Luciferase reporter gene, FoxH1/FAST1, and the vector control, wild type (WT) Smad3, or a phosphorylation mutant as indicated. Cells were treated with TGF-β for 24 h. Luciferase activity represents the average of four independent transfection experiments. One of the transfection experiments was also analyzed for the expression levels of the various Smad3 as shown in the lower panel. B, mutation of Smad3 linker sites increases its ability to up-regulate endogenous p15 expression. Smad3 or its phosphorylation mutants were introduced into Smad3–/– MEF by retroviral infection. Cells were treated with or without TGF-β for 8 h. Total RNA was prepared from cells and subjected to Northern blot analysis for p15 levels. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was analyzed as a control. C, mutation of the Smad3 linker sites increases its growth-inhibitory activity. Smad3 or its phosphorylation mutants were introduced into Smad3–/– MEF by retroviral infection. Cells were treated with or without TGF-β for 24 h. [3H]Thymidine incorporations were then analyzed. The results represent the average of four independent experiments.
Mutation of the Phosphorylation Sites Increases the Ability of Smad3 to Activate an Endogenous TGF-β/Smad Target Gene—To determine the effect of Smad3 linker phosphorylation on endogenous gene expression, Smad3 or its linker phosphorylation mutants were inserted into the retroviral vector pLZRSΔ-IRES-GFP, and introduced into primary Smad3–/– MEF by retroviral infection. We routinely achieve essentially 100% infection efficiency in primary MEF. Cells were treated with or without TGF-β. Total RNA was then isolated and analyzed for CDK inhibitor p15 levels by Northern blot analysis. p15, which inhibits CDK4 and CDK6 activity, is a Smad target gene and plays an important role in mediating the TGF-β/Smad growth-inhibitory effect (2). As shown in Fig. 7B, the Smad3 phosphorylation mutants have higher activity than the wild type Smad3 to activate the p15 expression.
Mutation of the Smad3 Phosphorylation Sites Increases Its Growth-inhibitory Activity—Primary Smad3–/– MEF were infected with the retroviral vector, retroviral wild type Smad3, or Smad3 phosphorylation mutants as described above, and then analyzed in [3H]thymidine incorporation assay. Smad3 is essential for the TGF-β growth inhibitory effects (15, 17, 58). Smad3–/– MEF have little response to the TGF-β growth-inhibitory signal (Fig. 7C and Refs. 15 and 58). Introduction of Smad3 restored the TGF-β growth inhibition response. Importantly, Smad3 phosphorylation mutants have higher activity than wild type Smad3 to inhibit cell proliferation (Fig. 7C).
DISCUSSION
Smads transduce the TGF-β signal at the cell surface into gene regulation in the nucleus (2–8). We have shown in this report that in addition to C-tail phosphorylation, TGF-β induces Smad3 phosphorylation in the linker region. Interestingly, C-tail phosphorylation by the receptor appears to be necessary for linker phosphorylation in the presence of TGF-β. Low doses of TGF-β are sufficient to induce linker phosphorylation, highly suggesting that phosphorylation occurs in living organisms at physiological concentrations of TGF-β. Mutation of the linker phosphorylation sites has no effect on TGF-β-induced nuclear accumulation of Smad3 (data not shown). Mutation of the linker phosphorylation sites increases the ability of Smad3 to activate a TGF-β/Smad target gene. Moreover, mutation of the linker phosphorylation sites increases the growth-inhibitory function of Smad3. These observations suggest that TGF-β-induced linker phosphorylation inhibits the growth-inhibitory response.
