Abstract
Transforming growth factor-β (TGFβ) superfamily ligands control a diverse set of cellular processes by activating type I and type II serine-threonine receptor kinases. Canonical TGFβ signaling is mediated via the TβRI/ALK5 type I receptor that phosphorylates Smad2 and Smad3 in their SXS motif to facilitate their activation and subsequent role in transcriptional regulation. Canonical bone morphogenic protein (BMP) signaling is mediated via the ALK1/2/3/6 type I receptors that phosphorylate Smad1, Smad5, and Smad8 in their SXS motif. However, studies in endothelial cells have shown that TGFβ can also lead to the phosphorylation of Smad1, dependent on ALK1 receptor activity. Here we present data showing that TGFβ can significantly induce Smad1 phosphorylation in several non-endothelial cell lineages. Additionally, by using chemical inhibitors specific for the TGFβ/activin/nodal (ALK4/5/7) and BMP (ALK1/2/3/6) type I receptors, we show that in some cell types TGFβ induces Smad1 phosphorylation independently of the BMP type I receptors. Thus, TGFβ-mediated Smad1 phosphorylation appears to occur via different receptor complexes in a cell type-specific manner.
Transforming growth factor (TGF)2 β superfamily members signal through heteromeric complexes of transmembrane serine/threonine kinase receptors to control diverse developmental processes and the pathogenesis of many diseases. The heteromeric receptor complex usually comprises two type II receptors and two type I receptors, and the receptors are classified based on their structural and functional properties. Type II receptors are constitutively active, and they phosphorylate type I receptors on serine/threonine residues in the GS domain in response to ligand binding. Activated type I receptors then phosphorylate specific downstream receptor-activated Smad (R-Smads) proteins in their distal C-terminal SXS motif, activating them to transduce the signal to the nucleus (1, 2).
There are five mammalian type II receptors: TβRII, ActR-II, ActR-IIB, BMPR-II, AMHR-II, and seven type I receptors, which are referred to as activin receptor-like kinases (ALK) 1–7 (1, 3). Of the ALKs, ALK5 is thought to be specific for TGFβ ligands, and ALK4 and ALK7 are thought to mediate signaling via nodal and activins (2). In canonical signaling, the activated ALK4/ALK5/ALK7 phosphorylate downstream R-Smads 2 and 3. The remaining ALKs, ALK1/2/3/6, phosphorylate R-Smads 1, 5, and 8 following specific activation by bone morphogenic proteins (BMP) (2). Once activated, R-Smads undergo heterooligomeric complex formation with Smad4, named the common Smad due to its role in all branches of TGFβ superfamily signaling, and this complex accumulates in the nucleus to regulate gene transcription in conjunction with a variety of transcriptional cofactors (1, 2).
The specificity of different ALKs for different Smad proteins is based on the L45 loop and phosphorylated GS motif in the specific type I receptor, and the L3 loop in the MH2 domain of the relevant R-Smad (4, 5). The L45 loops of ALK4/5/7 that phosphorylate Smad2/3 are completely conserved, but they differ by four amino acids from the L45 sequences in ALK3 and ALK6, and by seven amino acids from the L45 loops of ALK1 and ALK2 (4, 6). The L45 loop of the ALK interacts directly with the L3 loop in the MH2 domain of the relevant R-Smad (4, 5).
Although it is generally accepted that TGFβ induces Smad2/3 phosphorylation, several studies have shown that TGFβ can also induce Smad1/5 phosphorylation in endothelial cells (7, 8). The current model proposes that the type I receptor ALK1 is necessary for this phosphorylation event, in addition to ALK5 activity and the TGFβ transmembrane accessory receptor Endoglin (7–9). Similarly, in dermal fibroblasts TGFβ can induce Smad1 phosphorylation dependent on both ALK5 and ALK1 activity, although this also appears to require activation of the ERK1/2 pathway for a sustained response (10). It has recently been shown that TGFβ can also stimulate Smad1/5 phosphorylation in HaCaT keratinocytes dependent on ALK5 activity, and this is also thought to depend on a canonical activator of Smad1, most likely ALK2 (11). Thus, although there have been several reports that TGFβ can induce Smad1 phosphorylation, revealing cross-talk between the TGFβ ligand and the BMP R-Smads, this phosphorylation event is largely thought to be dependent on the activity of a BMP type I receptor.
During our studies we observed TGFβ-induced Smad1 in several cell lines for which this has previously not been reported, including C2C12 mouse mesenchymal stem cells and HepG2 human hepatocellular carcinoma cells. Significantly, we found that in these cell lines TGFβ leads to Smad1 phosphorylation independently of ALK1/2/3/6, exclusively relying on the ALK4/5/7 subset as a type I receptor. This is in line with a study published while our manuscript was in revision, which revealed ALK5-mediated TGFβ-induced Smad1 phosphorylation in mammary epithelial cells (12). Our data provide firm evidence that TGFβ-induced Smad1 phosphorylation can occur independently of involvement of one of its canonical BMP type I receptors.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment—C2C12 mouse mesenchymal cells, HEK293T human embryonic kidney cells, and COS monkey kidney cells were cultured in Dulbecco's modified Eagle's medium, 10% fetal bovine serum. Wild-type and Smad3 null mouse embryo fibroblast cells (13) were cultured in minimal essential medium, 15% fetal bovine serum. HepG2 human hepatocellular carcinoma cells and human HaCaT keratinocyte cells were cultured in minimal essential medium, 10% fetal bovine serum. The ALK4/5/7-specific inhibitor SB431542 (Calbiochem) (14) was dissolved as a 1 mm stock in dimethyl sulfoxide. The ALK2/3/6-specific inhibitor dorsomorphin (DM) was described previously (15). Where indicated, cells were simultaneously treated with ligand (TGFβ or BMP) and 5 μm SB431542 or 5 μm DM for 1.5 h prior to harvest. Treatments were carried out in reduced serum media (0.2% fetal bovine serum) with TGFβ1 (R&D Systems) added to a final concentration of 2 ng/ml, and BMP2 (R&D Systems) to a final concentration of 25 ng/ml, unless otherwise specified.
Expression Plasmids and Transfection—FLAG-Smads and His-tagged constitutively active ALK5 (caALK5; also called TβRI(T202D) for rat) have been previously described (16). HA-tagged caALK5 was generated by subcloning His-caALK5. HA-tagged constitutively active ALK1 (HA-caALK1) was a gift from Dr. Peter ten Dijke. HEK293T cells were transfected using Lipofectamine Plus (Invitrogen), HepG2 cells using Lipofectin (Invitrogen), and C2C12 cells using Lipofectamine 2000 (Invitrogen). C2C12 cells were also nucleofected using the Amaxa Biosystems Nucleofector™ as described below.
