Abstract
Salicylate (SA, 2-hydroxybenzoate) is a phytohormone best known for its role as a critical mediator of local and systemic plant defense responses. In response to pathogens such as Pseudomonas syringae, SA is synthesized and activates widespread gene expression. In gh3.12/pbs3 mutants of Arabidopsis thaliana, induced total SA accumulation is significantly compromised as is SA-dependent gene expression and plant defense. AtGH3 subfamily I and II members have been shown to conjugate phytohormone acyl substrates to amino acids in vitro, with this role supported by in planta analyses. Here we sought to determine the in vitro biochemical activity and kinetic properties of GH3.12/avrPphB susceptible 3 (PBS3), a member of the uncharacterized AtGH3 subfamily III. Using a novel high throughput adenylation assay, we characterized the acyl substrate preference of PBS3. We found PBS3 favors 4-substituted benzoates such as 4-aminobenzoate and 4-hydroxybenzoate, with moderate activity on benzoate and no observed activity with 2-substituted benzoates. Similar to known GH3 enzymes, PBS3 catalyzes the conjugation of specific amino acids (e.g. Glu) to its preferred acyl substrates. Kinetic analyses indicate 4-aminobenzoate and 4-hydroxybenzoate are preferred acyl substrates as PBS3 exhibits both higher affinities (apparent Km = 153 and 459 μm, respectively) and higher catalytic efficiencies (kcat/Km = 0.0179 and 0.0444 μm–1 min–1, respectively) with these acyl substrates compared with benzoate (apparent Km = 867 μm, kcat/Km = 0.0046 μm–1 min–1). Notably, SA specifically and reversibly inhibits PBS3 activity with an IC50 of 15 μm. This suggests a general mechanism for the rapid, reversible regulation of GH3 activity and small molecule cross-talk. For PBS3, this may allow for coordination of flux through diverse chorismate-derived pathways.
Mutant screens in Arabidopsis have successfully identified many components of plant disease resistance pathways, with a number of these mutants exhibiting altered phytohormone synthesis, perception, or signaling (1). The pbs3-1 ethyl methanesulfonate mutant was first isolated in a screen for enhanced susceptibility to avirulent Pseudomonas syringae pv. tomato DC3000, carrying the effector avrPphB, resistance to which is mediated by the plant resistance (R) gene RPS5 in Arabidopsis (2). Our subsequent analysis with Roger Innes revealed that the pbs3-1 mutant is also more susceptible to virulent P. syringae strains, including P. syringae pv. maculicola ES4326, suggesting a more general role for PBS3 than in directly mediating effector-R gene interactions (3). Furthermore, in response to P. syringae pathovars, the pbs3-1 EMS mutant and the pbs3-2 T-DNA insertion line exhibited reduced total SA accumulation and expression of the SA-dependent pathogenesis-related gene PR1 (At2g14610) in comparison with wild type plants (3). The mutation in pbs3-1 was then mapped and cloned, and PBS32 was found to be AtGH3.12 (At5g13320), a member of the AtGH3 multigene family (3).
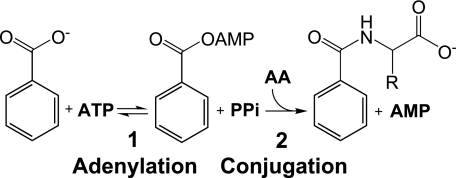
The first GH3 gene was characterized by Guilfoyle and co-workers (4) as an early auxin-responsive gene from Glycine max. Homologs have since been identified in many other plant species and in bacteria (5). The GH3 family is part of the broader acyl-adenylate/thioester-forming enzyme family, also called the firefly luciferase family (6). This superfamily consists of enzymes that catalyze a variety of reactions with a common first step as follows: the transfer of AMP from ATP to the carboxylic acid group of an acyl substrate, forming an activated acyl-adenylate intermediate. GH3 enzymes are unique within this superfamily in that amino acid conjugation occurs in the absence of a thioester intermediate. Fig. 1 shows the reaction catalyzed by GH3 enzymes, with benzoate as the acyl substrate. To date, biochemical activity has only been demonstrated for select plant GH3 enzymes (discussed below). Furthermore, mechanistic analyses are hampered by the lack of sequence conservation between GH3 and other acyl-adenylase/thioester-forming superfamily members and no structural information for any GH3 family member.
FIGURE 1.
Reaction catalyzed by GH3 enzymes, shown with benzoate as the acyl substrate. Adenylation of the acyl substrate is followed by formation of the amino acid conjugate.
The known acyl substrate specificity of the 19 Arabidopsis AtGH3 proteins corresponds to their phylogenetic relationship (6) that divides the family into three groups (supplemental Fig. S1). Group I includes JAR1, which catalyzes the formation of JA-Ile, an active form of the phytohormone jasmonate (JA) (7). JA-Ile promotes the interaction of the F-box protein COI1 with JAZ JA repressor family members (8). This interaction results in degradation of the JAZ repressor by the ubiquitin ligase-dependent 26 S proteasome followed by the activation of the associated JA-dependent responses (9–11). Group II is composed of several proteins that catalyze the formation of auxin-amino acid conjugates (12). Conjugation of indole-3-acetate (IAA) with Asp and Glu targets the phytohormone IAA for catabolism, whereas Ala and Leu conjugates of IAA appear to be inactive storage forms of IAA (13). IAA directly binds to TIR1, the F-box subunit of SCFTIR1, enabling its interaction with the Aux/IAA family of auxin transcriptional repressors, degradation of the repressor, and activation of IAA-associated responses (14, 15). In contrast, IAA-amino acid conjugates are too bulky to fit into the auxin binding pocket of TIR1 and do not promote complex formation between TIR1 and Aux/IAA repressors.3 Group III, which includes PBS3 (AtGH3.12), contains proteins for which the substrate specificity has not yet been reported, as these proteins were inactive on tested substrates (6).
The amino acid conjugation reaction catalyzed by characterized GH3 enzymes plays a critical role in plant hormone homeostasis and plant-microbe interactions (7, 13). Not only do plants regulate active phytohormone forms through amino acid conjugation, but some plant pathogens also manipulate phytohormone conjugation and response. For example, P. syringae pv. savastanoi encodes an IAA-AA synthetase that is required for full virulence on oleander (16). P. syringae pv. tomato DC3000 and P. syringae pv. maculicola ES4326 synthesize the phytotoxin coronatine, an analog of JA-Ile, that activates JA-Ile-dependent responses and is required for full virulence (17). Notably, the HopW1-1 effector of P. syringae pv. maculicola ES4326 targets PBS3, altering host SA signal transduction pathways (18).
Because PBS3 is a GH3 protein and pbs3 mutants exhibit SA-deficient phenotypes, we proposed that PBS3 acts upstream of SA, on SA, or on a compound that could compete with SA (3). In the last case, such a compound might bind to a metabolic or regulatory enzyme that normally binds SA (3). Here we systematically examine the acyl substrate and amino acid preference of PBS3 followed by determination of the kinetic and catalytic properties of PBS3. In addition, for the first time for any GH3 member, we assess the influence of putative inhibitors on enzymatic activity. The impact of each of the two point mutations in the lack of function pbs3-1 EMS mutant (3) is also ascertained. Together, these findings provide a framework to explore the role of PBS3 in pathogen-induced SA accumulation and disease resistance. Furthermore, they support an underexplored and significant role for 4-substituted benzoates in plant-pathogen interactions.
EXPERIMENTAL PROCEDURES
Materials and General Protocols—All specialty reagents and chemicals were purchased from Sigma unless otherwise specified. HPLC-grade solvents (EMD Biosciences) were employed in the HPLC analyses. TLC plates were silica gel 60 with F254 from EMD Biosciences. Commonly utilized protein and molecular biology reagents and protocols were prepared and used as described in Ref. 19.
AtPBS3 Cloning and Purification—The AtPBS3 coding sequence was amplified from cDNA isolated from Arabidopsis thaliana ecotype Col-0 and inserted into a pET-28a vector (Novagen) as an NdeI/BamHI fragment using forward primer 5′tcatgacatatgaagccaatcttcgata and reverse primer 5′cacgtggtgggatcctcaaatactgaagaatt. The resulting construct, pET-His-PBS3, contains an N-terminal histidine tag fused to the coding region of AtPBS3. Crude cell extracts were prepared from a 2-liter culture of Escherichia coli Rosetta2 (DE3) cells transformed with pET-His-PBS3 grown in TB media containing 0.2% glucose, 50 μg/ml kanamycin, and 30 μg/ml chloramphenicol. Cultures were grown at 37 °C to mid-log phase and shifted to 18 °C, and 0.1 mm isopropyl 1-thio-β-d-galactopyranoside was added to induce His-PBS3 synthesis. Cells were harvested after shaking overnight (∼25 g wet weight) and stored at –20 °C. His-PBS3 was then purified using nickel-nitrilotriacetic acid His-Bind resin (Novagen) according to manufacturer's directions. His-PBS3 was the only visible protein when 10 μg of this eluant was resolved by SDS-PAGE and visualized using Coomassie Blue. Thrombin (Novagen) was used to cleave the histidine tag. As His-PBS3 and thrombin-cleaved PBS3 displayed similar activity on a set of five acyl substrates, His-PBS3 was utilized unless specified. Protein concentrations were determined by a Bradford assay modified for 96-well plate format using Coomassie Blue G-250 (EM Biosciences) with bovine serum albumin as the standard. Aliquots of the purified recombinant PBS3 proteins (5.3–9.5 mg/ml in 100 mm Tris, pH 7.7, 10% glycerol, 1 mm DTT) were stored at –80 °C. The same batch of enzyme was used for each set of assays reported in a single figure or table, with assays repeated with enzyme from at least one other batch to confirm results. In all assays, the appropriate enzyme concentrations were utilized such that velocity was linear with increasing enzyme concentration.