We identified TGF-β-inducible phosphorylation sites in the Smad3 linker region at Ser208, Ser204, and Thr179. The Ser208 and Ser204 phosphopeptide antibodies do not recognize the analogous positions in Smad2. The Thr179 phosphopeptide antibody also recognizes the analogous position (Thr220) in Smad2, and identified that Smad2 is phosphorylated at Thr220 in response to TGF-β. Carefully reviewing published work revealed that a previous study had shown that in response to TGF-β treatment, a phosphopeptide containing the linker region of Smad2 is phosphorylated as analyzed by two-dimensional phosphopeptide mapping (12). Smad3 and Smad2 are highly homologous. They have overlapping as well as distinct functions (65). It is not clear whether the TGF-β-induced phosphorylation of the Smad2 linker region restricts only to those sites that are analogous to the Smad3 sites. Future studies are necessary to identify all TGF-β-induced phosphorylation sites in Smad2.
Because ERK phosphorylates the same three sites Ser208, Ser204, and Thr179 in response to EGF (33), we analyzed whether ERK is the kinase that phosphorylates these three sites in the presence of TGF-β. Previous studies have shown that TGF-β can significantly activate ERK in a cell type-dependent manner. For example, TGF-β stimulates articular chondrocyte proliferation (66). In response to TGF-β, ERK is rapidly and markedly activated in articular chondrocytes (66). Its activity peaks 5 min after TGF-β treatment and then declines to basal level in 4 h. In Mv1Lu cells, TGF-β potently inhibits their proliferation. TGF-β treatment causes a transient and modest activation of ERK in Mv1Lu cells: the activation is within 2-fold; ERK activity peaks ∼5 min after TGF-β treatment and then rapidly declines to basal level within minutes. This is consistent with a previously reported time course and extent of ERK activation by TGF-β in Mv1Lu cells (59). Pretreatment of Mv1Lu cells with a MEK1 inhibitor has little effect on Smad3 linker phosphorylation in the presence of TGF-β (Fig. 4A). In addition, MKP1, which effectively dephosphorylates and inactivates ERK and other members of the MAP kinase superfamily, including JNK and p38, has essentially no effect on TGF-β-induced phosphorylation of the linker sites in Smad3 (Fig. 4B). Thus, although ERK phosphorylates Ser208, Ser204, and Thr179 in response to EGF treatment (15, 33), ERK is not responsible for TGF-β-dependent phosphorylation of the same three sites.
One interesting observation is that although both EGF and TGF-β induce phosphorylation of the Smad3 linker region at the same sites, they have different requirements for the C-tail. Whereas our results suggest that TGF-β-induced linker phosphorylation is dependent on C-tail phosphorylation (Fig. 2B), the EGF-induced linker phosphorylation does not have such a requirement (Fig. 2B). Thus, EGF-induced Smad3 and TGF-β-induced Smad3 may adapt different conformations to carry out distinct tasks.
GSK3 plays an important role in insulin and Wnt signaling pathways (47). Recent studies have shown that GSK3 also regulates TGF-β family signaling. It was shown that ERK-mediated phosphorylation of Smad1 in the linker region primes the Smad1 linker for phosphorylation by GSK-3β (39), thus enabling ubiquitin ligase Smurf1 binding to Smad1. This binding inhibits the interaction between Smad1 and its nuclear translocation factor, leading to cytoplasmic retention. Smurf1 interaction with Smad1 can also trigger its ubiquitination and degradation (39). Further studies indicate that ERK and GSK phosphorylation of the Smad1 linker sites leads Smad1 to be transported to the centrosomal region, polyubiquitinated, and degraded by proteasomal machinery (40, 41). In Xenopus embryos, Wnt signaling decreases the levels of GSK3-phosphorylated Smad1 and redistributes it from the centrosome to the cytoplasmic LRP6 signalosomes (40, 41). Importantly, a recent study showed that Axin facilitates GSK3β phosphorylation of Smad3 at Thr66 in the N-terminal domain at the basal state, triggering Smad3 ubiquitination and degradation (42). It also showed that GSK3β can phosphorylate the Ser204 site in vitro and at the basal state in vivo (42). We have shown that GSK3 is responsible to phosphorylate the Ser204 site in response to TGF-β. Our study also suggests that Ser208 phosphorylation serves as the priming site for Ser204 phosphorylation by GSK3. Interestingly, GSK3 phosphorylation of Ser204 appears to be unique to the Smad3 linker region, because sequence alignment indicates that the linker regions of other Smads do not fit the p + 4 rule for the consensus GSK3 phosphorylation site (data not shown). Thus, GSK3 phosphorylation of Ser204 may have a unique role in the regulation of Smad3 activity. We have shown in this study that mutation of Ser204 increases the growth-inhibitory function of Smad3. The stability of Smad3 is not affected by Ser204 phosphorylation, based on published studies and our mutagenesis results (42) (Fig. 7A and data not shown). In addition, there appears to be no biochemical relationship between Thr66 and Ser204 phosphorylation. Thr66 phosphorylation is diminished in the presence of TGF-β (42), whereas Ser204 phosphorylation is increased in the presence of TGF-β. Mutation of Thr66 into T66V or T66D does not affect the TGF-β-induced phosphorylation of Ser204 (data not shown). Thus, although both Thr66 and Ser204 are phosphorylated by GSK3, they appear to be independently regulated.