Western Blotting and Immunoprecipitation—For direct Western blotting of whole cell lysates, cells were harvested in 2× Laemmli sample buffer. Immunoprecipitation for the detection of exogenous Smad phosphorylation, and caALK5-Smad interactions, was performed essentially as described previously (17). Western blotting was performed using anti-Smad and anti-phospho-Smad antibodies. Specifically, total Smad levels were assessed by using anti-Smad1 (Zymed Laboratories Inc.), anti-Smad2 (Zymed Laboratories Inc.), anti-Smad3 (Zymed Laboratories Inc.), or anti-Smad5 (Santa-Cruz) antibodies. Smad SXS motif phosphorylation was assessed by using anti-P-Smad1 (Cell Signaling), anti-P-Smad2 (Cell Signaling) antibodies, or anti-P-Smad3 sera (a kind gift from Dr. Ed Leof, Mayo Clinic). Epitope-tagged proteins were detected using anti-HA (Mouse 1.1; Babco), anti-FLAG (M2; Sigma), anti-His (Serotec), and anti-GST antibodies (Babco). Equal protein loading was confirmed where appropriate using anti-β-actin (Sigma).
RNA Interference—Nucleofection was used to examine the effect of siRNA-mediated Smad1 knockdown on TGFβ-induced phospho-Smad1. C2C12 cells were nucleofected with siControl or siSmad1 (RNA: CCG GAA GGG ACU ACC UCA UGU GAU U) (Invitrogen; Stealth™ siRNA) using the Amaxa Biosystems Nucleofector™ with solution V and program B-032 (Amaxa Inc.). After 48 h, cells were cultured in the presence or absence of TGFβ or BMP for 1 h prior to harvest for Western blotting.
Transcription Reporter Assays—To assess the effect of Smad1 knockdown on TGFβ-induced transcription, C2C12 cells were nucleofected with siControl or siSmad1 as described above. After 24 h cells were transfected with the reporter 3TP-lux, which contains TGFβ-responsive elements of the plasminogen activator inhibitor-1 and collagenase promoters (18). Following a further 24-h incubation, cells were cultured in the presence or absence of TGFβ for 9.5 h prior to harvest for luciferase assay, as previously described (17), and Western blotting.
To assess the ability of DM to inhibit ALK1 kinase activity, COS cells grown to 50% confluence were transiently co-transfected with plasmids encoding an Id1 promoter luciferase reporter (kindly provided by Dr. Peter ten Dijke, Leiden University Medical Center, Netherlands) (19), and the constitutively active form of BMP type I receptor ALK1 (caALK1), using FuGENE HD (Roche). Transfection with pRL-TK Renilla luciferase (Promega) was used to control for transfection efficiency. Cells were incubated with DM (0–8 μm) or vehicle starting 1 h after transfection. Cell extracts were harvested after 24 h, and relative promoter activity quantitated by the ratio of firefly to Renilla luciferase activity using the dual luciferase assay kit (Promega).
In Vitro Kinase Assays—Assays were performed largely as previously described (12). In brief, His-caALK5 was immunoprecipitated from transfected HEK293T cells after lysis in FLAG lysis buffer (25 mm Tris-HCl (pH 7.5), 300 mm NaCl, 1% Triton X-100, with protease inhibitors (aprotinin, leupeptin, and phenylmethylsulfonyl fluoride) and phosphatase inhibitors (sodium fluoride and Na3VO4)). His-caALK5 bound protein-A beads were washed three times in wash buffer (10 mm Tris, pH 8, 150 mm NaCl, 1% Nonidet P-40) and twice in kinase buffer (5 mm Tris, pH 7.4, 1 mm magnesium chloride, and 0.1 mm calcium chloride). Precipitated His-caALK was subsequently incubated for 45 min at ambient temperature with GST-Smad1 or GST-Smad3 (a gift from Dr. Xiao-Fan Wang, Duke University), in kinase buffer containing 50 μm ATP. Products were analyzed by Western blotting.
RESULTS
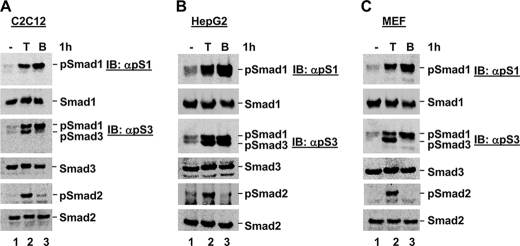
TGFβ Stimulates Smad1 Phosphorylation in a Variety of Cell Lines—During routine studies in our laboratory, we observed that TGFβ may induce Smad1 phosphorylation in various cell types, in-line with several previous reports that it can do so in endothelial cells (7, 8), dermal fibroblasts (10), and HaCaT keratinocytes (11). In an effort to examine this in more detail, we treated several cell lines in the presence or absence of TGFβ for 1 h, in parallel with BMP as a control for Smad1 phosphorylation, and harvested cells for Western blot analysis of total Smad and phospho-Smad proteins. As shown in Fig. 1, we detected a protein species migrating at the size of phospho-Smad1 in response to both TGFβ and BMP treatment in C2C12 (Fig. 1A), HepG2 (Fig. 1B), and mouse embryonic fibroblast (MEF) cells (Fig. 1C). This band was detected using an anti-phospho-Smad1 antibody, as well as an anti-phospho-Smad3 sera, which can recognize both phospho-Smad3 and phospho-Smad1. These data suggest that TGFβ might induce Smad1 phosphorylation, as well as the anticipated phosphorylation of Smad2 and Smad3, in the cell lines tested.
FIGURE 1.
TGFβ stimulates Smad1 phosphorylation in a variety of cell lines. A, TGFβ induces Smad1 phosphorylation in C2C12 cells. C2C12 cells were cultured in the presence or absence of TGFβ or BMP for 1 h, and harvested for Western blot analysis with anti-Smad and anti-phospho-Smad antibodies as detailed under “Experimental Procedures.” B, TGFβ induces Smad1 phosphorylation in HepG2 cells. HepG2 cells were cultured in the presence or absence of TGFβ or BMP for 1 h, and harvested for Western blot analysis as in A. C, TGFβ induces Smad1 phosphorylation in MEF cells. MEF cells were cultured in the presence or absence of TGFβ or BMP for 1 h, and harvested for Western blot analysis as in A. IB, immunoblot.