Generation and Purification of PBS3 Mutants—Amino acid substitutions were introduced into the pET-His-PBS3 plasmid using the QuikChange II site-directed mutagenesis kit (Stratagene) with forward primer 5′ccacgtgttgtttggtaatgaaggagtcgcttgataatgtttac and reverse primer 5′gtaaacattatcaagcgactccttcattaccaaacaacacgtgg to make the E502R mutation and forward primer 5′gatgtcgattcaaagacggatcgaccgggcctctcgagataagagtggtg and reverse primer 5′caccactcttatctcgagaggcccggtcgatccgtctttgaatcgacatc to make the I519T mutation. The double mutant was made using the pET-His-I519K plasmid with the E502R forward and reverse primers. The His-tagged single and double mutant proteins were purified as described for wild type PBS3, above.
Adenylation Reaction—His-PBS3 adenylation activity was monitored spectrophotometrically at 340 nm by coupling the production of pyrophosphate to oxidation of NADH using pyrophosphate reagent (Sigma) (20). The reagent contained the coupling enzymes fructose-6-phosphate kinase, aldolase, triose-phosphate isomerase, glycerophosphate dehydrogenase, and appropriate substrates and cofactors, including NADH. The pyrophosphate reagent was reconstituted in 4 ml of double distilled H2O and used at a volume of 65 μl per 200-μl reaction. For the screen of substrates, reaction mixtures (200 μl) were in 45 mm imidazole, pH 7.4, and contained 5.0 mm MgCl2, 2.5 mm ATP, 1 mm DTT, 0.1–10 mm substrate, and 100 μg of His-PBS3 enzyme in addition to the pyrophosphate reagent. For determination of kinetic parameters for ATP, reactions contained 50 μg/ml His-PBS3 enzyme, pyrophosphate reagent, 1 mm DTT, and 10 mm acyl substrate. The concentration of ATP was varied while either (a) maintaining a 1.5:1 Mg2+ to ATP ratio or (b) with 10 mm MgCl2. Three replicates were performed for each substrate or concentration value. The reactions were initiated with the addition of substrate to a 96-well plate preheated to 30 °C and analyzed using a Spectromax Plus microplate spectrophotometer (Molecular Devices). The change in absorbance at 340 nm was measured every 10–15 s for at least 20 min and converted to velocity by least squares fitting of each curve using the accompanying program SOFTmax PRO 3.0 with manual assessment/confirmation of the linear range. The velocity of a no His-PBS3 control was subtracted; this velocity was on average 0.16 μm/min. An extinction coefficient of 6.22 μm–1 cm–1 for NADH was used to convert velocity values from milli-absorbance units/min to micromolar/min. Kinetic parameters were estimated by fitting initial velocity values to the Hanes equation (21).
Amino Acid Conjugation Reaction (Full Reaction)—Reaction mixtures (200 μl) were in 50 mm Tris-HCl, pH 8.5, and contained 5.0 mm MgCl2, 2.5 mm ATP, 1.0 mm DTT, 1.0 mm acyl substrate, 1.0 mm amino acid, and 100 μg/ml His-PBS3 enzyme. Reactions were incubated at 30 °C for 2 h and monitored by TLC and/or HPLC. Reaction mixtures and standards were spotted on silica gel 60 F254 plates (EMD Biosciences) and developed in either 30:60:10 dichloromethane/ethyl acetate/formic acid or 8:1:1 isopropyl alcohol/ammonium hydroxide/water (for basic and hydrophilic amino acids). The method used to detect the conjugates varied by substrate. pABA conjugates were detected by staining TLC plates with vanillin reagent (6% (w/v) vanillic acid and 1% sulfuric acid in ethanol) and by visualizing TLC plates under short wavelength UV. 4-HBA, vanillic acid, and trans-cinnamic acid conjugates were detected by visualizing TLC plates under short wavelength UV light. BA conjugates were detected by HPLC using a UV detector at 233 nm. HPLC conditions are the same as those used in the Km determination (see below). SA was analyzed by TLC visualized under short wavelength UV light and by HPLC using a fluorescence detector (ex305/em407 nm); however, no spots or peaks associated with SA-conjugates were detected.
Km Determinations—Reaction mixtures (200 μl) were in 50 mm Tris-HCl, pH 8.5, and contained 5.0 mm MgCl2, 5.0 mm ATP, 1 mm DTT, 10 mm glutamate, varying concentrations of 4-HBA, pABA, or BA, and 50 μg/ml His-PBS3 (reaction with 4-HBA) or 125 μg/ml His-PBS3 (reactions with pABA and BA). Acyl substrate concentrations ranged from 50 μm to 5 mm. Reactions were quenched every 2 min by diluting the reaction mixture 10-fold into 2.5% HCl. Samples were filtered through a 0.2-μm Millex-LG syringe filter (Millipore), and a 50-μl aliquot was injected into a Shimadzu SCL-10AVP series HPLC system equipped with a Shimadzu SPD-10AVP photodiode array detector and a Shimadzu RF-10AXL fluorescence detector. A 5-μm, 15 cm × 4.6-mm inner diameter Supelcosil LC-ABZPlus column (Supelco) preceded by a LC-ABZ-Plus guard column was pre-equilibrated in 5% acetonitrile with 25 mm potassium phosphate buffer (initial HPLC buffer), pH 2.5, at a flow rate of 1.0 ml/min. The elution program and detection method varied by substrate. For BA, the initial HPLC buffer was run for 10 min and then changed to 52% acetonitrile and 48% 25 mm potassium phosphate buffer over 12 min. Under these conditions, BA-Glu eluted at 15.5 min and BA at 19 min. Elution was monitored by UV absorbance at 233 nm. Authentic samples were used to generate calibration curves for BA (y = 0.0397x) and BA-Glu (TCI America; y = 0.0407x), with x in absorbance units and y in nanomolar compound. For pABA, pABA eluted at 7.5 min and pABA-Glu eluted at 5.0 min using the BA program. Elution was monitored by fluorescence at ex290/em340 nm. Authentic samples were used to generate calibration curves for pABA (y = 0.2585x) and pABA-Glu (y = 0.1045x), with x in fluorescence units and y in nanomolar compound). For 4-HBA, the initial HPLC buffer was run for 4 min and then changed to 52% acetonitrile and 48% 25 mm potassium phosphate buffer over 7 min. Elution was monitored by UV absorbance at 254 nm. An authentic sample of 4-HBA was used to generate a calibration curve (y = 0.0274x), and synthesized 4-HBA-Glu (see below) was used to generate a calibration curve (y = 0.0318x) with x in UV absorbance area units and y in nanomolar compound. Under these conditions, 4-HBA eluted at 9.7 min and 4-HBA-Glu eluted at 8.3 min. The reaction velocity with each substrate concentration was determined from a linear fit of four time points. Kinetic parameters were estimated by fitting initial velocity values to the Hanes equation as above, with data from at least six substrate concentrations. Nonlinear curve fitting using Synergy KaleidaGraph version 4.0 gave very similar results.
Mass Spectrometry to Verify 4-HBA-Glu Formation—A 5-μm, 15-cm × 4.6-mm inner diameter Prevail C18 column (Alltech) preceded by a 7.5-cm × 4.6-mm guard column was pre-equilibrated in 5% acetonitrile with 25 mm formate buffer, pH 2.8 (initial HPLC buffer), at a flow rate of 1.0 ml/min. The initial buffer was run for 7 min and then changed to 52% acetonitrile over 15 min. The putative 4-HBA-Glu peak eluting at 11.3–11.9 min was collected and dried down under vacuum. This sample was rerun using our standard HPLC assay for verification. Samples for LC-MS analysis were resuspended in a 50:50 mixture of 25 mm ammonium formate and acetonitrile and analyzed using a Thermo Fisher Surveyor HPLC with a Phenomenex C18 2 × 150-mm column and a Thermo Fisher LCQ classic mass spectrometer at the Vincent Coates Foundation Mass Spectrometry Laboratory (Stanford University). LC-MS with electrospray ionization-positive ion analysis employed a C18 column with similar elution scheme. MS-MS fragmentation was performed on the putative dominant ion peak for further confirmation of compound identity. The sample was also analyzed by nanospray infusion on the Q-TOF mass spectrometer (Waters Micromass Q-TOF hybrid quadrupole time of flight) with MS-MS performed on 268.1 m/z.
Synthesis of 4-HBA-Glu Standard—As 4-HBA-Glu is not commercially available, it was synthesized for calibration of the HPLC. Briefly, 4-acetoxybenzoic acid was converted to the acyl chloride and condensed with diethyl l-glutamate to form diethyl 4-acetoxybenzoyl-l-glutamate. The protecting groups were removed in base to yield 4-HBA-Glu as a white crystalline solid with 99% purity by HPLC. The structure was confirmed by 1H NMR and 13C NMR. See supplemental Method S1 for complete procedures and spectral data.