We have shown that flavopiridol, a chemical inhibitor that potently inhibits all CDK family members and GSK3 (Refs. 49–56 and references therein), essentially abolishes TGF-β-mediated phosphorylation of all three sites in the linker region of Smad3. This suggests that CDK family members are responsible for TGF-β-induced phosphorylation of Thr179 and Ser208. Ideally, the flavopiridol data should be complemented by analysis in CDK knock-out cells and by the knockdown approach. However, this is very difficult. We have previously shown that both CDK2 and CDK4 can phosphorylate Thr179 at the basal state (15). CDK6, which is homologous to CDK4, can also phosphorylate Thr179 (data not shown). Moreover, CDK1, which is similar to CDK2, can also phosphorylate Smad3 (67). Mouse knock-out studies of all three cyclin D isoforms, both CDK4 and CDK6, both cyclin E isoforms, or CDK2 revealed the compensatory mechanisms that operate in the absence of a cyclin or CDK (68–75). Most strikingly, a recent study showed that in CDK2–/–CDK4–/–CDK6–/– triple knock-out cells, CDK1 activity is increased and it compensates for all the activities of CDK2, CDK4, and CDK6 (76). One cannot use RNA interference, antisense, or chemical inhibitor to inhibit CDK1 activity in the triple knock-out cells, because triple knock-out cells completely rely on the single CDK, CDK1, to proliferate. If the CDK1 activity is inhibited in triple knock-out cells, triple knock-out cells will not be able to survive. In addition, there is another complexity. Besides CDK2, CDK4, CDK6, and CDK1, there are several other members in the CDK family. Although these technical challenges prevent us from making further conclusions, the strong flavopiridol data and the in vitro kinase assay results by CDK2 and CDK4 highly suggest that the CDK family members mediate the TGF-β-induced phosphorylation of Thr179 and Ser208. The mechanism by which TGF-β treatment leads to CDK phosphorylation of Smad3 linker sites remains to be elucidated. We cannot detect an increase in CDK activities after cells are treated with TGF-β for 30 min, 1 h, or 1.5 h (data not shown). Because CDKs are very abundant kinases, the activation of CDKs can be masked if only a very small fraction of CDKs are activated. In addition, the mechanism may not involve CDK activation. For example, CDKs may phosphorylate the Smad3 linker region once its conformation is changed in response to C-tail phosphorylation. In addition, it is not clear why the Ser213 site in Smad3, which is phosphorylated by CDKs at the basal state (15), is not phosphorylated in response to TGF-β. The ultimate understanding of the dynamic regulation of Smad3 linker phosphorylation will be dependent on successful structural studies.
In conclusion, we have identified three sites in the Smad3 linker region that are rapidly phosphorylated by TGF-β treatment. We have provided evidence suggesting that the C-tail phosphorylation by the receptor is necessary for TGF-β-induced phosphorylation of the linker region. Our observations reveal that the main scheme of the TGF-β signal transduction pathways are much more complex than previously envisioned. Our findings provide an important foundation for further identification of key components in the TGF-β signal transduction pathways.