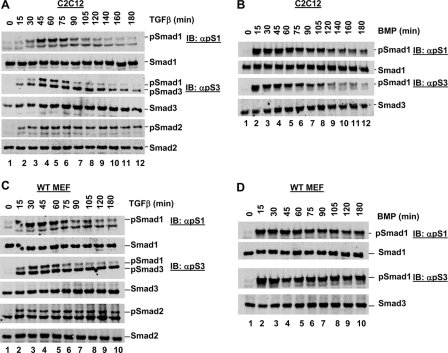
TGFβ-induced Smad1 Phosphorylation Is Time-dependent—We next examined the kinetics of TGFβ-induced Smad1 phosphorylation as compared with those of TGFβ-induced Smad3 phosphorylation and BMP-induced Smad1 phosphorylation. C2C12 cells were treated with TGFβ over a time course of up to 180 min, and cell lysates were analyzed by Western blotting. Smad3 phosphorylation was observed after just 15 min of TGFβ stimulation (Fig. 2A, lane 2), with the level increasing until around the 105-min time point. The TGFβ-induced Smad3 phosphorylation level began to decline at around 120 min (Fig. 2A, lane 9), although a significant amount remained for the duration of the time course. In contrast, TGFβ-induced Smad1 phosphorylation was not detected until 45 min post-treatment (Fig. 2A, lane 4), and no significant level was detected after the 75-min time point (Fig. 2A, lane 6). Thus, TGFβ-induced Smad1 phosphorylation is transient and first occurs at a later time than TGFβ-induced Smad3 phosphorylation, which is sustained over the treatment time course. BMP-induced Smad1 phosphorylation in C2C12 cells occurred earlier than TGFβ-induced Smad1, after just 15 min stimulation (Fig. 2B, lane 2), and was also more sustained only decreasing to a low level after around 120 min (Fig. 2B, lane 9). We observed a similar response in MEFs where TGFβ-induced Smad1 phosphorylation was only apparent between 30 and 75 min post-TGFβ stimulation (Fig. 2C), as compared with TGFβ-induced Smad3 phosphorylation, which remained robust from 15 to 180 min (Fig. 2C). BMP-induced Smad1 phosphorylation peaked at just 15 min BMP treatment in MEFs, remaining fairly high for the duration of the time course (Fig. 2D). Thus in C2C12 cells and MEFs, TGFβ-induced Smad1 phosphorylation peaks at a later time than both TGFβ-induced Smad3 phosphorylation and BMP-induced Smad1 phosphorylation, and is apparently more transient in nature.
FIGURE 2.
TGFβ-induced Smad1 phosphorylation is time-dependent. A, TGFβ time course of C2C12 cells. C2C12 cells were cultured in the presence of TGFβ for the indicated time, prior to harvest for Western blot analysis as in described in the legend to Fig. 1A. B, BMP time course of C2C12 cells. C2C12 cells were cultured in the presence of BMP for the indicated time, prior to harvest for Western blot analysis as described in the legend to Fig. 1A. C, TGFβ time course of MEF cells. MEF cells were cultured in the presence of TGFβ for the indicated time, prior to harvest for Western blot analysis as described in the legend to Fig. 1A. D, BMP time course of MEF cells. MEF cells were cultured in the presence of BMP for the indicated time, prior to harvest for Western blot analysis as described in the legend to Fig. 1A. WT, wild type. IB, immunoblot.
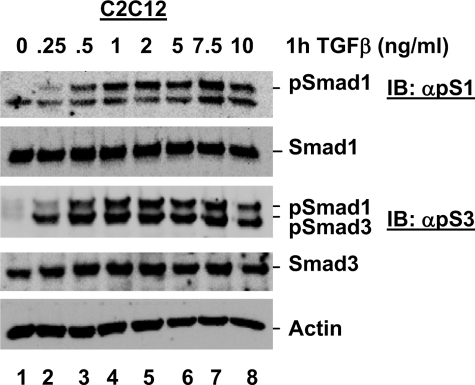
We next assessed the concentration of TGFβ required to stimulate Smad1 and Smad3 phosphorylation in C2C12 cells. Smad3 phosphorylation was induced by 0.25 ng/ml TGFβ, with levels reaching their maximum at 0.5 ng/ml (Fig. 3). However, significant Smad1 phosphorylation was not observed until treatment with 0.5 ng/ml TGFβ, with levels reaching their maximum at 1 ng/ml (Fig. 3). Thus, TGFβ-induced Smad1 phosphorylation requires a higher concentration of TGFβ than TGFβ-induced Smad3 phosphorylation in C2C12 cells. Taken together these kinetic data suggest that, due to the different timing, duration length, and minimal TGFβ concentration required for TGFβ-induced Smad1 phosphorylation, it may play a distinct role in TGFβ-induced phospho-Smad2/3 and BMP-induced phospho-Smad1 in cells.
FIGURE 3.
TGFβ-induced Smad1 phosphorylation is dose-dependent. C2C12 cells were cultured in the presence of varying concentrations of TGFβ, as indicated, and harvested for Western blot analysis as described in the legend to Fig. 1A. IB, immunoblot.
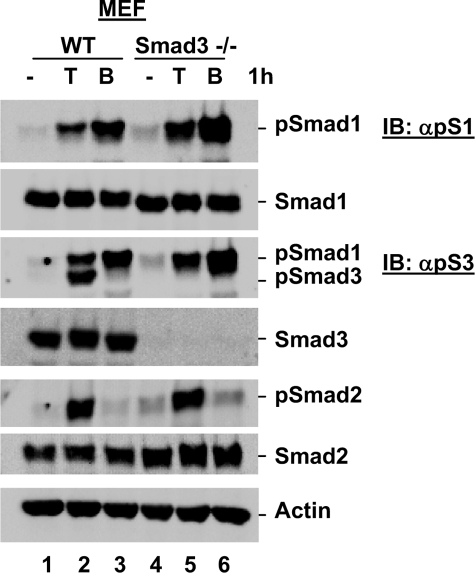
Genetic Depletion of Smads Confirms Smad1 Is Phosphorylated in Response to TGFβ—We next sought to confirm that the suspected phospho-Smad1 band detected by anti-phospho-Smad3 and anti-phospho-Smad1 antibodies in response to TGFβ does indeed correspond to phospho-Smad1. As this protein species can be detected by the anti-phospho-Smad3 antibody, we first utilized Smad3-null MEFs (13) to rule out that this protein corresponds to a modified version of Smad3. Wild-type and Smad3-null MEFs were cultured in the presence or absence of TGFβ or BMP for 1 h prior to harvest for Western blot analysis. As shown in Fig. 4, TGFβ induced Smad3 phosphorylation, Smad2 phosphorylation, and the suspected Smad1 phosphorylation in wild-type MEFs (Fig. 4, lane 2), as we previously observed for this cell line. Although no TGFβ-induced Smad3 phosphorylation was observed in Smad3-null MEFs, in line with the complete loss of total Smad3 protein, Smad2 phosphorylation and the proposed TGFβ-induced phospho-Smad1 protein species remained robust in response to TGFβ treatment (Fig. 4, lane 5). These data completely rule out that the additional phospho-species detected in response to TGFβ simply corresponds to a modification of Smad3, further suggesting that this protein species is phospho-Smad1.