Inhibition Studies—Inhibition of PBS3 amino acid conjugation was assessed with SA using our HPLC assay to monitor pABA-Glu formation. Reaction conditions were the same as for the Km experiment (above), with the addition of SA (5–600 μm). A pABA concentration equivalent to the Km (150 μm) was used. The adenylation assay was then used to measure the effect of potential inhibitors on adenylation velocity using pABA as the acyl substrate. Reaction mixtures (200 μl) were in 45 mm imidazole, pH 7.4, and contained 5.0 mm MgCl2, 5.0 mm ATP, 1 mm DTT, 150 μm pABA, 100 μg/ml His-PBS3 enzyme, 65 μl of pyrophosphate reagent reconstituted in 4.0 ml of double distilled H2O, and potential inhibitors. SA and MeSA were tested over a range of concentrations from 10 to 600 μm. We verified that the decreased velocity with SA was due to inhibition of PBS3 rather than of the coupled reactions by testing the effect of SA with PPi as a substrate; we observed no change in activity. Other potential inhibitors (INA, 2,4-DHBA, 3-HBA, IAA, and JA) were tested at 30 and 300 μm only.
Potential Inhibition of ICS1-catalyzed SA Biosynthesis—To evaluate compounds as inhibitors of the AtICS1-catalyzed conversion of chorismate to isochorismate, a coupled spectrophotometric assay was used (22). The conversion of chorismate to isochorismate was coupled to the oxidation of NADH through the coupling enzymes isochorismate pyruvate lyase (recombinant PchB), which converts isochorismate to pyruvate and SA, and lactic dehydrogenase (Sigma L1254), which converts pyruvate to lactate in an NADH-dependent fashion. The 200-μl reaction contained an effective chorismate concentration of 90 μm (apparent Km of AtICS1 for chorismate), 0.4 mm NADH, 0.833 μg/ml l-lactic dehydrogenase, 32.0 μg/ml PchB, and 10 μg/ml AtICS1 in 100 mm Tris, pH 7.7, with 10% glycerol, and 10 mm MgCl2. We assessed AtICS1 activity in duplicate for reactions with or without 200 μm 4-HBA, pABA, or pABA-Glu and for the appropriate no enzyme controls. We also performed these assays with 1 mm 4-HBA, pABA, or pABA-Glu. The Student's t test (α = 0.1) was used to evaluate whether any observed small changes in activity were statistically significant.
Potential Inhibition of Isochorismate Pyruvate Lyase—To evaluate compounds as inhibitors of the isochorismate pyruvate lyase (IPL) reaction in which isochorismate is converted to SA, we employed a modified version of our coupled ICS spectrophotometric assay, described above. In this case, 50 μg/reaction of recombinant PchB was employed with 60 μm isochorismate, the approximate Km of our recombinant enzyme for isochorismate under our assay conditions. The reported apparent Km for purified P. aeruginosa PchB for isochorismate is 12.5 μm (23). We assessed IPL activity in duplicate for reactions with or without 100 μm 4-HBA, pABA, or pABA-Glu and for the appropriate no enzyme controls. The Student's t test (α = 0.1) was used to evaluate whether any observed small changes in activity were statistically significant. Isochorismate was prepared and purified as in Ref. 22.
RESULTS
Determination of Acyl Substrates of PBS3—To explore the biochemical activity of PBS3, we overexpressed and purified recombinant His-tagged PBS3. We also developed a novel high throughput, 96-well format adenylation assay for use in determining putative acyl substrate(s) of PBS3. In this assay, the release of pyrophosphate, which occurs when the acyl substrate is adenylated to form the acyl substrate-AMP conjugate (Fig. 1), is coupled to the oxidation of NADH that can be measured spectrophotometrically (20). This allows for rapid kinetic assessment of the GH3 adenylation reaction that is not readily performed using standard radiolabeled PPi exchange assays. We report our findings using 1 mm acyl substrate to allow for comparison with previously reported GH3 acyl substrate surveys (6, 12). Assessments with 100 μm and/or 10 mm substrate were performed for substrates with moderate and low activity to ensure we had not missed the linear range of activity. We did not observed significant differences in the relative activities of PBS3 with these acyl substrates compared with our results using 1 mm substrate.
The reduced accumulation of SA and expression of PR1 in response to P. syringae in pbs3 mutants suggested PBS3 might act upstream of SA, on SA, or on a compound that could compete with SA in binding (either as a substrate or modulator of activity) to an SA-binding metabolic or regulatory enzyme (3). Therefore, our initial screen consisted of three overlapping classes of commercially available compounds as follows: 1) phytohormones, including SA and the SA functional analog INA; 2) compounds that are structurally similar to SA (i.e. benzoates); and 3) compounds in the chorismate pathway. We found that recombinant PBS3 did not exhibit significant activity on any of the phytohormones tested (Table 1), including SA and the functional SA analog 2,6-dichloroisonicotinic acid (INA). In contrast, recombinant PBS3 was very active on BA and 4-HBA; respective activities were 59.6 and 135.2 nmol/min/mg for BA and 4-HBA. Moderate activity was observed for the chorismate pathway compounds chorismate (32.9 nmol/min/mg), a precursor of SA, and trans-cinnamate (21.3 nmol/min/mg).
TABLE 1.
Initial acyl substrate screen
| Activitya | |
|---|---|
| nmol/min/mg | |
| Phytohormones | |
| Indole-3-acetate | 1.5 ± 0.1 |
| Jasmonate | 2.9 ± 0.1 |
| Gibberellate | 0.1 ± 0.1 |
| Abscisate | 0.8 ± 0.1 |
| Salicylate | 1.7 ± 0.1 |
| INA | 0.1 ± 0.2 |
| Benzoates | |
| Benzoate | 59.6 ± 1.1 |
| Phenylacetate | 2.3 ± 0.1 |
| Salicylate (2-hydroxybenzoate) | 1.7 ± 0.1 |
| 4-Hydroxybenzoate | 135.2 ± 8.6 |
| Chorismate pathway compounds | |
| Chorismateb | 32.9 ± 2.0 |
| Prephenate | 10.1 ± 0.4 |
| Anthranilate (2-aminobenzoate) | 0.7 ± 1.1 |
| Phenylalanine | 0.0 ± 0.0 |
| Tyrosine | 0.0 ± 0.0 |
| Tryptophan | 0.0 ± 0.0 |
| trans-Cinnamate | 21.3 ± 1.8 |
Activities are the mean of three replicates ± 1 S.D. and are determined for 1 mm acyl substrate using the coupled adenylation assay. Activities below 10 nmol/min/mg are minimal. Additional experiments yielded similar results.
Activity is due to 4-HBA in chorismate preparation.
Except for chorismate and prephenate, the acyl substrates on which PBS3 showed activity contain aromatic rings. Therefore, we sought to establish whether PBS3 is truly active on chorismate (and prephenate) or is active on breakdown products of these compounds. Known breakdown products of chorismate are phenyl pyruvate, prephenate, and 4-HBA (24–26). As shown in Table 1, PBS3 was highly active on 4-HBA with low activity on the nonaromatic prephenate (10.1 nmol/min/mg), and no significant activity on the aromatic breakdown product phenyl pyruvate. To determine whether the observed activity on chorismate and prephenate was because of 4-HBA contamination, we performed HPLC analysis on our freshly prepared chorismate and prephenate solutions. 4-HBA was present in our chorismate solution at a level sufficient to account for the observed PBS3 activity on chorismate (33.5 μm 4-HBA in 1 mm chorismate). Therefore, PBS3 does not appear to be active on chorismate. On the other hand, we did not detect 4-HBA in our 1 mm prephenate solution, even after incubation for 1 h under assay conditions (30 °C, pH 7.4). This suggests that the low adenylation activity of PBS3 with 1 mm prephenate is valid.
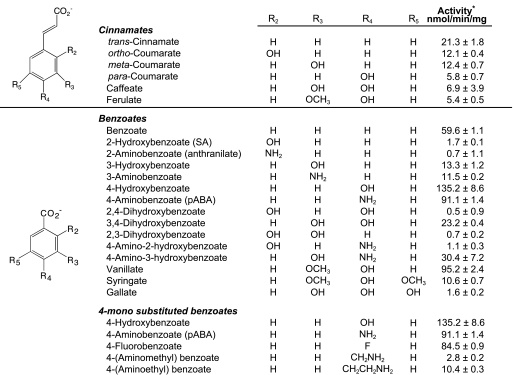
To further examine the acyl substrate preference and specificity of PBS3, compounds structurally related to the active acyl substrates identified above were examined (Table 2). As PBS3 exhibited moderate activity on trans-cinnamate, we tested an additional series of cinnamic acids. We found that the parent compound trans-cinnamate PBS3 exhibited higher activity with than with any of the tested derivatives. As PBS3 exhibited significantly greater adenylation activity with 4-HBA and BA than with trans-cinnamate, we further investigated its substrate preference on substituted benzoates.
TABLE 2.
Acyl substrate preference and specificity of PBS3
* Activities are the mean of three replicates ± 1 S.D. and are determined for 1 mm acyl substrate using the coupled adenylation assay. Activities below 10 nmol/min/mg are minimal. Some compounds are repeated from Table 1 for ease of comparison. Additional experiments yielded similar results.
PBS3 Strongly Prefers para-Substituted Benzoates as the Acyl Substrate—We first examined the adenylation activity of PBS3 on mono-substituted benzoates substituted with hydroxyl (–OH) or amino (–NH2) groups at either the 2-, 3-, or 4-position (Table 2). The compounds on which PBS3 exhibited the highest velocity, 4-HBA (135.2 nmol/min/mg) and pABA (91.1 nmol/min/mg), are substituted at the para-position and were favored over BA. In contrast, substitution at the meta- position with either –OH or –NH2 resulted in significantly lowered PBS3 adenylation activity compared with BA, with adenylation activities of 13.3 and 11.5 nmol/min/mg for 3-HBA and 3-ABA, respectively. Finally, substitution at the ortho (2–) position was strongly disfavored with PBS3 exhibiting essentially no activity on SA or 2-ABA.