Acknowledgments
We are very grateful to Dr. Y. E. Zhang for communications on GSK3 phosphorylation of Ser204 prior to publication and for many insightful discussions on Smad3 linker phosphorylation. We thank Drs. E. B. Leof, J. R. Woodgett, and X.-F. Wang for reagents and K. Cohen-Solal, A. K. Kamaraju, M. Reiss, A. B. Roberts, and W. Xie for helpful discussions.
This work was supported by National Institutes of Health Grant CA93771 (to F. L.).
Footnotes
The abbreviations used are: TGF-β, transforming growth factor-β; EGF, epidermal growth factor; ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; CDK, cyclin-dependent kinase; GSK, glycogen synthase kinase; R-Smads, receptor-regulated Smads; JNK, c-Jun N-terminal kinase; MEF, mouse embryonic fibroblast; HA, hemagglutinin; GST, glutathione S-transferase; MKP1, MAP kinase phosphatase 1.
G. Wang, I. Matsuura, D. He, and F. Liu, unpublished results.
References
- 1.Roberts, A. B., and Sporn, M. B. (1990) in Peptide Growth Factors and Their Receptors (Sporn, M. B., and Roberts, A. B., eds) pp. 419–472, Springer, Heidelberg
- 2.Massagué, J., Blain, S. W., and Lo, R. S. (2000) Cell 103 295–309 [DOI] [PubMed] [Google Scholar]
- 3.Roberts, A. B., Russo, A., Felici, A., and Flanders, K. C. (2003) Ann. N. Y. Acad. Sci. 995 1–10 [DOI] [PubMed] [Google Scholar]
- 4.Heldin, C.-H., Miyazono, K., and ten Dijke, P. (1997) Nature 390 465–471 [DOI] [PubMed] [Google Scholar]
- 5.Shi, Y., and Massagué, J. (2003) Cell 113 685–700 [DOI] [PubMed] [Google Scholar]
- 6.Derynck, R., and Zhang, Y. E. (2003) Nature 425 577–584 [DOI] [PubMed] [Google Scholar]
- 7.ten Dijke, P., and Hill, C. S. (2004) Trends Biochem. Sci. 29 265–273 [DOI] [PubMed] [Google Scholar]
- 8.Feng, X. H., and Derynck, R. (2005) Annu. Rev. Cell. Dev. Biol. 21 659–693 [DOI] [PubMed] [Google Scholar]
- 9.Macias-Silva, M., Abdollah, S., Hoodless, P. A., Pirone, R., Attisano, L., and Wrana, J. L. (1996) Cell 87 1215–1224 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, Y., Feng, X., We, R., and Derynck, R. (1996) Nature 383 168–172 [DOI] [PubMed] [Google Scholar]
- 11.Abdollah, S., Macias-Silva, M., Tsukazaki, T., Hayashi, H., Attisano, L., and Wrana, J. L. (1997) J. Biol. Chem. 272 27678–27685 [DOI] [PubMed] [Google Scholar]
- 12.Souchelnytskyi, S., Tamaki, K., Engstrom, U., Wernstedt, C., ten Dijke, P., and Heldin, C. H. (1997) J. Biol. Chem. 272 28107–28115 [DOI] [PubMed] [Google Scholar]
- 13.Hoodless, P. A., Haerry, T., Abdollah, S., Stapleton, M., O'Connor, M. B., Attisano, L., and Wrana, J. L. (1996) Cell 85 489–500 [DOI] [PubMed] [Google Scholar]
- 14.Kretzschmar, M., Liu, F., Hata, A., Doody, J., and Massague, J. (1997) Genes Dev. 11 984–995 [DOI] [PubMed] [Google Scholar]
- 15.Matsuura, I., Denissova, N. G., Wang, G., He, D., Long, J., and Liu, F. (2004) Nature 430 226–231 [DOI] [PubMed] [Google Scholar]