FIGURE 4.
TGFβ-induced Smad1 phosphorylation is observed in Smad3 null MEF cells. TGFβ-induced phospho-Smad1 is present in Smad3 null MEF cells. Wild type (WT) and Smad3 null (Smad3–/–) MEF cells were cultured in the presence or absence of TGFβ or BMP for 1 h and harvested for Western blot analysis with anti-Smad and anti-phospho-Smad antibodies. IB, immunoblot.
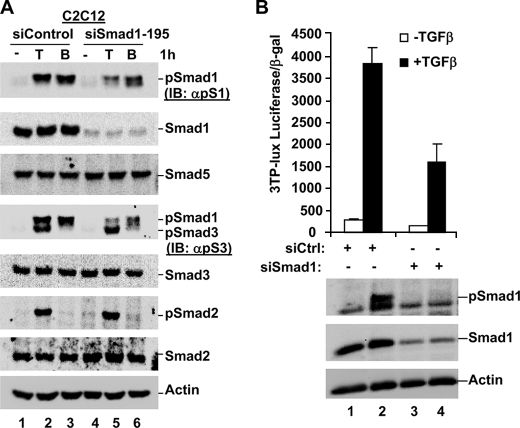
We next sought to determine whether the TGFβ-induced phospho-Smad1 band is eliminated in cells depleted of total Smad1 protein. To do this, we nucleofected C2C12 cells with siRNA against Smad1, and cultured cells in the absence or presence of TGFβ or BMP for 1 h prior to harvest for Western blot analysis. As shown in Fig. 5A, TGFβ treatment was able to induce phospho-Smad2, phospho-Smad3, and phospho-Smad1 in siControl nucleofected cells (Fig. 5A, lane 2). Expression of siSmad1 resulted in a reduction in Smad1 protein expression by almost 100%. Importantly, although phospho-Smad2 and phospho-Smad3 were still detected in siSmad1-expressing cells in response to TGFβ, the additional phospho-species we have routinely observed is lost in C2C12 cells depleted of Smad1 protein (Fig. 5A, lane 5). Furthermore, BMP-induced Smad1 phosphorylation is also significantly reduced in cells expressing siSmad1 (Fig. 5A, lane 6). These data confirm that TGFβ induces Smad1 phosphorylation in C2C12 cells, as detected by both our anti-phospho-Smad1 and anti-phospho-Smad3 antibodies, and suggest that these antibodies are most likely detecting TGFβ-induced Smad1 phosphorylation in the other cell lines we have tested.
FIGURE 5.
Knockdown of Smad1 using siRNA abolishes TGFβ-induced Smad1 phosphorylation and reduces TGFβ-induced transcription. A, TGFβ-induced phospho-Smad1 is lost in cells expressing siSmad1. C2C12 cells were nucleofected with siControl or siSmad1. After 48 h, cells were cultured in the presence or absence of TGFβ or BMP for 1 h prior to harvest for Western blotting. B, TGFβ-induced transcription is significantly reduced in cells expressing siSmad1. C2C12 cells were nucleofected with siControl or siSmad1, and transfected 24 h later with the TGFβ-responsive 3TP-lux transcriptional reporter. After an additional 24 h, cells were cultured with/without TGFβ for 9.5 h, prior to harvest for luciferase assay and Western blot analysis as described under “Experimental Procedures.” IB, immunoblot.
We wondered what effect Smad1 depletion would have on TGFβ-induced transcription. To assess this, C2C12 cells were nucleofected with siRNA against Smad1, or siControl, and cultured for 24 h prior to transfection with the TGFβ-responsive 3TP-lux luciferase reporter, which is widely used to assess TGFβ signaling requirements (18). Western blot analysis of cell lysates confirmed efficient depletion of Smad1 (∼90%) and a corresponding loss of Smad1 phosphorylation in response to TGFβ (Fig. 5B). Treatment of siControl cells with TGFβ resulted in a high degree of TGFβ-induced transcription, as expected (Fig. 5B). Significantly, depletion of Smad1 reduced the level of TGFβ-induced transcription by ∼2.5-fold as compared with siControl cells. This suggests that, at least in C2C12 cells, Smad1 may be involved in TGFβ-induced transcription in addition to the anticipated Smad2/3 proteins.
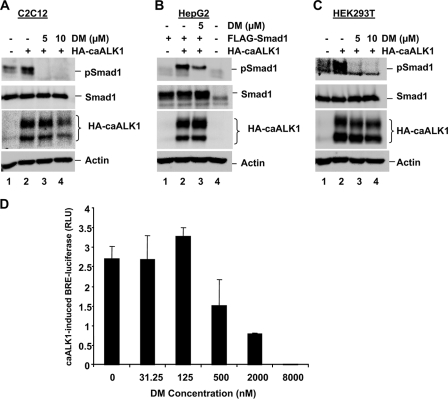
TGFβ-induced Smad1 Phosphorylation Is Independent of BMP Type I Receptors in Some Cell Lines—Previous reports of TGFβ-induced Smad1 phosphorylation in endothelial cells and dermal fibroblasts suggest a model where both the BMP type I receptor ALK1, and the TGFβ type I receptor ALK5, are required for this phenomenon (7, 8, 10). In HaCaT cells it is proposed that ALK5, and most likely the BMP type I receptor ALK2, are required for TGFβ-induced Smad1 phosphorylation (11). To help determine the ALK requirements of TGFβ-induced Smad1 phosphorylation in the cell lines under our investigation, we planned to take advantage of the established and specific ALK inhibitors SB431542 (against ALK4/5/7) (14) and DM (against ALK2/3/6) (15). However, as ALK1 is a key candidate for involvement in TGFβ-induced Smad1 phosphorylation, and the ability of DM to inhibit ALK1 is unclear,3 we first sought to determine whether DM could also inhibit ALK1 kinase activity. We tested the ability of 5 and 10 μm DM to block Smad1 phosphorylation induced by caALK1. C2C12 cells were transfected with caALK1 and cultured in the presence or absence of DM for 1.5 h prior to harvest. As shown in Fig. 6A, caALK1-induced endogenous Smad1 phosphorylation in C2C12 cells (lane 2). This likely reflects caALK1 activity as no TGFβ or BMP was added to stimulate the activity of additional receptors. Significantly, caALK1-induced Smad1 phosphorylation was completely abolished in cells treated with just 5 μm DM (Fig. 6A, lane 3), suggesting DM can inhibit ALK1 activity in C2C12 cells. We performed a similar experiment in HepG2 cells co-transfected with caALK1 and Smad1. The caALK1 was able to induce phosphorylation of exogenous Smad1 (Fig. 6B, lane 2) and this was also inhibited by culture of cells in 5 μm DM (Fig. 6B, lane 3), further suggesting DM can effectively inhibit ALK1 kinase activity. We also found DM could inhibit caALK1-induced endogenous Smad1 phosphorylation in HEK293T cells (Fig. 6C, lane 3). We next assessed the ability of caALK1 to induce transcription from BRE-luc, an Id1 promoter luciferase reporter gene dependent on BMP R-Smad activation, in the presence of increasing concentrations of DM. As shown in Fig. 6D, transcriptional activity was increased ∼2.7-fold over baseline on transfection with HA-caALK1, and culture in just 2 μm DM reduced caALK1-induced BRE-luc activity by ∼80% (Fig. 6D). Together the data in Fig. 6 suggest 5 μm DM is more than capable of inhibiting ALK1 receptor activity, as well as the anticipated ALK2/3/6 receptor activity (15), in certain cell lines.