We then investigated the impact of additional hydroxyl or methoxy (–MeO) substitutions on PBS3 adenylation activity (Table 2). Again para-hydroxyl-substituted benzoates were favored with ortho-hydroxyl-substituted benzoates strongly disfavored as follows: 4-HBA > BA > 3,4-DHBA > 3-HBA ≫ (SA; 3,4,5-THBA; 2,4-DHBA; and 2,3-DHBA). At the meta- position, MeO substitutions were preferred over additional hydroxyl substitutions. For both vanillate (4-hydroxy-3-methoxy benzoate) versus 3,4-DHBA and syringate (4-hydroxy-3,5-dimethoxybenzoate) versus gallate (3,4,5-trihydroxybenzoate), PBS3 exhibited at least 4-fold higher adenylation activity with the MeO compared with the hydroxyl substitutions. Additional modifications (hydroxyl substituents) to pABA also reduced PBS3 adenylation activity, with loss of activity associated with substitution at the 2-position. PBS3 activity was 91.1 nmol/min/mg with pABA (4-ABA) compared with 30.4 nmol/min/mg with 4-A-3HBA and 1.11 nmol/min/mg with 4-A-2HBA.
To further examine the effect of different substituents at the para-position of benzoate, we compared the adenylation activities of PBS3 on 4-HBA, 4-fluorobenzoate, pABA, 4-amino-methylbenzoate (4-AMeBA), and 4-aminoethylbenzoate (4-AEtBA) (Table 2). The highest activity was observed on 4-HBA, with activity on pABA and 4-fluorobenzoate somewhat lower but still high, followed by a dramatic reduction in activity with the bulkier acyl substrates 4-AMeBA and 4-AEtBA.
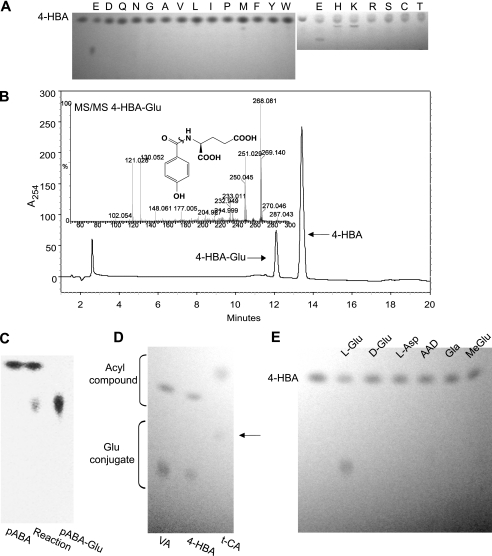
PBS3 Catalyzes the Formation of Amino Acid Conjugates—Once potential acyl substrates were identified, a subset was tested for conjugation to amino acids by recombinant PBS3. Because 4-HBA displayed the highest velocity in the adenylation assay (Tables 1 and 2), we first tested the ability of PBS3 to catalyze the conjugation of amino acids to 4-HBA. Of the 20 amino acids, TLC spots indicative of the 4-HBA-AA conjugate were reproducibly detected for reactions with Glu, His, and Lys, with a faint spot visible for Met (Fig. 2A). Very faint spots indicative of the amino acid conjugate were sometimes visible for reactions with Ile, Leu, Val, Thr, Ser, and Trp. A subset of these PBS3 reaction mixtures was then analyzed by HPLC to verify the presence of novel peaks consistent with the formation of specific 4-HBA-amino acid conjugates. Novel peaks indicative of 4-HBA-AA conjugate formation were observed by HPLC for reactions with Glu, His, Met, Thr, and Ser. Fig. 2B shows the chromatogram for the PBS3 reaction mixture with 4-HBA and Glu. The putative 4-HBA-Glu conjugate was only formed in the presence of PBS3 and required Mg2+ and ATP in the reaction mixture (data not shown). To verify the identity of 4-HBA-Glu, which is not commercially available, the fraction corresponding to the putative 4-HBA-Glu conjugate was collected and analyzed by LC-MS with electrospray ionization-positive ion analysis and by nanospray infusion Q-TOF MS with MS/MS. The mass spectrum of the putative 4-HBA-Glu conjugate is consistent with it being the protonated 4-HBA-Glu conjugate (C12 H14NO6) with m/z = 268.1. MS/MS analyses of the dominant ion peak (M + H) at 268.1 m/z result in major ion intensities at m/z = 251.0 consistent with loss of –OH, and fission products m/z = 130 and 121 likely associated with cleavage of the amide bond (Fig. 2B, inset). Our mass spectra data are similar to those reported for other benzoyl-amino acid conjugates in which fragments consistent with cleavage of the amide bond were also observed (27, 28). In addition, 4-HBA-Glu was chemically synthesized and analyzed by 1H and 13C NMR (see supplemental Methods S1). The chemically synthesized 4-HBA-Glu displayed identical HPLC retention time and UV spectral properties to the 4-HBA-Glu from enzymatic reactions catalyzed by PBS3. This further confirms that PBS3 catalyzes the formation of 4-HBA-Glu in vitro.
FIGURE 2.
PBS3 catalyzes the formation of amino acid conjugates with preferred acyl substrates. A, TLC analysis of 4-HBA amino acid conjugate formation by PBS3. B, HPLC analysis of 4-HBA-Glu formation by PBS3. Inset, verification of putative 4-HBA-Glu peak by LC-MS. Shown is Q-TOF MS/MS on m/z = 268.1 with 4-HBA-Glu structure. Likely fission site is indicated. C, TLC analysis of pABA-Glu formation by PBS3 as follows: pABA standard (left), reaction mixture (middle), and pABA-Glu standard (right). D, TLC analysis of the PBS3 reactions with vanillic acid (VA), 4-HBA, or trans-cinnamic acid (t-CA) as the acyl substrate and Glu. The arrow indicates a faint spot corresponding to trans-cinnamic acid-Glu. E, TLC of the PBS3 reaction with 4-HBA and Glu derivatives. AAD, l-2-aminoadipic acid; Gla, γ-carboxyl l-glutamic acid; MeGlu, l-gutamic acid γ-methyl ester. Images of TLC plates were converted to grayscale and adjustments of “levels” in Photoshop were performed. Independent experiments with different batches of purified, recombinant PBS3 produced similar results.
Conjugation of pABA by PBS3 was also tested with the 20 amino acids and monitored by TLC. Unlike the results with 4-HBA, only a spot for pABA-Glu was detected either by UV light (similarly to 4-HBA) or vanillin staining (Fig. 2C). The Rf value of this putative pABA-Glu conjugate was similar to that of the commercially available standard. pABA-Glu formation by PBS3 was also followed by HPLC. The conversion of pABA to pABA-Glu occurred only with PBS3 in the reaction mixture. As PBS3 also formed additional amino acid conjugates with 4-HBA, we confirmed that similar amino acids conjugates were not formed with pABA by analyzing a subset of these reactions by HPLC. We did not detect putative pABA-His, pABA-Met, or pABA-Ser peaks by HPLC (data not shown).
Other compounds with high velocity (BA and vanillate), moderate velocity (trans-cinnamate), and minimal velocity (SA) were also tested as acyl substrates in the Glu conjugation reaction. TLC analyses showed that recombinant PBS3 catalyzed the conjugation of Glu to vanillate and trans-cinnamate (Fig. 2D) but not to SA. As none of a variety of stains tested worked satisfactorily for BA, we assessed the BA reaction mixture by HPLC. We detected formation of the BA-Glu conjugate from BA and Glu by PBS3, with commercially available BA and BA-Glu used as standards. To further confirm that PBS3 was unable to form the SA-Glu conjugate, we assessed the SA reaction mixture by HPLC. Although SA was readily detected by fluorescence, no novel peaks indicative of SA-Glu were apparent. These results validate the use of the adenylation assay to study substrate specificity by confirming that those acyl substrates with high velocity in the adenylation assay were also capable of forming amino acid conjugates, whereas those substrates with essentially no activity using the adenylation assay (e.g. SA) were not.
We found that 4-HBA and pABA conjugates were formed with Glu (l-glutamic acid) but not Asp. In addition, with pABA as the acyl substrate, pABA-Glu was the only amino acid conjugate detected. Therefore, we wanted to further examine the specificity of PBS3 for Glu. We tested the related compounds d-glutamic acid, l-2-aminoadipic acid, l-glutamic acid γ-methyl ester, and γ-carboxyl l-glutamic acid in our conjugation assay. l-2-Aminoadipic acid contains an additional carbon between the α-carbon and the γ-carboxyl group; l-glutamic acid γ-methyl ester lacks a second carboxylic acid group, and γ-carboxyl l-glutamic acid contains a third carboxylic acid group. PBS3 did not form 4-HBA (Fig. 2E) or pABA conjugates (not shown) with these glutamic acid derivatives.