- 16.Liu, F., and Matsuura, I. (2005) Cell Cycle 4 63–66 [DOI] [PubMed] [Google Scholar]
- 17.Liu, F. (2005) Cytokine Growth Factor Rev. 17 9–17 [DOI] [PubMed] [Google Scholar]
- 18.Kretzschmar, M., Doody, J., and Massague J. (1997) Nature 389 618–622 [DOI] [PubMed] [Google Scholar]
- 19.de Caestecker, M. P., Parks, W. T., Frank, C. J., Castagnino, P., Bottaro, D. P., Roberts, A. B., and Lechleider, R. J. (1998) Genes Dev. 12 1587–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretzschmar, M., Doody, J., Timokhina, I., and Massague, J. (1999) Genes Dev. 13 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, P. P., Shen, X., Huang, D., Liu, Y., Counter, C., and Wang, X. F. (1999) J. Biol. Chem. 274 35381–35387 [DOI] [PubMed] [Google Scholar]
- 22.Yue, J., Frey, R. S., and Mulder, K. M. (1999) Oncogene 18 2033–2037 [DOI] [PubMed] [Google Scholar]
- 23.Mulder, K. M. (2000) Cytokine Growth Factor Rev. 11 23–35 [DOI] [PubMed] [Google Scholar]
- 24.Lehmann, K., Janda, E., Pierreux, C. E., Rytomaa, M., Schulze, A., McMahon, M., Hill, C. S., Beug, H., and Downward, J. (2000) Genes Dev. 14 2610–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanchette, F., Rivard, N., Rudd, P., Grondin, F., Attisano, L., and Dubois, C. M. (2001) J. Biol. Chem. 276 33986–33994 [DOI] [PubMed] [Google Scholar]
- 26.Grimm, O. H., and Gurdon, J. B. (2002) Nat. Cell. Biol. 4 519–522 [DOI] [PubMed] [Google Scholar]
- 27.Funaba, M., Zimmerman, C. M., and Mathews, L. S. (2002) J. Biol. Chem. 277 41361–41368 [DOI] [PubMed] [Google Scholar]
- 28.Sater, A. K., El-Hodiri, H. M., Goswami, M., Alexander, T. B., Al-Sheikh, O., Etkin, L. D., and Akif Uzman, J. (2003) Differentiation 71 434–444 [DOI] [PubMed] [Google Scholar]
- 29.Pera, E. M., Ikeda, A., Eivers, E., and De Robertis, E. M. (2003) Genes Dev. 17 3023–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massagué, J. (2003) Genes Dev. 17 2993–2997 [DOI] [PubMed] [Google Scholar]
- 31.Aubin, J., Davy, A., and Soriano, P. (2004) Genes Dev. 18 1482–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Robertis, E. M., and Kuroda, H. (2004) Annu. Rev. Cell. Dev. Biol. 20 285–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuura, I., Wang, G., He, D., and Liu, F. (2005) Biochemistry 44 12546- [DOI] [PubMed] [Google Scholar]
- 34.Engel, M. E., McDonnell, M. A., Law, B. K., and Moses, H. L. (1999) J. Biol. Chem. 274 37413–37420 [DOI] [PubMed] [Google Scholar]
- 35.Mori, S., Matsuzaki, K., Yoshida, K., Furukawa, F., Tahashi, Y., Yamagata, H., Sekimoto, G., Seki, T., Matsui, H., Nishizawa, M., Fujisawa, J., and Okazaki, K. (2004) Oncogene 23 7416–7429 [DOI] [PubMed] [Google Scholar]
- 36.Yamagata, H., Matsuzaki, K., Mori, S., Yoshida, K., Tahashi, Y., Furukawa, F., Sekimoto, G., Watanabe, T., Uemura, Y., Sakaida, N., Yoshioka, K., Kamiyama, Y., Seki, T., and Okazaki, K. (2005) Cancer Res. 65 157–165 [PubMed] [Google Scholar]