FIGURE 6.
Smad1 phosphorylation induced by constitutively active ALK1 is inhibited by DM. A, caALK1-induced phospho-Smad1 is inhibited by DM in C2C12 cells. C2C12 cells were nucleofected with green fluorescent protein or constitutively active ALK1 receptor (HA-caALK1). After 48 h cells were cultured in the presence or absence of 5 or 10 μm DM for 1.5 h prior to harvest for Western blotting with anti-Smad1, anti-phospho-Smad1, anti-HA, and anti-actin antibodies. B, caALK1-induced phospho-Smad1 is inhibited by DM in HepG2 cells. HepG2 cells were co-transfected with HA-caALK1 and FLAG-Smad1. After 48 h cells were cultured in the presence or absence of 5 or 10 μm DM for 1.5 h prior to harvest for Western blotting as in A. C, caALK1-induced phospho-Smad1 is inhibited by DM in HEK293T cells. HEK293T cells were transfected with green fluorescent protein or HA-caALK1. After 48 h cells were cultured in the presence or absence of 5 or 10 μm DM for 1.5 h prior to harvest for Western blotting as in A. D, caALK1-induced Id1 promoter activity is inhibited by DM in COS cells. COS cells were co-transfected with plasmids encoding an Id1 promoter luciferase reporter (BRE-Luc), caALK1, and pRL-TK Renilla luciferase. Cell Id1 promoter activity, in the presence of caALK1 and increasing concentrations of DM, is reflected by the expression of BRE-luciferase activity divided by Renilla luciferase activity (DM, IC50 ≤ 1 μm, n = 3 measurements, results expressed as mean ± S.D.).
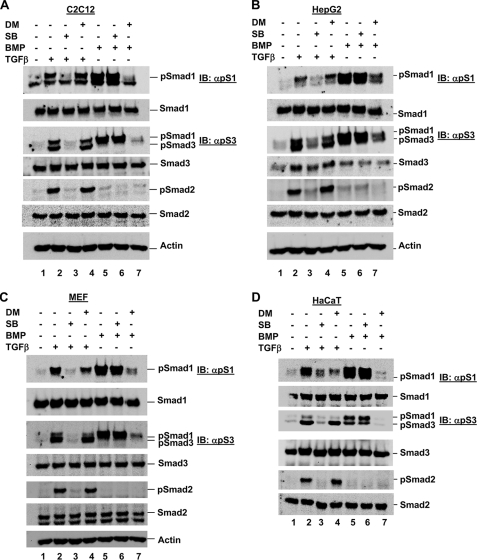
Next, to assess the involvement of different ALKs in TGFβ-induced Smad1 phosphorylation in the cell lines under our investigation, we cultured cells for 1.5 h in the presence of TGFβ or BMP in combination with the established and specific ALK inhibitor SB431542 (against ALK4/5/7) (14) or DM, which we propose can inhibit ALK1/2/3/6 in certain cell lines (15) (Fig. 6). We first assessed C2C12 cells and found that TGFβ induced Smad3 and Smad1 phosphorylation at fairly similar levels (Fig. 7A, lane 2), as we have consistently been able to observe. Furthermore, inhibition of ALK4/5/7 using SB431542 resulted in a complete loss of TGFβ-induced Smad1, Smad2, and Smad3 phosphorylation (Fig. 7A, lane 3), suggesting that these TGFβ-induced phosphorylation events are dependent on ALK4/5/7 kinase activity. However, in contrast, co-culture with SB431542 treatment had no effect on BMP-induced Smad1 phosphorylation, confirming that there is no dependence on ALK4/5/7 activity for this event (Fig. 7A, lane 6). Most significantly, inhibition of ALK1/2/3/6 by DM did not alter the traditional TGFβ-induced Smad2 or Smad3 phosphorylation, or the TGFβ-induced Smad1 phosphorylation (Fig. 7A, lane 4), suggesting that all three of these TGFβ-induced phosphorylation events are independent of BMP type I receptor ALKs. The same DM treatment, however, was able to fully abolish BMP-induced Smad1 phosphorylation in C2C12 cells (Fig. 7A, compare lanes 4 and 7), demonstrating that DM is fully active at the concentration and treatment times used in this experiment. These data suggest that BMP type I receptors may not be needed for TGFβ-induced Smad1 phosphorylation in all cell lines.
FIGURE 7.
TGFβ-induced Smad1 phosphorylation is independent of BMP type I receptors in certain cell lines. A, C2C12 cells were co-treated with ligand (TGFβ or BMP) and ALK inhibitor (SB431542 or DM), as indicated, for 1.5 h prior to harvest for Western blot analysis as described in the legend to Fig. 1A. B, HepG2 cells were co-treated with ligand (TGFβ or BMP) and ALK inhibitor (SB431542 or DM), as indicated, for 1.5 h prior to harvest for Western blot analysis as described in the legend to Fig. 1A. C, MEF cells were co-treated with ligand (TGFβ or BMP) and ALK inhibitor (SB431542 or DM), as indicated, for 1.5 h prior to harvest for Western blot analysis as described in the legend to Fig. 1A. D, HaCaT cells were co-treated with ligand (TGFβ or BMP) and ALK inhibitor (SB431542 or DM), as indicated, for 1.5 h prior to harvest for Western blot analysis as described in the legend to Fig. 1A. IB, immunoblot.