Point Mutations in the C Terminus of PBS3 Present in the pbs3-1 Mutant Result in Loss of Biochemical Activity—The pbs3-1 EMS mutant contains two amino acid substitutions in highly conserved amino acids at the C terminus (E502K and I519T) (3). This portion of the PBS3 protein has no known motifs and is contained within the region of PBS3 (amino acids 335–575) bound by the P. syringae HopW1-1 effector (18). Glu-502 is predicted to be within an α-helix, whereas residue Ile-519 is predicted to reside in a loop region following the aforementioned α-helix (supplemental Fig. S2). The high degree of conservation of Ile-519 in all Arabidopsis GH3 proteins (Ile in all but two GH3s which have a Val at this position) suggests it does not confer substrate specificity; however, it could play a more general role in substrate binding or in catalysis. To ascertain the impact of these mutations on PBS3 activity, we expressed and purified recombinant enzyme with either or both of the mutations and compared its activity with the wild type recombinant enzyme. We found that the single and double mutations resulted in a complete loss of activity (<0.5 nmol/min/mg) as measured by the adenylation assay with 4-HBA, pABA, or BA as acyl substrates. Wild type recombinant PBS3 exhibited activity similar to that found with previous enzyme preparations (see Table 2). The loss of biochemical activity associated with these mutations is consistent with the similar phenotypes we observed for the recessive pbs3-1 EMS mutant and the null pbs3-2 T-DNA insertion mutant (3).
Kinetic and Catalytic Properties of PBS3—Similar to other characterized GH3 enzymes, the exclusion of Mg2+ or ATP from the reaction mixture resulted in loss of PBS3 activity (data not shown). To determine the pH requirements of the reaction, we performed TLC analyses on the reaction mixtures using either pABA or vanillate as the acyl substrate with Glu as the amino acid at pH 7.5, 8.0, and 8.5. Clearly visible products were observed for the reactions at pH 8.0 and 8.5 but not at pH 7.5 (data not shown).
To further characterize the PBS3 benzoyl amino acid synthetase reaction, we determined its kinetic properties using the following three biologically relevant acyl substrates with the highest adenylation activity: 4-HBA, pABA, and BA. For these analyses, we first determined the apparent Km value for ATP using 4-HBA and pABA as acyl substrates to ensure we used sufficient ATP in our kinetic assays. Using the coupled adenylation assay, we found PBS3 exhibited standard Michaelis-Menten kinetics with an apparent Km for ATP of 791 μm (with 4-HBA) and 636 μm (with pABA). We then determined the apparent Km of PBS3 for the acyl substrates 4-HBA, pABA, and BA at pH 8.5 with saturating Mg2+, ATP, and Glu. The concentration of the Glu conjugate produced in the reactions was quantified by HPLC over time, with the results summarized in Table 3. With all three acyl substrates, we found PBS3 exhibited standard Michaelis-Menten kinetics. The corresponding Hanes plots indicated that PBS3 had the highest affinity for pABA (Km = 153 μm), followed by 4-HBA (Km = 459 μm) and BA (Km = 867 μm). The kcat values for pABA and BA were similar (2.74 and 3.96 min–1), whereas that for 4-HBA was 5–7-fold higher (20.36 min–1). PBS3 exhibited similar catalytic efficiencies for 4-HBA and pABA (0.0444 and 0.0179 μm–1 min–1, respectively) with reduced catalytic efficiency with BA as the acyl substrate (0.0046 μm–1 min–1). Nonlinear curve fitting of our kinetic data gave similar results with apparent Km values of 460, 120, and 931 μm and calculated kcat values of 20.83, 2.70, and 4.06 min–1 for 4-HBA, pABA, and BA, respectively.
TABLE 3.
Kinetic parameters of PBS3 substrates
Kinetic constants were determined by following acyl substrate-Glu formation by HPLC. Similar results were obtained for experiments with an independent enzyme preparation.
| Acyl substrate | kcat | Km | kcat/Km |
|---|---|---|---|
| min-1 | μm | min-1 μm-1 | |
| 4-HBA | 20.36 | 459 | 0.0444 |
| pABA | 2.74 | 153 | 0.0179 |
| BA | 3.96 | 867 | 0.0046 |
Does PBS3 Impact SA Metabolism by Altering the Enzymatic Activity of SA Biosynthetic Enzymes?—One potential manner by which PBS3 could impact SA metabolism is by altering the enzymatic activity of SA biosynthetic enzymes. As the pbs3 mutants appear to be compromised in SA biosynthesis, if this hypothesis is correct, we would expect the following: 1) PBS3 substrate(s) inhibit SA biosynthetic activity and/or 2) PBS3 product(s) activate SA biosynthetic enzymes. In A. thaliana, pathogen-induced SA biosynthesis requires the inducible isochorismate synthase AtICS1 (29). AtICS1 catalyzes the reversible conversion of chorismate to isochorismate (22). Isochorismate is likely then converted to SA by an IPL with the concomitant release of pyruvate, as it is in bacteria that synthesize SA (23, 30). Therefore, we wanted to determine whether the PBS3 substrates 4-HBA or pABA or the PBS3 product pABA-Glu impacts the activity of known SA biosynthetic enzymes (AtICS1 and the P. aeruginosa isochorismate pyruvate lyase PchB). We used the P. aeruginosa PchB enzyme as a surrogate for a putative, yet-to-be identified, Arabidopsis IPL. All assessments were performed with substrate concentrations at the Km value for the respective enzyme. We employed a coupled spectrophotometric assay to assess the conversion of chorismate to isochorismate by AtICS1 (22) and found no statistically significant difference in activity with the addition of 200 μm 4-HBA, pABA, or pABA-Glu. This was also true when assessed with 1 mm 4-HBA, pABA, or pABA-Glu. Furthermore, we found that the conversion of isochorismate to SA by PchB was also unaffected by these compounds.
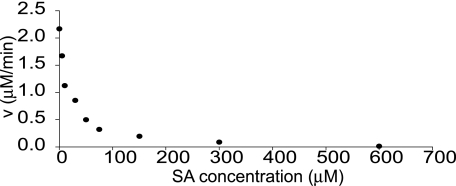
PBS3 Activity Is Inhibited by SA but Not by Other Phytohormones—As PBS3 impacts SA metabolism and the most active acyl substrates of PBS3 are structurally similar to SA, we then sought to determine whether SA could inhibit PBS3 activity. To test this, we assayed the effect of increasing concentrations of SA on PBS3-catalyzed pABA-Glu formation. As shown in Fig. 3, SA dramatically reduced the amino acid conjugation activity of PBS3 with an estimated IC50 of 15 μm. To determine whether the inhibition of PBS3 by SA was competitive, we added excess (saturating) pABA to the reaction mixture with 15 μm SA (the IC50). Full activity with maximal velocity was restored (data not shown). This suggests that SA acts as a competitive inhibitor of PBS3 activity.
FIGURE 3.
Inhibition of PBS3 activity by SA. SA inhibits the formation of pABA-Glu catalyzed by PBS3. The reaction velocities were determined by monitoring the formation of pABA-Glu by HPLC of PBS3 reactions with 150 μm pABA and 10 mm Glu. Independent experiments yielded similar results.
We then sought to determine whether 3-HBA or phytohormones, including MeSA (the mobile ester of SA) and the SA functional analog INA, could modulate PBS3 activity. For this, we used our high throughput adenylation assay. With this assay, 30 μm SA inhibited PBS3 activity by 33% and 300 μm SA inhibited it by 75% (Table 4). Although this observed inhibition was of reduced magnitude compared with that found using the full HPLC assay, we verified that the inhibition was due to the impact of SA on PBS3 activity by confirming that SA had no effect on activity of the coupling enzymes with PPi as the substrate. Of the six compounds tested, we found that only SA and the SA functional analog INA dramatically inhibited PBS3 activity (Table 4). IAA, JA, MeSA, and 3-HBA did not substantially inhibit PBS3 activity.
TABLE 4.
Fractional inhibition of pABA-AMP formation
| Compound | Fractional inhibitiona(30 μm) | Fractional inhibition (300 μm) |
|---|---|---|
| SA | 0.33 | 0.75 |
| MeSA | 0.02 | 0.00 |
| INA | 0.09 | 0.70 |
| IAA | 0.04 | 0.19 |
| JA | 0.02 | 0.10 |
| 3-HBA | 0.04 | 0.22 |
Fractional inhibition compares activity with additional compound to control reaction. Inhibition was assessed using the coupled adenylation assay with 150 μm pABA as the acyl substrate and 30 or 300 μm of the compound being tested. Repeat experiments with a separate enzyme batch gave similar results.
DISCUSSION
Insights into PBS3 Active Site and Reaction Mechanism—The GH3 enzymes differ from other members of the acyl-adenylate/thioester-forming superfamily in that amino acid conjugation occurs directly to the acyl-adenylate without the requirement for a thiol ester intermediate. Adenylation reactions, such as those catalyzed by this superfamily, require Mg2+ and ATP to complex with the phosphoester oxygens and enhance the electrophilicity of the phosphorus atoms. As expected, we found PBS3 activity requires Mg2+ and ATP. We also found alkaline pH is necessary for amino acid conjugation, but not the adenylation reaction, probably to deprotonate the attacking nucleophile, the α-NH+3 group of the amino acid substrate (pKa ∼9.5). This is consistent with the pH requirements of other characterized GH3 enzymes (12) but is not a hallmark of all superfamily members. The reactions catalyzed by other superfamily family members, such as CoA ligases, do not use an amino acid as the nucleophile, and thus an alkaline pH would not be required as it is for PBS3.