- 37.Furukawa, F., Matsuzaki, K., Mori, S., Tahashi, Y., Yoshida, K., Sugano, Y., Yamagata, H., Matsushita, M., Seki, T., Inagaki, Y., Nishizawa, M., Fujisawa, J., and Inoue, K. (2003) Hepatology 38 879–889 [DOI] [PubMed] [Google Scholar]
- 38.Kamaraju, A. K., and Roberts, A. B. (2005) J. Biol. Chem. 280 1024–1036 [DOI] [PubMed] [Google Scholar]
- 39.Sapkota, G., Alarcon, C., Spagnoli, F. M., Brivanlou, A. H., and Massague, J. (2007) Mol. Cell. 25 441–454 [DOI] [PubMed] [Google Scholar]
- 40.Fuentealba, L. C., Eivers, E., Ikeda, A., Hurtado, C., Kuroda, H., Pera, E. M., and De Robertis, E. M. (2007) Cell 131 980–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eivers, E., Fuentealba, L. C., and De Robertis, E. M. (2008) Curr. Opin. Genet. Dev. 18 304–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo, X., Ramirez, A., Waddell, D. S., Li, Z., Liu, X., and Wang, X. F. (2008) Genes Dev. 22 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wicks, S. J., Lui, S., Abdel-Wahab, N., Mason, R. M., and Chantry, A. (2000) Mol. Cell. Biol. 20 8103–8111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Javelaud, D., and Mauviel, A. (2005) Oncogene 24 5742–5750 [DOI] [PubMed] [Google Scholar]
- 45.Wang, G., Long, J., Matsuura, I., He, D., and Liu, F. (2005) Biochem. J. 386 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prokova, V., Mavridou, S., Papakosta, P., and Kardassis, D. (2005) Nucleic Acids Res. 33 3708–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frame, S., and Cohen, P. (2001) Biochem. J. 359 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manning, G., Whyte, D. B., Martinez, R., Hunter, T., and Sudarsanam, S. (2002) Science 298 1912–1934 [DOI] [PubMed] [Google Scholar]
- 49.De Azevedo, W. F., Jr., Mueller-Dieckmann, H. J., Schulze-Gahmen, U., Worland, P. J., Sausville, E., and Kim, S. H. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 2735–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlson, B. A., Dubay, M. M., Sausville, E. A., Brizuela, L., and Worland, P. J. (1996) Cancer Res. 56 2973–2978 [PubMed] [Google Scholar]
- 51.Meijer, L., Leclerc, S., and Leost, M. (1999) Pharmacol. Ther. 82 279–284 [DOI] [PubMed] [Google Scholar]
- 52.Losiewicz, M. D., Carlson, B. A., Kaur, G., Sausville, E. A., and Worland, P. J. (1994) Biochem. Biophys. Res. Commun. 201 589–595 [DOI] [PubMed] [Google Scholar]
- 53.Chao, S. H., and Price, D. H. (2001) J. Biol. Chem. 276 31793–31799 [DOI] [PubMed] [Google Scholar]
- 54.Senderowicz, A. M. (2005) Prog. Drug Res. 63 183–206 [DOI] [PubMed] [Google Scholar]
- 55.Shchemelinin, I., Sefc, L., and Necas, E. (2006) Folia Biol. (Prague) 52 137–148 [PubMed] [Google Scholar]
- 56.Leclerc, S., Garnier, M., Hoessel, R., Marko, D., Bibb, J. A., Snyder, G. L., Greengard, P., Biernat, J., Wu, Y. Z., Mandelkow, E. M., Eisenbrand, G., and Meijer, L. (2001) J. Biol. Chem. 276 251–260 [DOI] [PubMed] [Google Scholar]