To investigate if this phenomenon is specific to C2C12 cells, or also holds true for other cells, we performed a similar experiment using the specific ALK inhibitors with three other cell lines. We observed that SB431542 also abolished TGFβ-induced Smad1 phosphorylation in HepG2 cells (Fig. 7B, lane 3) as effectively as it abolished Smad2 and Smad3 phosphorylation. However, as for C2C12 cells, DM did not abolish TGFβ-induced Smad1 phosphorylation (Fig. 7B, lane 4), although it was fully able to abolish BMP-induced Smad1 phosphorylation (Fig. 7B, lanes 7).
In MEFs, SB431542 could completely abolish TGFβ-induced Smad1 phosphorylation (Fig. 7C, lane 3). DM was able to partially reduce TGFβ-induced Smad1 phosphorylation, although it had no effect on TGFβ-induced Smad2 and Smad3 phosphorylation (Fig. 7C, lane 4). Again, in these cells DM was able to abolish BMP-induced Smad1 phosphorylation (Fig. 7C, lane 7). Interestingly, in HaCaT keratinocytes both SB431542 and DM were able to completely abolish TGFβ-induced Smad1 phosphorylation (Fig. 7D, lanes 3 and 4). This suggests that ALK1/2/3/6 may play a role in TGFβ-induced Smad1 phosphorylation in MEFs and HaCaT cells.
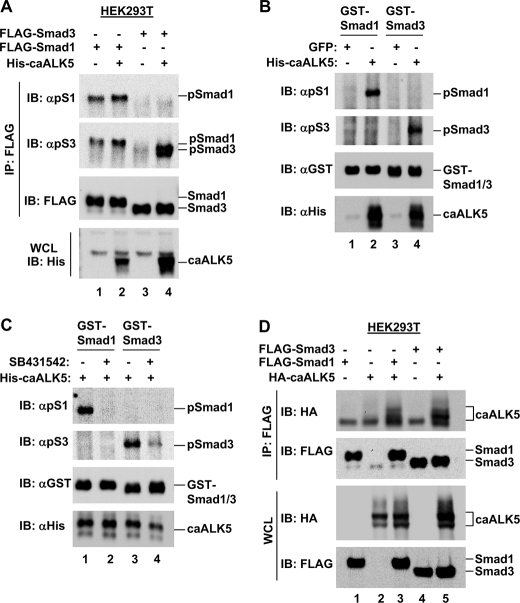
ALK5 Can Phosphorylate Smad1—As ALK5 is thought to be specific for Smad2/3 phosphorylation, we investigated whether ALK5 could also initiate Smad1 phosphorylation. HEK293T cells were transfected with caALK5 and FLAG-Smad1 and FLAG-Smad3, and Smad SXS phosphorylation was assessed following immunoprecipitation of FLAG-Smad proteins. We observed phosphorylation of Smad1 (lane 2) and the positive control Smad3 (lane 4) in the presence of caALK5 (Fig. 8A). These phosphorylation events should be a direct reflection of caALK5 activity as no TGFβ or BMP was added to stimulate the activity of additional receptors. Thus, this suggests ALK5 may have the intrinsic ability to phosphorylate Smad1 in addition to the anticipated Smad2 and Smad3 in vivo.
FIGURE 8.
ALK5 can induce Smad1 phosphorylation both in vivo and in vitro. A, caALK5 can induce Smad1 phosphorylation in HEK293T cells. HEK293T cells were co-transfected with FLAG-Smad1 or FLAG-Smad3 and His-caALK5, a constitutively active form of the ALK5 receptor, as indicated. After 48 h cells were harvested for anti-FLAG immunoprecipitation (IP) and Western blotting using anti-phospho Smad1, anti-phospho-Smad3, anti-FLAG, and anti-His antibodies. WCL, whole cell lysate. B, caALK5 can induce Smad1 phosphorylation in vitro. HEK293T cells were transfected with His-caALK5 or GFP as a negative control, and harvested for anti-His IP. Immunoprecipitated His-caALK5 was tested for its ability to phosphorylate recombinant GST-Smad1 and GST-Smad3 in an in vitro kinase assay. Assay products were analyzed by Western blotting using anti-phospho-Smad1, anti-phospho-Smad3, anti-GST, and anti-His antibodies. C, caALK5 induces Smad1 phosphorylation in vitro dependent on its kinase activity. HEK293T cells were transfected with His-caALK5, and harvested for anti-His IP. Immunoprecipitated His-caALK5 was tested for its ability to phosphorylate recombinant GST-Smad1 and GST-Smad3, in vitro, in the presence or absence of the ALK4/5/7 kinase inhibitor SB431542. Assay products were analyzed as in B. D, caALK5 interacts with Smad1 and Smad3 in HEK293T cells. HEK293T cells were co-transfected with FLAG-Smad1 or FLAG-Smad3, and HA-caALK5 as indicated. FLAG-Smad bound caALK5 was identified by anti-FLAG immunoprecipitation and anti-HA Western blotting. IB, immunoblot.
We also assessed the ability of caALK5 to initiate phosphorylation of recombinant GST-Smad1 in an in vitro kinase assay. His-caALK5 was precipitated from HEK293T cells, and incubated with GST-Smad1 or GST-Smad3 in the presence of ATP. Western blot analysis of assay products revealed strong phosphorylation of both GST-Smad1 and GST-Smad3 on incubation with caALK5 (Fig. 8B, lanes 2 and 4). To further confirm that the in vitro phosphorylation observed was due to His-caALK5 kinase activity, and not an additional factor in the His-immunoprecipitation product, we performed a similar assay in the presence or absence of the ALK4/5/7 kinase inhibitor SB431542. Precipitated His-caALK5 was again able to initiate phosphorylation of both GST-Smad1 and GST-Smad3 (Fig. 8C, lanes 1 and 3). However, phosphorylation was completely abolished in the presence of SB431542 (Fig. 8C, lanes 2 and 4). Taken together, the data in Fig. 8 provide firm evidence that ALK5 is able to phosphorylate Smad1, in addition to the anticipated Smad3.
Finally, if ALK5 can directly phosphorylate Smad1, we predicted that these proteins would interact. To test this hypothesis, HEK293T cells were co-transfected with HA-caALK5 and FLAG-Smad1 or FLAG-Smad3, and lysates subjected to FLAG immunoprecipitation. The positive control FLAG-Smad3 was able to co-precipitate HA-caALK5, in line with its role as a canonical ALK5 phosphorylation target (Fig. 8D, lane 5). Importantly, FLAG-Smad1 was also able to co-precipitate HA-caALK5 (Fig. 8D, lane 3), suggesting these proteins indeed interact and providing further evidence that Smad1 may also be an ALK5 phosphorylation target.
In short, our data suggest that TGFβ-induced Smad1 phosphorylation does not always require a functional BMP type I receptor, although ALK1/2/3/6 activity is clearly necessary for this phenomenon to occur in some cell lines. Thus, the mechanism of TGFβ-induced Smad1 phosphorylation may be largely cell line specific.
