Abstract
A second dermatan sulfate epimerase (DS-epi2) was identified as a homolog of the first epimerase (DS-epi1), which was previously described by our group. DS-epi2 is 1,222 amino acids long and has an ∼700-amino acid N-terminal epimerase domain that is highly conserved between the two enzymes. In addition, the C-terminal portion is predicted to be an O-sulfotransferase domain. In this study we found that DS-epi2 has epimerase activity, which involves conversion of d-glucuronic acid to l-iduronic acid (EC 5.1.3.19), but no O-sulfotransferase activity was detected. In dermatan sulfate, iduronic acid residues are either clustered together in blocks or alternating with glucuronic acid, forming hybrid structures. By using a short interfering RNA approach, we found that DS-epi2 and DS-epi1 are both involved in the biosynthesis of the iduronic acid blocks in fibroblasts and that DS-epi2 can also synthesize the hybrid structures. Both iduronic acid-containing domains have been shown to bind to several growth factors, many of which have biological roles in brain development. DS-epi2 has been genetically linked to bipolar disorder, which suggests that the dermatan sulfate domains generated by a defective enzyme may be involved in the etiology of the disease.
Chondroitin/dermatan sulfate (CS/DS)2 proteoglycans are widely distributed in the extracellular matrix and at the cell surface. They are thought to be involved in neurobiological processes, cartilage function, cancer, and coagulation (1, 2). A complex biosynthetic machinery in the Golgi is responsible for the production and for the final fine structure of CS/DS chains (3). Five enzymes form a tetrasaccharide linker region attached to a serine residue of the core protein. Six additional CS/DS-specific enzymes produce the polymeric backbone, which consists of alternating glucuronic acid (GlcUA) and N-acetylgalactosamine (GalNAc) residues. Two types of modifications of the polymer occur during elongation. One modification consists of the transformation, to different extents, of glucuronic acid into its epimer iduronic acid (IdoUA). The second modification is the sulfation of hydroxyl groups in different positions as follows: GalNAc can be sulfated at C-4 and C-6, and GlcUA/IdoUA can be sulfated only at C-2, at least in mammals. The presence of four sulfotransferases specific for addition of 4-O-sulfate, three for 6-O-sulfate, and one for 2-O-sulfate suggests that there is tight regulation of this process. As these modifications are incomplete, i.e. not all the substrate available is modified, the final CS/DS chains are structurally heterogeneous.
During DS biosynthesis, epimerization precedes O-sulfation (4) and creates different domains in the chain. One domain consists of the structure (IdoUA-GalNAc)n ≥ 4, where IdoUA residues are clustered together to form long stretches called iduronic acid blocks. There is a second type of domain consisting of unmodified (GlcUA-GalNAc) stretches. A third domain consists of hybrid structures, where an isolated IdoUA residue or a few clustered IdoUAs alternate with GlcUA, such as in GlcUA-GalNAc-(IdoUA-GalNAc)n = 1–3 structures. We recently identified the gene encoding the DS-epimerase (5). When searching data bases, a single homolog called DSEL (dermatan sulfate epimerase-like) was found. DSEL was originally cloned as c18orf4, a gene of unknown function that was genetically associated with type II bipolar disorder (6). In this study, we show that the protein encoded by DSEL has epimerase activity.
In light of this, we suggest renaming of the two proteins to DS-epimerase 1 (DS-epi1, encoded by the DSE gene; NCBI NM_013352; NP_037484) and to DS-epimerase 2 (DS-epi2, encoded by the DSEL gene; NCBI NM_032160; NP_115536).