We are limited in our analysis of PBS3 by the lack of a crystal structure for a GH3 enzyme and by the limited sequence conservation with other acyl-adenylate/thioester-forming superfamily members. However, the AMP-binding motifs of PBS3 are evident, and the importance of these motifs has been clearly demonstrated for GH3 enzymes. For example, in JAR1, mutation of a conserved residue of AMP-binding motif I results in a complete loss of activity (7). To date, no other known motifs have been identified in PBS3 or other GH3 enzymes. However, our study of the impact of the point mutations contained in pbs3-1 on PBS3 activity, with loss of adenylation activity for both E502K and I519T PBS3 mutants, suggests that the highly conserved C-terminal region of PBS3, including loop residue Ile-519 (see supplemental Fig. S2), may participate in substrate binding or catalysis.
In terms of acyl substrate specificity, PBS3 had the highest velocity with 4-HBA and pABA (4-ABA) of the compounds tested (Table 2). However, bulkier groups at the para-position, such as 4-AMeBA or 4-AEtBA, were not well tolerated. Substitution at the meta- and ortho-positions led to substantially lower reaction velocity with ortho substitutions resulting in lack of activity, perhaps due to steric hindrance. Meta substitutions were tolerated and more electronegative substituents such as MeO provided a 4-fold enhancement of activity compared with –OH substitutions. In addition, our results comparing PBS3 activity with benzoate versus phenylacetate suggest that it is important for the carboxylate group to be attached directly to the aromatic ring, likely for resonance stabilization. The fact that PBS3 exhibited moderate activity with trans-cinnamate, which has a double bond between the carboxylic acid and aromatic ring, supports this conclusion. However, another explanation for the lack of activity with phenylacetate is that although the cinnamates and benzoates display a planar geometry, phenylacetate does not, and thus may be perceived quite differently by the active site.
The observed substrate preference of PBS3 for subsets of benzoates is similar to that observed for glycosyltransferases and CoA ligases active on benzoate substrates (31). Despite the clear preference for benzoates, PBS3 was also moderately active on trans-cinnamate (Table 2). This is unusual, as glycosyltransferases and CoA ligases active on benzoates have not been reported to have significant activity on cinnamates (32, 33). Similar to PBS3, the few characterized GH3 proteins have exhibited clear substrate preferences while remaining active on a fairly broad range of substrates. For example, Arabidopsis Group II GH3 proteins are capable not only of adenylating IAA in vitro but also adenylating other compounds with auxin activity, including indole-3-pyruvate, indole-3-butyrate, and the structurally distinct phenylacetate and naphthalene acetate (12). We also observed that SA inhibits both the adenylation and full (adenylation and amino acid conjugation) PBS3 reactions. Furthermore, the restoration of maximal PBS3 velocity when excess pABA is added to the reaction mixture with 15 μm SA suggests SA acts as a competitive inhibitor of PBS3. In this capacity, SA most likely competes with the acyl substrate (e.g. pABA) for the active site in a mutually exclusive fashion. Although less frequently observed, it is also possible that SA could act as an allosteric competitive inhibitor, binding a distinct site on PBS3.
Although PBS3 tolerates a range of benzoates as substrates, there is a strict requirement for Glu as the amino acid substrate with pABA. No activity was observed with the other 19 amino acids, including Asp which differs from Glu by only one methylene group. In addition, modified forms of Glu were inactive. This suggests a precise configuration of the active site for l-glutamic acid. The strict requirement for Glu is unusual among the few characterized GH3 enzymes, which are capable of conjugating the addition of multiple amino acids to the acyl substrate in vitro. For example, JAR1 catalyzes the addition of jasmonate to at least 11 amino acids and the ethylene precursor 1-amino-cyclopropane-1-carboxylate (7). JAR1 does not, however, appear to form Glu or Asp conjugates. The Arabidopsis Group II GH3 proteins also catalyze the formation of multiple IAA-amino acid conjugates in vitro, including those with Glu and Asp (12). Some of these IAA-conjugating proteins, such as GH3.17, exhibit a strong preference for either Glu or Asp (12). Surprisingly, with 4-HBA as the acyl substrate, PBS3 can utilize a number of amino acids, including Glu but not Asp (Fig. 2). What accounts for this difference in amino acid specificity? PBS3 exhibits ∼3-fold higher affinity for pABA than for 4-HBA (Table 3). Perhaps the pABA-AMP intermediate more precisely fits the active site, constraining acceptable amino acids for use in the conjugation reaction. In terms of in planta activity, this would depend upon the relative availability of acyl and amino acid substrates.
Kinetic Parameters of PBS3 and Physiological Relevance of Substrates—We found that PBS3 had the highest affinity for pABA (Km = 153 μm), compared with 4-HBA (459 μm) and BA (867 μm) (Table 3). One Km value that has been reported previously for a GH3 family member is that of JAR1 for (–)-jasmonic acid (34). This value, 190 μm, is similar to what we found for PBS3. The kcat for PBS3 with 4-HBA (20.36 min–1) was 7- and 5-fold higher than the kcat with pABA or BA, respectively. This resulted in similar catalytic efficiencies for 4-HBA and pABA, compared with that of BA, which was much lower. Taken together, these findings suggest 4-HBA and pABA as likely substrates in planta. However, to assess the relevance of our findings, we must consider the physiological concentrations of the substrates of PBS3. How do the Km values we have determined compare with physiological concentrations of these substrates?
In Arabidopsis, pathogen-induced BA levels have been rarely reported, although BA levels of ∼4 nmol/g FW, assessed using gas chromatography-MS, were detected at 2 days post-inoculation with a virulent P. syringae strain (35). As pathogen-induced BA levels have not been measured with sufficient temporal (or spatial) resolution to be confident in physiological concentrations, we also reviewed reports of the affinity of pathogen-inducible Arabidopsis enzymes for BA to provide insight into relevant physiological concentrations. The Arabidopsis glucosyltransferases with the highest activity and affinity for BA have reported apparent Km values for BA of 260 μm (31) and 540 μm (32) for UGT74F1 and 80 μm (31) for UGT74F2. The apparent Km value of the Arabidopsis SA/BA methyltransferase AtBSMT1 for BA is reported to be 65 μm (36). These affinities suggest that physiological concentrations of BA are insufficient to support efficient catalysis by PBS3 using BA as the acyl substrate. Therefore, given the low affinity (Km = 867 μm) and catalytic efficiency of PBS3 with BA, it appears that BA may not be the favored substrate of PBS3 in planta.
pABA-Glu has been detected in Arabidopsis leaves at 0.3 nmol/g FW and is thought to be present as a breakdown product of folates (37). Folates consist of three subunits as follows: a pterin moiety, pABA, and varying numbers of glutamic acids. They are essential cofactors in one-carbon transfer reactions, which are central to plant metabolism, and are involved in the synthesis of methionine, pantothenate, purines, pyrimidines, and thymidylate (38). It may be that pABA-Glu is not just a breakdown product of folate but is actually synthesized de novo in response to biotic stress. Although an increase in pABA or pABA-Glu has not been detected in response to pathogen attack in Arabidopsis, pABA-Glu is not likely to be efficiently isolated using standard procedures for the measurement of metabolites like SA (3, 39, 40). The apparent Km value of the pABA-glucosyltransferase for pABA was found to be 120 μm (41), which is quite similar to the value we found for PBS3. Perhaps the formation of pABA-Glu conjugates is a way for plants to restrict access of the pathogen to free pABA. Bacteria can commonly import pABA but few have been found that import pABA-Glu (42). Furthermore, there is precedent for pABA limitations on the growth of phytopathogens, as observed for growth of a folate auxotroph of Ralstonia solanacearum on tobacco (43). Therefore, pABA remains an attractive candidate as a potential substrate for PBS3 in planta.
4-HBA is typically known as an intermediate in the synthesis of ubiquinone, a lipid-soluble electron carrier of the mitochondrial electron transport chain (44). Therefore, all plants normally produce 4-HBA in small quantities. However, some plants use 4-HBA in the synthesis of specialized metabolites such as shikonin (45), and 4-HBA is often found associated with the plant cell wall, typically as a component of lignin (46). Pathogen (or elicitor-)-induced synthesis of 4-HBA and incorporation into the plant cell wall has been reported for a number of plants, including Arabidopsis (47) and carrot (48). For example, in Arabidopsis in response to virulent and avirulent P. syringae pv. tomato DC3000, 4-HBA accumulated to 0.2 nmol/mg cell wall by 72 hours post-infection (47) or ∼5 nmol/g FW (calculated using our milligrams of cell wall/g FW conversion factor obtained with this cell wall preparation protocol). Enzymes that use 4-HBA as a substrate include five putative Arabidopsis 4-HBA glucosyltransferases, which exhibit Km values of 210–290 μm for 4-HBA (31) and Lithospermum erythrorhizon geranyldiphosphate, 4-HBA geranyltransferases involved in shikonin synthesis, with reported Km values for 4-HBA ranging from 18.4 to 45.9 μm (45, 49). Therefore, 4-HBA remains an attractive candidate for the in planta substrate for PBS3, with 4-HBA/4-HBA-Glu perhaps playing a role in pathogen-induced cell wall remodeling.