DISCUSSION
The receptors for TGFβ superfamily ligands and the specific Smads downstream of these have been identified and well characterized, and a canonical scheme for signaling from the cell surface to the nucleus in response to the various ligands has been established. However, unexpected signaling outcomes, or even opposing biological outcomes to TGFβ signaling pathways in the same cells, suggest that the signaling model is not quite as simple as first thought.
For example, in addition to the canonical TβRII/ALK5-mediated Smad2/3 phosphorylation, TβRII can form a complex with ALK1 in endothelial cells, dependent on ALK5 kinase activity and Endoglin, to result in Smad1/5 phosphorylation in response to TGFβ (7–9). In this case, activation of canonical TGFβ signaling inhibits proliferation and migration of endothelial cells, whereas TGFβ-mediated Smad1/5 phosphorylation stimulates proliferation and migration (7). It has even been proposed that ALK1 activation in response to TGFβ signaling may inhibit ALK5/Smad signaling (7), perhaps as an indirect effect of high Endoglin levels that favor TGFβ/ALK1 signaling and not TGFβ/ALK5 signaling (9). TGFβ-induced Smad1 phosphorylation has also been observed in HaCaT keratinocytes (11). In keeping with the endothelial cell model, this signaling is thought to require a BMP type I receptor, in addition to ALK5 kinase activity, with ALK2 being the most likely candidate because HaCaT cells were found not to express ALK1 (11). While this manuscript was in preparation, it was also shown that TGFβ can induce Smad1 phosphorylation in a number of epithelial cells and fibroblasts via ALK5 and ALK2 and/or ALK3 (20), which again suggests a dependence on a BMP type I receptor for TGFβ ligand to BMP-Smad cross-activation. We have identified TGFβ-induced Smad1 in different cell lines to those tested by any of these groups. Additionally, our data suggest that TGFβ-induced Smad1 phosphorylation can occur independently of BMP type I receptors in some cell lines (Fig. 7). Specifically, culture of cells in the presence of the ALK inhibitor DM, which we show can inhibit ALK1 kinase activity (Fig. 6) in addition to its published ability to inhibit ALK2/3/6, was unable to reduce the level of TGFβ-induced Smad1 phosphorylation in C2C12 and HepG2 cells. However, this was completely abolished by the ALK4/5/7 inhibitor SB431542 (Figs. 7, A and B, and 9). This differs to data obtained in the study by Daly et al. (20), where both SB431542 and DM could abolish TGFβ-induced Smad1/5 phosphorylation in EpH4 mouse mammary gland epithelial cells. Importantly, DM was able to inhibit BMP-induced Smad1 phosphorylation in HepG2 and C2C12 cells (Fig. 7, A and B), as well as TGFβ-induced Smad1 phosphorylation in HaCaT cells (Fig. 7D) in agreement with the idea that HaCaT cells rely on ALK2/3/6 for this phenomenon (11, 20). Thus, DM was active and fully capable of inhibiting ALK1/2/3/6 under the conditions employed in our study.
FIGURE 9.
TGFβ promotes Smad1 phosphorylation independently of ALK1/2/3/6 in certain cell lines. In C2C12 and HepG2 cells (left panel) TGFβ-induced Smad1/2/3 phosphorylation can be blocked by SB431542, suggesting these phosphorylation events depend on ALK4/5/7 activity. However, these phosphorylation events appear to be independent of ALK1/2/3/6 activity because they are not blocked by DM. As expected, BMP-induced Smad1 phosphorylation is blocked by DM in C2C12 and HepG2 cells. In HaCaT cells (right panel) TGFβ-induced Smad1/2/3 phosphorylation can also be blocked by SB431542, suggesting these events also depend on ALK4/5/7 activity. However, in contrast to C2C12 and HepG2 cells, both TGFβ and BMP-induced Smad1 phosphorylation can be blocked by DM, suggesting that both events depend on ALK1/2/3/6 activity in HaCaT cells.
While our manuscript was under revision, Liu et al. (12) reported that TGFβ could induce Smad1 phosphorylation in the mouse mammary epithelial cell line 4T1. This phosphorylation event was strictly dependent on ALK5 kinase activity, and the authors rule out involvement of BMP receptors using short hairpin RNA. Our data resulting from the use of ALK inhibitors in Fig. 7 further support the idea that TGFβ-induced Smad1 phosphorylation is independent of BMP receptors in some cells. Critically, ALK5-mediated direct phosphorylation of Smad1 was demonstrated by in vitro kinase assays in both studies (Fig. 8) (12).
It is intriguing as to how ALK4/5/7, presumably specifically ALK5 as this is thought to be responsible for TGFβ-induced signaling, may lead to the phosphorylation of Smad1 independently of BMP-specific type I receptors. Specificity of type I receptors for the canonical Smad is usually based on their L45 loop and phosphorylated GS motif, and on the L3 loop in the MH2 domain of the partner Smad (4–6). However, our data suggest that ALK5 can directly contact Smad1 despite the assumed incompatible L45–L3 pairing (Fig. 8D). Furthermore, constitutively active ALK5 could initiate exogenous Smad1 phosphorylation in HEK293T cells (Fig. 8A) and GST-Smad1 phosphorylation in vitro (Fig. 8, B and C). It is important to point out that most of the early studies made their conclusions on receptor-Smad specificity based on functional assays carried out in rather limited cell lines, such as Mv1Lu and COS1 cells. Thus, it is conceivable that a cell-specific intermediate factor may also be involved in the cross-activation of Smad1 by ALK5.
In C2C12 cells, TGFβ-induced Smad1 phosphorylation could be completely abolished using siSmad1 (Fig. 5). As siSmad1 had no effect on the protein level of closely related Smad5 (Fig. 5), this suggests our phospho-antibodies specifically recognize TGFβ-induced phospho-Smad1. However, it cannot be ruled out that Smad5 is also phosphorylated by TGFβ signaling in the cells we investigated and that it is just undetectable by our antibodies.
Our observation that TGFβ-induced Smad1 phosphorylation peaks at a later time than TGFβ-induced Smad3 phosphorylation in C2C12 and MEF cells, and is significantly more transient in nature (Fig. 2), completely agrees with the kinetic data recently published for EpH4 mouse mammary gland epithelial cells (20). Our finding that a higher concentration of TGFβ is required to induce Smad1 phosphorylation than Smad3 phosphorylation in C2C12 cells (Fig. 3) is also in accordance with this finding for EpH4 cells (20). Thus, despite the apparent difference in ALK requirement for the TGFβ-induced Smad1 phosphorylation events between C2C12 and EpH4 cells, the overall kinetics appear to be similar.