- 57.Hoeflich, K. P., Luo, J., Rubie, E. A., Tsao, M. S., Jin, O., and Woodgett, J. R. (2000) Nature 406 86–90 [DOI] [PubMed] [Google Scholar]
- 58.Datto, M. B., Frederick, J. P., Pan, L., Borton, A. J., Zhuang, Y., and Wang, X. F. (1999) Mol. Cell. Biol. 19 2495–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hartsough, M. T., and Mulder, K. M. (1995) J. Biol. Chem. 270 7117–7124 [DOI] [PubMed] [Google Scholar]
- 60.Chu, Y., Solski, P. A., Khosravi-Far, R., Der, C. J., and Kelly, K. (1996) J. Biol. Chem. 271 6497–6501 [DOI] [PubMed] [Google Scholar]
- 61.Yu, L., Hébert, M. C., and Zhang, Y. E. (2002) EMBO J. 21 3749–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryves, W. J., Fryer, L., Dale, T., and Harwood, A. J. (1998) Anal. Biochem. 264 124–127 [DOI] [PubMed] [Google Scholar]
- 63.Chen, X., Weisberg, E., Fridmacher, V., Watanabe, M., Naco, G., and Whitman, M. (1997) Nature 389 85–89 [DOI] [PubMed] [Google Scholar]
- 64.Liu, F., Pouponnot, C., and Massague, J. (1997) Genes Dev. 11 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu, F. (2003) Front. Biosci. 8 S1280–1303 [DOI] [PubMed] [Google Scholar]
- 66.Yonekura, A., Osaki, M., Hirota, Y., Tsukazaki, T., Miyazaki, Y., Matsumoto, T., Ohtsuru, A., Namba, H., Shindo, H., and Yamashita, S. (1999) Endocr. J. 46 545–553 [DOI] [PubMed] [Google Scholar]
- 67.Li, L., Iwamoto, Y., Berezovskaya, A., and Boussiotis, V. A. (2006) Nat. Immunol. 7 1157–1165 [DOI] [PubMed] [Google Scholar]
- 68.Kozar, K., Ciemerych, M. A., Rebel, V. I., Shigematsu, H., Zagozdzon, A., Sicinska, E., Geng, Y., Yu, Q., Bhattacharya, S., Bronson, R. T., Akashi, K., and Sicinski, P. (2004) Cell 118 477–491 [DOI] [PubMed] [Google Scholar]
- 69.Malumbres, M., Sotillo, R., Santamaria, D., Galan, J., Cerezo, A., Ortega, S., Dubus, P., and Barbacid, M. (2004) Cell 118 493–504 [DOI] [PubMed] [Google Scholar]
- 70.Geng, Y., Yu, Q., Sicinska, E., Das, M., Schneider, J. E., Bhattacharya, S., Rideout, W. M., Bronson, R. T., Gardner, H., and Sicinski, P. (2003) Cell 114 431–443 [DOI] [PubMed] [Google Scholar]
- 71.Parisi, T., Beck, A. R., Rougier, N., McNeil, T., Lucian, L., Werb, Z., and Amati, B. (2003) EMBO J. 22 4794–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ortega, S., Prieto, I., Odajima, J., Martin, A., Dubus, P., Sotillo, R., Barbero, J. L., Malumbres, M., and Barbacid, M. (2003) Nat. Genet. 35 25–31 [DOI] [PubMed] [Google Scholar]
- 73.Berthet, C., Aleem, E., Coppola, V., Tessarollo, L., and Kaldis, P. (2003) Curr. Biol. 13 1775–1785 [DOI] [PubMed] [Google Scholar]
- 74.Pagano, M., and Jackson, P. K. (2004) Cell 118 535–538 [DOI] [PubMed] [Google Scholar]
- 75.Sherr, C. J., and Roberts, J. M. (2004) Genes. Dev. 18 2699–2711 [DOI] [PubMed] [Google Scholar]
- 76.Santamaría, D., Barrière, C., Cerqueira, A., Hunt, S., Tardy, C., Newton, K., Cáceres, J. F., Dubus, P., Malumbres, M., and Barbacid, M. (2007) Nature 448 811–815 [DOI] [PubMed] [Google Scholar]