As the analytical methods required to extract, enrich, and detect these putative conjugates are highly specific, our in vitro analysis now allows us to employ methods specific to these compounds and to the analysis of compounds incorporated into the cell wall. To date, in planta activities of GH3 enzymes appear consistent with in vitro findings. For example, IAA-Asp is one of the dominant IAA-amino acid conjugates formed by GH3.6 as evaluated by TLC, and overexpression of GH3.6 results in a 4.5-fold increase in IAA-Asp in Arabidopsis leaves (12). Similarly, jar1 mutants exhibit >7-fold reduction in JA-Ile levels, and JA-Ile is a dominant JA-amino acid conjugate observed by TLC (7). Consistent with the preference of PBS3 for Glu, the only endogenous benzoyl-amino acid conjugates detected in planta have been either Glu or Asp conjugates (e.g. pABA-Glu (37), 4-HBA-Glu (50), SA-Asp (51), and BA-Asp (28)). As PBS3 is not active with Asp, it will be interesting to determine whether another Arabidopsis GH3 family member exhibits overlapping acyl substrate specificity but altered AA preference, as is the case for the Arabidopsis GH3 family members acting on IAA (12). This possibility may also require the use of a PBS3 overexpresser to detect significantly altered levels of putative Glu conjugates.
How Does PBS3 Function Impact SA Metabolism and SA-dependent Gene Expression?—Previous analyses of pbs3/gdg1/win3 mutants by us and others showed that the lack of function of AtGH3.12 (PBS3) reduced total (free and glucose-conjugated SA (SAG)) pathogen-induced SA accumulation, activation of SA-dependent gene expression, and disease resistance (3, 52, 53). Furthermore, exogenous application of SA to gh3.12 mutants induced the expression of SA-dependent genes such as PR1 and rescued the enhanced disease susceptibility phenotype (3, 52). Therefore, the most likely position for PBS3 function would be upstream of SA biosynthesis. The action of PBS3 upstream of SA rather than on SA itself is further supported by the observed substrate specificity of PBS3, with PBS3 being inactive on SA (Tables 1 and 2). It is not likely that PBS3 functions through modulation of the activity of SA biosynthetic enzymes, as we found the activity of AtICS1 and a surrogate IPL were not affected by favored PBS3 substrates or products. Furthermore, if PBS3 acted in this way to promote SA biosynthesis and subsequent formation of the SAG conjugate, it would not make sense for PBS3 activity to then be inhibited by low concentrations of SA (IC50 = 15 μm SA). Of interest, AtBSMT1, which catalyzes the formation of MeSA from SA, functions at these low concentrations of SA (Km = 16 μm for SA) (36). In contrast, the Km value of the pathogen-induced Arabidopsis SA glucosyltransferase 1 (UGT 74F1; At2g43840), which converts free SA to SAG, is 190–230 μm for SA (31, 32), suggesting higher concentrations of SA are required for SAG formation. In situ quantification of free SA has not been reported in Arabidopsis. In tobacco, tobacco mosaic virus inoculation resulted in free SA accumulation of 6.5 μm at 16 hpi with maximal detected local concentrations of >200 μm at 40 hpi (54).
Therefore, it is likely that PBS3 functions upstream of SA, early in the defense response when SA levels are low. In this scenario, PBS3 activity leads to conjugate formation (e.g. 4HBA-Glu), which signals or primes SA biosynthesis. PBS3 activity is then no longer required once SA synthesis has been sufficiently initiated. The inhibition of PBS3 by SA serves as a mechanism to rapidly reduce synthesis of the conjugate when it is no longer necessary for SA priming. In this scenario, we would expect a reduced conjugate (e.g. 4-HBA-Glu) concentration as the defense response progresses. A role for PBS3 upstream of SA in a regulatory capacity is also supported by recent expression profiling comparing expression of wild type and a variety of Arabidopsis mutants in defense signaling at 24 hpi with P. syringae pv. maculicola ES4326 using a custom mini array (55). In this study, PBS3 was placed upstream of SA and NPR1, as the pbs3 mutation impacted the expression of many more genes than the standard SA biosynthesis and signaling mutants ics1(sid2), eds5, and npr1 (55). Of interest, the expression of chalcone synthase, a key enzyme directing flux to flavonoid biosynthesis from 4-coumaroyl-CoA, was strongly down-regulated at 24 hpi with P. syringae pv. maculicola in wild type and ics1 but was up-regulated in the pbs3 mutant (55). In other systems such as elicited carrot cell cultures, up-regulation of 4-HBA synthesis and enhanced flux through the general phenylpropanoid pathway was associated with down-regulation of flavonoid biosynthesis from 4-coumaroyl-CoA (56). This suggests that flux from 4-coumaroyl-CoA to phenylpropanoid cell wall precursors may be up-regulated to facilitate the observed cell wall remodeling. Therefore, it is possible that PBS3 coordinately modulates pathogen-induced SA, 4-HBA, and their associated responses with 4-HBA playing a role in cell wall remodeling and the direction of chorismate flux (from 4-coumaroyl-CoA) for this purpose. Future work will address this possibility and potential mechanisms of action. For example, given the known function of phytohormone-AA conjugates in promoting or inhibiting the ubiquitin ligase-dependent proteasomic degradation of the associated transcriptional repressor, it is tempting to speculate that 4-HBA-Glu may act in a similar manner. With our knowledge of the in vitro specificity of PBS3 gained through this study, potential PBS3 products could be used in assays with candidate F-box proteins and repressors to identify F-box-repressor interactions mediating SA and/or 4-HBA-associated signaling (similar to Ref. 8 for JA). This is critical as these interactions require binding of the appropriate molecule, e.g. JA-Ile for JA signaling.
In summary, this study further supports the critical role of amino acid conjugation of small molecules in phytohormone regulation and homeostasis. To date, this conjugation is catalyzed by members of the GH3 enzyme family. Here we characterize the in vitro activity of the first GH3 subfamily III member to be analyzed, PBS3 (AtGH3.12). In the process, we developed a high throughput adenylation assay to screen for preferred acyl substrates of GH3 enzymes. This high throughput assay can be applied to the identification of substrates for other GH3 enzymes for which no acyl substrates have been identified or inferred through analysis of mutant phenotypes. We also determine kinetic parameters for recombinant PBS3 and demonstrate that PBS3 is specifically inhibited by low concentrations of SA. This is important as it presents a novel mechanism for rapidly and reversibly fine-tuning the activity of small molecules, including phytohormones. It could also be a mechanism for rapid and reversible cross-talk between phytohormones or other small molecules, such as SA and 4-HBA. Our in vitro analyses of PBS3 suggest a potential, underexplored role for 4-HBA (and/or pABA) and their respective amino acid conjugates in plantmicrobe interactions and provide a framework to explore the mechanism by which PBS3 impacts pathogen-induced SA accumulation and disease resistance. In addition, using knowledge of the substrate preferences and biochemical and kinetic properties of PBS3, one could test and optimize PBS3 for use in biotechnological applications. For example, PBS3 might be used to remove and purify 4-HBA, an inhibitory by-product of cellulosic ethanol production (57, 58). 4-HBA, hydrolyzed from the 4-HBA-AA conjugate, could then be sold to the liquid crystal polymer industry, as it is a preferred monomer used in the manufacture of liquid crystal polymers (59).
Acknowledgments
We thank Marcus Strawn (Wildermuth laboratory) for purified isochorismate, recombinant Arabidopsis isochorismate synthase (AtICS1), recombinant P. aeruginosa isochorismate pyruvate lyase (PchB), and valuable discussions regarding this research. We thank Eric Simmons and Dr. Richmond Sarpong (University of California, Berkeley) for their generous assistance with synthesis of 4-HBA-Glu. We thank Dr. Allis Chien and Theresa McLaughlin of the Vincent Coates Foundation Mass Spectrometry Laboratory (Stanford University) for mass spectrometry analyses, Dr. Chloe Zubieta for creating the single PBS3 mutant constructs, and Dr. Krishna Niyogi (University of California, Berkeley) for use of the Spectramax Plus microplate spectrophotometer (Molecular Devices). We also thank M. Strawn and Drs. K. Niyogi and D. Burgess for their comments on the manuscript.
Author's Choice—Final version full access.
This work was supported by National Science Foundation Arabidopsis 2010 Grant MCB-0420267 (to M. C. W.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods S1 and Figs. S1 and S2.
Footnotes
The abbreviations used are: PBS3, avrPphB susceptible 3 (GH3.12); AA, amino acid; BA, benzoate; 4-HBA, 4-hydroxybenzoate; FW, fresh weight; IAA, indole-3-acetate; JA, jasmonate; pABA, para-aminobenzoate; SA, salicylate; MeSA, methyl salicylate; SAG, SA glucoside; ICS1, isochorismate synthase 1; IPL, isochorismate pyruvate lyase; Q-TOF, quadrupole time of flight; MS/MS, tandem mass spectrometry; DTT, dithiothreitol; HPLC, high pressure liquid chromatography; 4-AEtBA, 4-aminoethylbenzoate; LC, liquid chromatography; INA, 2,6-dichloroisonicotinate.