There are several potential outcomes to the TGFβ-induced Smad1 phosphorylation that we observed. First, TGFβ-induced phospho-Smad1 could play a role at promoters usually activated by BMP (i.e. where phospho-Smad1 is traditionally expected to act), in keeping with the idea of phospho-Smad1 binding the DNA at the usually expected GC-rich sequence. Second, TGFβ-induced phospho-Smad1 could play a role at promoters usually activated by TGFβ (i.e. at GTCT sequences where phospho-Smad3 is traditionally expected to act). We screened a panel of TGFβ and BMP responsive luciferase reporters in response to TGFβ, in several different cell lines. We were unable to identify a canonical BMP-regulated gene reporter that could respond to TGFβ (data not shown). This supports recent data showing that TGFβ-induced phosphorylation of Smad1/5 cannot initiate transcription via BMP-responsive elements in epithelial cells (20). Additionally, we were unable to rescue TGFβ-induced reporter responses in Smad3 null MEF cells by overexpression of Smad1 (data not shown), suggesting that TGFβ-induced phospho-Smad1 does not simply substitute for phospho-Smad3 in this system. However, we did observe that depletion of Smad1 significantly reduced the level of TGFβ-induced transcription from the 3TP-lux luciferase reporter in C2C12 cells, suggesting Smad1 may play a role in TGFβ-induced transcription in addition to Smad2/3 proteins. It seems possible that involvement of Smad1 in TGFβ-induced transcription could be limited to those cell lines (C2C12; HepG2) that are able to induce Smad1 phosphorylation in an ALK5-dependent, and BMP receptor-independent, manner.
In short, we provide firm evidence that TGFβ can activate Smad1 independently of BMP type I receptor activity in certain cells. Future studies are likely to determine whether TGFβ-induced ALK1/2/3/6-independent Smad1 phosphorylation can occur in additional cell types, and further determine whether this has a unique function as compared with the TGFβ-induced Smad1 phosphorylation dependent on ALK1/2/3/6.
Acknowledgments
We thank Dr. Ed Leof for anti-phospho-Smad3 antibody, Dr. Peter ten Dijke for Id1-luc and caALK1, and Dr. Xiao-Fan Wang for Smad3–/– MEFs, and recombinant GST-Smad1 and GST-Smad3 proteins. We thank Carol Lai, Dr. Irwin Liu, and Dr. Fangyan Dai for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK073932 (to X. L.) and R01CA108454, R01AR053591, and P50HL083794 (X.-H. F.).
Footnotes
The abbreviations used are: TGF, transforming growth factor; DM, dorsomorphin; MEF, mouse embryonic fibroblast; ALK, activin receptor-like kinases; caALK, constitutively active ALK; HA, hemagglutinin; GST, glutathione S-transferase; siRNA, small interfering RNA; BMP, bone morphogenic protein.
P. Yu, unpublished data.
References
- 1.Feng, X.-H., and Derynck, R. (2005) Annu. Rev. Cell Dev. Biol. 21 659–693 [DOI] [PubMed] [Google Scholar]
- 2.Schmierer, B., and Hill, C. S. (2007) Nat. Rev. Mol. Cell Biol. 8 970–982 [DOI] [PubMed] [Google Scholar]
- 3.Chang, H., Brown, C. W., and Matzuk, M. M. (2002) Endocr. Rev. 23 787–823 [DOI] [PubMed] [Google Scholar]
- 4.Chen, Y. G., Hata, A., Lo, R. S., Wotton, D., Shi, Y., Pavletich, N., and Massague, J. (1998) Genes Dev. 12 2144–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo, R. S., Chen, Y. G., Shi, Y., Pavletich, N. P., and Massague, J. (1998) EMBO J. 17 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng, X.-H., and Derynck, R. (1997) EMBO J. 16 3912–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goumans, M. J., Valdimarsdottir, G., Itoh, S., Lebrin, F., Larsson, J., Mummery, C., Karlsson, S., and ten Dijke, P. (2003) Mol. Cell 12 817–828 [DOI] [PubMed] [Google Scholar]
- 8.Goumans, M. J., Valdimarsdottir, G., Itoh, S., Rosendahl, A., Sideras, P., and ten Dijke, P. (2002) EMBO J. 21 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebrin, F., Goumans, M. J., Jonker, L., Carvalho, R. L., Valdimarsdottir, G., Thorikay, M., Mummery, C., Arthur, H. M., and ten Dijke, P. (2004) EMBO J. 23 4018–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pannu, J., Nakerakanti, S., Smith, E., ten Dijke, P., and Trojanowska, M. (2007) J. Biol. Chem. 282 10405–10413 [DOI] [PubMed] [Google Scholar]
- 11.Bharathy, S., Xie, W., Yingling, J. M., and Reiss, M. (2008) Cancer Res. 68 1656–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, I. M., Schilling, S. H., Knouse, K. A., Choy, L., Derynck, R., and Wang, X. F. (2009) EMBO J. 28 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piek, E., Ju, W. J., Heyer, J., Escalante-Alcalde, D., Stewart, C. L., Weinstein, M., Deng, C., Kucherlapati, R., Bottinger, E. P., and Roberts, A. B. (2001) J. Biol. Chem. 276 19945–19953 [DOI] [PubMed] [Google Scholar]
- 14.Inman, G. J., Nicolas, F. J., Callahan, J. F., Harling, J. D., Gaster, L. M., Reith, A. D., Laping, N. J., and Hill, C. S. (2002) Mol. Pharmacol. 62 65–74 [DOI] [PubMed] [Google Scholar]
- 15.Yu, P. B., Hong, C. C., Sachidanandan, C., Babitt, J. L., Deng, D. Y., Hoyng, S. A., Lin, H. Y., Bloch, K. D., and Peterson, R. T. (2008) Nat. Chem. Biol. 4 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng, X.-H., Lin, X., and Derynck, R. (2000) EMBO J. 19 5178–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrighton, K. H., Willis, D., Long, J., Liu, F., Lin, X., and Feng, X.-H. (2006) J. Biol. Chem. 281 38365–38375 [DOI] [PubMed] [Google Scholar]
- 18.Wrana, J. L., Attisano, L., Carcamo, J., Zentella, A., Doody, J., Laiho, M., Wang, X. F., and Massague, J. (1992) Cell 71 1003–1014 [DOI] [PubMed] [Google Scholar]
- 19.Korchynskyi, O., and ten Dijke, P. (2002) J. Biol. Chem. 277 4883–4891 [DOI] [PubMed] [Google Scholar]
- 20.Daly, A. C., Randall, R. A., and Hill, C. S. (2008) Mol. Cell. Biol. 28 6889–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]