EXPERIMENTAL PROCEDURES
Plasmid Constructs—A genomic clone (RZPDB737E111024D) containing an uninterrupted open reading frame of human dermatan sulfate epimerase-like (DSEL; NCBI NM_032160; also named C18orf4 and NCAG1) was obtained from RZPD, Germany. The 3,666-nucleotide coding sequence was subcloned into a pcDNA3.1/myc-His vector (Invitrogen) using BamHI and AgeI restriction sites that had been introduced using the primers 5′-TAGGATCCATGCCTAAGGGAGGA-3′ and 5′-GACGACCGGTGTCCATAAACTTTGGATA-3′ (DNA Technology A/S, Denmark). The coding sequence corresponding to amino acids 31–1,222, excluding a predicted signal peptide, was subcloned from pcDNA3.1-DS-epi2 into the pCS2 vector in-frame with an N-terminal signal peptide from chordin and a FLAG tag, using primers 5′-ACATCTCGAGACTTTTGAGGAATCTG-3′ and 5′-GCAGTCTAGATTAGTCCATAAACTTTG-3′ to introduce XhoI and XbaI sites, respectively (7). From the DS-epi2/pCS2 vector, the sequences spanning amino acids residues 31–734 and 722–1,222, corresponding to the epimerase and O-sulfotransferase domains, respectively (numbers as in NCBI NP_115536), were amplified using the primer pairs 5′-ACATCTCGAGACTTTTGAGGAATCTG-3′/5′-GCAGTCTAGATTACAGGTTATGGTAATCAGT-3′ and 5′-ACATCTCGAGGAACATGTTGTTTCTATT-3′/5′-GCAGTCTAGATTAGTCCATAAACTTTG-3′, respectively (XhoI and XbaI sites introduced), and subcloned into the pCS2 vector. DS-epi1/His in pcDNA3.1 was prepared as described previously (5). All expression constructs were sequenced by MWG Biotec AG, Germany.
Transient Overexpression of DS-epi1 and DS-epi2 in 293HEK Cells—Cells were obtained from the ATCC and cultured in minimum Eagle's medium with 10% fetal bovine serum. Expression vectors were transfected using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. After 48 h, the cells were labeled as described below or, alternatively, washed with phosphate-buffered saline and lysed in 20 mm MES, pH 6.5, 150 mm NaCl, 10% glycerol, 2 mm dithiothreitol, 1 mm EDTA, 1% Triton X-100, and protease inhibitors, i.e. 1 mm phenylmethylsulfonyl fluoride, and aprotinin, leupeptin, and pepstatin, all at 1 μg/ml (lysis buffer). Two hundred microliters of cleared lysates were desalted using the dialysis buffer 20 mm MES (pH 5.5 at 37 °C), 10% glycerol, 0.5 mm EDTA, 0.1% Triton X-100, 1 mm dithiothreitol, and protease inhibitors. Protein content was determined by Bradford reagent (Bio-Rad) with BSA as standard, and equal amounts of protein were assayed either for epimerase or O-sulfotransferase activity. Alternatively, 2 mg of cell lysate, the pH of which was brought to 7.5 by addition of HEPES to final 50 mm, was immunopurified by adding 20 μl of anti-FLAG M2-agarose gel (Sigma). After 4 h of incubation at 4 °C, the gel was washed with lysis buffer, pH 7.5, and finally resuspended in 200 μl of dialysis buffer. One-fifth of resuspended gel-bound immunopurified material was used for activity assays.
siRNA-mediated Down-regulation of DS-epi1 and DS-epi2 in HFL-1—Primary human lung fibroblasts (HFL-1) were cultivated in minimum Eagles medium with 10% donor calf serum and used until passage 25. Negative control siRNA was from Qiagen (catalog number 1027281), whereas three DS-epi1 siRNAs and two DS-epi2 siRNAs were from Ambion. The most effective siRNAs were evaluated by qRT-PCR mRNA measurements. The DS-epi1 siRNA chosen had the sense sequence 5-GCUACACCACUAGAUCACUtt-3′ (Ambion 134073) and the DS-epi2 siRNA had the sense sequence 5′-GGAUGGUUGGCUACAAAGAtt-3′ (Ambion 33832). HFL-1 cells were trypsinized and first reverse-transfected in the presence of a final concentration of 50 nm siRNA complexed with Lipofectamine 2000, according to the manufacturer's instructions. A second round of conventional transfection was performed after 48 h by changing to labeling medium (see below) containing freshly added siRNAs and Lipofectamine 2000 complexes.