M. Estelle, personal communication.
References
- 1.Glazebrook, J. (2001) Curr. Opin. Plant Biol. 4 301–308 [DOI] [PubMed] [Google Scholar]
- 2.Warren, R. F., Merritt, P. M., Holub, E., and Innes, R. W. (1999) Genetics 152 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nobuta, K., Okrent, R. A., Stoutemyer, M., Rodibaugh, N., Kempema, L., Wildermuth, M. C., and Innes, R. W. (2007) Plant Physiol. 144 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagen, G., Kleinschmidt, A., and Guilfoyle, T. (1984) Planta 162 147–153 [DOI] [PubMed] [Google Scholar]
- 5.Terol, J., Domingo, C., and Talon, M. (2006) Gene (Amst.) 371 279–290 [DOI] [PubMed] [Google Scholar]
- 6.Staswick, P. E., Tiryaki, I., and Rowe, M. L. (2002) Plant Cell 14 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staswick, P. E., and Tiryaki, I. (2004) Plant Cell 16 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., Nomura, K., He, S. Y., Howe, G. A., and Browse, J. (2007) Nature 448 661–665 [DOI] [PubMed] [Google Scholar]
- 9.Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., Garcia-Casado, G., Lopez-Vidriero, I., Lozano, F. M., Ponce, M. R., Micol, J. L., and Solano, R. (2007) Nature 448 666–671 [DOI] [PubMed] [Google Scholar]
- 10.Yan, Y., Stolz, S., Chetelat, A., Reymond, P., Pagni, M., Dubugnon, L., and Farmer, E. E. (2007) Plant Cell 19 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, H. S., Koo, A. J., Gao, X., Jayanty, S., Thines, B., Jones, A. D., and Howe, G. A. (2008) Plant Physiol. 146 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M. T., Maldonado, M. C., and Suza, W. (2005) Plant Cell 17 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodward, A. W., and Bartel, B. (2005) Ann. Bot. 95 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005) Nature 435 441–445 [DOI] [PubMed] [Google Scholar]
- 15.Kepinski, S., and Leyser, O. (2005) Nature 435 446–451 [DOI] [PubMed] [Google Scholar]
- 16.Glass, N. L., and Kosuge, T. (1988) J. Bacteriol. 170 2367–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brooks, D. M., Hernandez-Guzman, G., Kloek, A. P., Alarcon-Chaidez, F., Sreedharan, A., Rangaswamy, V., Penaloza-Vazquez, A., Bender, C. L., and Kunkel, B. N. (2004) Mol. Plant-Microbe Interact. 17 162–174 [DOI] [PubMed] [Google Scholar]
- 18.Lee, M. W., Jelenska, J., and Greenberg, J. T. (2008) Plant J. 54 452–465 [DOI] [PubMed] [Google Scholar]
- 19.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (eds) (2005) Current Protocols in Molecular Biology, John Wiley & Sons, Inc., New York
- 20.O'Brien, W. E. (1976) Anal. Biochem. 76 423–430 [DOI] [PubMed] [Google Scholar]
- 21.Rudolph, F. B., and Fromm, H. J. (1979) Methods Enzymol. 63 138–159 [DOI] [PubMed] [Google Scholar]
- 22.Strawn, M. A., Marr, S. K., Inoue, K., Inada, N., Zubieta, C., and Wildermuth, M. C. (2007) J. Biol. Chem. 282 5919–5933 [DOI] [PubMed] [Google Scholar]
- 23.Gaille, C., Kast, P., and Haas, D. (2002) J. Biol. Chem. 277 21768–21775 [DOI] [PubMed] [Google Scholar]
- 24.Gajewski, J. J., Jurayj, J., Kimbrough, D. R., Gande, M. E., Ganem, B., and Carpenter, B. K. (1987) J. Am. Chem. Soc. 109 1170–1186 [Google Scholar]
- 25.Gibson, F. (1964) Biochem. J. 90 256–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holden, M. J., Mayhew, M. P., Gallagher, D. T., and Vilker, V. L. (2002) Biochim. Biophys. Acta 1594 160–167 [DOI] [PubMed] [Google Scholar]
- 27.Bourne, D. J., Barrow, K. D., and Milborrow, B. V. (1991) Phytochemistry 30 4041–4044 [Google Scholar]
- 28.Suzuki, Y., Yamaguchi, I., Murofushi, N., and Takahashi, N. (1988) Plant Cell Physiol. 29 439–444 [Google Scholar]
- 29.Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001) Nature 414 562–565 [DOI] [PubMed] [Google Scholar]
- 30.Serino, L., Reimmann, C., Baur, H., Beyeler, M., Visca, P., and Haas, D. (1995) Mol. Gen. Genet. 249 217–228 [DOI] [PubMed] [Google Scholar]
- 31.Lim, E. K., Doucet, C. J., Li, Y., Elias, L., Worrall, D., Spencer, S. P., Ross, J., and Bowles, D. J. (2002) J. Biol. Chem. 277 586–592 [DOI] [PubMed] [Google Scholar]
- 32.Song, J. T. (2006) Mol. Cells 22 233–238 [PubMed] [Google Scholar]
- 33.Beuerle, T., and Pichersky, E. (2002) Arch. Biochem. Biophys. 400 258–264 [DOI] [PubMed] [Google Scholar]
- 34.Suza, W. P., and Staswick, P. E. (2008) Planta 227 1221–1232 [DOI] [PubMed] [Google Scholar]
- 35.Schmelz, E. A., Engelberth, J., Alborn, H. T., O'Donnell, P., Sammons, M., Toshima, H., and Tumlinson, J. H., III. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10552–10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen, F., D'Auria, J. C., Tholl, D., Ross, J. R., Gershenzon, J., Noel, J. P., and Pichersky, E. (2003) Plant J. 36 577–588 [DOI] [PubMed] [Google Scholar]
- 37.Orsomando, G., Bozzo, G. G., de la Garza, R. D., Basset, G. J., Quinlivan, E. P., Naponelli, V., Rebeille, F., Ravanel, S., Gregory, J. F., III, and Hanson, A. D. (2006) Plant J. 46 426–435 [DOI] [PubMed] [Google Scholar]
- 38.Hanson, A. D., and Roje, S. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 119–137 [DOI] [PubMed] [Google Scholar]
- 39.Zhang, G.-F., Mortier, K. A., Storozhenko, S., Van De Steene, J., Van Der Straeten, D., and Lambert, W. E. (2005) Rapid Commun. Mass Spectrom. 19 963–969 [DOI] [PubMed] [Google Scholar]
- 40.Quinlivan, E. P., Hanson, A. D., and Gregory, J. F. (2006) Anal. Biochem. 348 163–184 [DOI] [PubMed] [Google Scholar]
- 41.Eudes, A., Bozzo, G. G., Waller, J. C., Naponelli, V., Lim, E. K., Bowles, D. J., Gregory, J. F., III, and Hanson, A. D. (2008) J. Biol. Chem. 283 15451–15459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hussein, M. J., Green, J. M., and Nichols, B. P. (1998) J. Bacteriol. 180 6260–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shinohara, R., Kanda, A., Ohnishi, K., Kiba, A., and Hikichi, Y. (2005) Appl. Environ. Microbiol. 71 417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swiezewska, E. (2004) Methods Enzymol. 378 124–131 [DOI] [PubMed] [Google Scholar]
- 45.Yazaki, K., Kunihisa, M., Fujisaki, T., and Sato, F. (2002) J. Biol. Chem. 277 6240–6246 [DOI] [PubMed] [Google Scholar]
- 46.Lu, F., Ralph, J., Morreel, K., Messens, E., and Boerjan, W. (2004) Org. Biomol. Chem. 2 2888–2890 [DOI] [PubMed] [Google Scholar]
- 47.Tan, J., Bednarek, P., Liu, J., Schneider, B., Svatos, A., and Hahlbrock, K. (2004) Phytochemistry 65 691–699 [DOI] [PubMed] [Google Scholar]
- 48.Veit, S., Woerle, J. M., Nuernberger, T., Koch, W., and Seitz, H. U. (2001) Plant Physiol. 127 832–841 [PMC free article] [PubMed] [Google Scholar]
- 49.Muhlenweg, A., Melzer, M., Li, S. M., and Heide, L. (1998) Planta 205 407–413 [DOI] [PubMed] [Google Scholar]
- 50.Trennheuser, F., Burkhard, G., and Becker, H. (1994) Phytochemistry 37 899–903 [Google Scholar]
- 51.Steffan, H., Ziegler, A., and Rapp, A. (1988) Vitis 27 79–86 [Google Scholar]
- 52.Jagadeeswaran, G., Raina, S., Acharya, B. R., Maqbool, S. B., Mosher, S. L., Appel, H. M., Schultz, J. C., Klessig, D. F., and Raina, R. (2007) Plant J. 51 234–246 [DOI] [PubMed] [Google Scholar]
- 53.Lee, M. W., Lu, H., Jung, H. W., and Greenberg, J. T. (2007) Mol. Plant-Microbe Interact. 20 1192–1200 [DOI] [PubMed] [Google Scholar]
- 54.Huang, W. E., Huang, L., Preston, G. M., Naylor, M., Carr, J. P., Li, Y., Singer, A. C., Whiteley, A. S., and Wang, H. (2006) Plant J. 46 1073–1083 [DOI] [PubMed] [Google Scholar]
- 55.Wang, L., Mitra, R. M., Hasselmann, K. D., Sato, M., Lenarz-Wyatt, L., Cohen, J. D., Katagiri, F., and Glazebrook, J. (2008) Mol. Plant-Microbe Interact. 21 1408–1420 [DOI] [PubMed] [Google Scholar]
- 56.Gleitz, J., Schnitzler, J. P., Steimle, D., and Seitz, H. U. (1991) Planta 184 362–367 [DOI] [PubMed] [Google Scholar]
- 57.Palmqvist, E., and Hahn-Hagerdal, B. (2000) Bioresource Technol. 74 25–33 [Google Scholar]
- 58.Palmqvist, E., Grage, H., Meinander, N. Q., and Hahn-Hagerdal, B. (1999) Biotechnol. Bioeng. 63 46–55 [DOI] [PubMed] [Google Scholar]
- 59.Figuly, G. (1996) in Encyclopedia (Salamone, J., ed) CRC Press, Boca Raton, FL