Microsome Preparation—293 human embryo kidney (HEK) cells were transiently transfected in 75-cm2 culture flasks with either empty vector, pcDNA3.1-DS-epi1/His, or pCS2-DS-epi2/FLAG or co-transfected with the two epimerase-containing vectors. After 48 h, microsomes were prepared at 4 °C as follows. The cells were washed with phosphate-buffered saline, then washed with homogenization buffer (20 mm MES, pH 6.5, 250 mm sucrose, 5 mm EDTA, and protease inhibitors), and scraped in the presence of 2.5 ml of the latter buffer. They were homogenized by 10 manual strokes of a Potter device, and debris and nuclei were pelleted for 10 min at 1,000 × g. The mitochondria were pelleted at 10,000 × g for 10 min, and finally microsomes were pelleted for 1 h at 50,000 × g and resuspended in 1.5 ml of detergent-free dialysis buffer (see above) containing BSA at 10 μg/ml. The microsomes were assayed with detergent or alternatively sonicated and assayed without detergent (see below).
Epimerase Assay—Epimerase activity assays were performed in a final volume of 100 μl of 0.8× dialysis buffer (see above), 0.2 mg of BSA, 2 mm MnCl2, 0.5% Nonidet P-40, and 30,000 dpm of the labeled chondroitin substrate (5-3H-defructosylated K4, prepared according to Ref. 8). Incubations were carried out at 37 °C for 14–20 h, and the tritium released was quantified as described previously (5).
O-Sulfotransferase Assay—O-Sulfotransferase activity assays were performed in a final volume of 50 μl of 10% glycerol, 0.5 mm EDTA, 1% Triton X-100 (unless otherwise stated), 1 mm dithiothreitol, 5 mm MnCl2, protease inhibitors, 1 μCi of [35S]PAPS, and 1 μm unlabeled PAPS, containing 10 μg of polysaccharide substrate. Two separate incubations were performed at 37 °C for 2 h in the above solution buffered either with 20 mm MES at pH 5.5, which is the optimal pH for the epimerase activity in DS-epi2, or with 50 mm imidazole, pH 6.8, which is optimal for assay of CS O-sulfotransferases (9). Dermatan was prepared from pig skin dermatan sulfate, which was completely desulfated by HCl/methanol treatment for 5 h according to Ref. 10. The residual glucuronic acid moieties and heparan sulfate were eliminated by chondroitinase AC-I treatment and deamination at pH 1.5, respectively. Chondroitin was prepared as above from horse nasal septum. To obtain polysaccharide substrates still retaining a fraction of GalNAc that was either 4- or 6-sulfated, CS-4-sulfated from bovine nasal cartilage and CS-6-sulfated from horse nasal septum were partially desulfated in 90% DMSO and 10% methanol for 1 or 2 h, according to Ref. 11. Two preparations of partially desulfated CS-4 were obtained, retaining 10 and 1% of the GalNAc in the 4-sulfated form, and two partially desulfated CS-6 preparations retained 15 and 5% of GalNAc as 6-sulfated form. Polysaccharide products with 35S radioactivity incorporated were re-isolated after ethanol precipitation and recovered in the void volume of a Sephadex G-25 column (GE Healthcare), as described in Ref. 12. Lysates from control 293HEK gave 2,000–15,000 dpm 35S incorporated into all substrates tested, with a blank of 50–100 dpm. The assay was linear with time and protein concentration of the enzyme preparations.
Western Blot—DS-epi2 was detected using an anti-FLAG M2 antibody conjugated with horseradish peroxidase (Sigma) at 1:500 dilution. DS-epi1 was detected using an immunopurified anti-DS-epi1 rabbit polyclonal antibody (at 1 μg/ml) obtained after immunization with the peptide KWSKYKHDLAAS, corresponding to residues 509–520 of the human/murine sequence (Innovagen, Sweden). In both cases, ECL-Plus (GE Healthcare) was used as horseradish peroxidase substrate.
Cell Labeling and CS/DS Chain Preparation—After transfection with plasmid or siRNA as above, 293HEK or HFL-1 cells were starved of sulfate for 2 h with sulfate-free minimum Eagle's medium, with 10% serum, followed by labeling for 24 h by addition of Na35SO4 (PerkinElmer Life Sciences, 1500 Ci/mmol; 100 μCi/ml final). Labeled medium was removed and diluted 1:3 with denaturing buffer containing 9 m urea, 0.1% Triton X-100, 50 mm acetate, pH 5.5, 0.25 m NaCl, protease inhibitors, and proteoglycans purified on an anion-exchange DE52 column. The column was washed with 50 bed volumes of 6 m urea, 0.1% Triton X-100, 50 mm acetate, pH 5.5, 0.25 m NaCl, and protease inhibitors, and eluted with 4 m guanidine, 50 mm acetate, pH 5.8, 0.1% Triton X-100 (dissociating buffer), with 50 μg of dextran T500 added as carrier. DE52-purified proteoglycans were separated on a Superose 6 column (10/300; GE Healthcare) run at 0.1 ml/min in dissociating buffer, and 0.5-ml fractions were collected. Pooled fractions contained either versican or a mixture of biglycan and decorin, as judged from SDS-PAGE of radioactive material (13). Ten μg amounts of CS4 were added to the pooled proteoglycans as carrier, and they were desalted using PD10 columns (GE Healthcare). GAG chains were cleaved by β-elimination in 50 mm KOH with 100 mm NaBH4 at 45 °C for 16–18 h, and they were re-isolated on DE52 columns, run as above. Heparan sulfate chains were depolymerized by deamination at pH 1.5 (14), followed by reisolation of CS/DS chains on Superose 6 run in 0.2 m NH4HCO3.
Chondroitinase Treatment of CS/DS—Labeled CS/DS chains (50,000-100,000 dpm) were resuspended in 50 μl of chondroitinase B buffer (20 mm Tris, pH 7.5, 50 mm NaCl, 4 mm CaCl2, and 0.1 mg/ml BSA) or chondroitinase AC-I buffer (20 mm Tris, pH 7.5, 50 mm sodium acetate) together with 1 milliunit of the respective enzymes (from Seikagaku) and 100 ng each of CS4, CS6, and DS. After 90 min at 37 °C, the reaction was stopped by boiling, and the split products were separated on a Superdex Peptide size permeation column (10/300; GE Healthcare) using 0.2 m NH4HCO3 as running buffer. The flow was set at 0.3 ml/min, and 300 μl was collected in each fraction. Disaccharide CS/DS fingerprint was performed by 2-h cleavage of 0.5 million dpm CD/DS chains by 50 milliunits of chondroitinase ABC in 50 mm Tris, pH 8.0, 50 mm sodium acetate. Split products were reduced with 25 mm NaBH4 for 15 min at 37 °C, the disaccharides were re-isolated after Superdex Peptide column run as above, and finally separated on a CarboPac PA1 high pressure liquid chromatography column according to Ref. 15.
qRT-PCR—HFL-1 cells were transfected as above with siRNAs targeting DS-epi1 or DS-epi2, and after the labeling period, total RNA was purified. cDNA was subsequently synthesized from 1 μg of total RNA using the iScript cDNA synthesis kit (Bio-Rad). cDNA corresponding to an amount of 100 ng of RNA was used thereafter for PCR on an ABI PRISM platform with iTAQ SYBR green supermix with ROX (Bio-Rad). Relative quantification was performed according to the ΔCt method using 18S RNA gene as a housekeeping gene. The forward and reverse primer pairs used were as follows: 18 S RNA (5′-cgaacgtctgccctatcaac-3′,5′-tgccttccttggatgtggta-3′), DS-epi1 (5′-gcagcagaggagaaaaatgg-3′,5′-aagaagctcgctgctgtctc-3′), and DS-epi2 (5′-ttacttatggggctgggttg-3′,5′-tgggggcaaaagtaaatgag-3′).
RESULTS
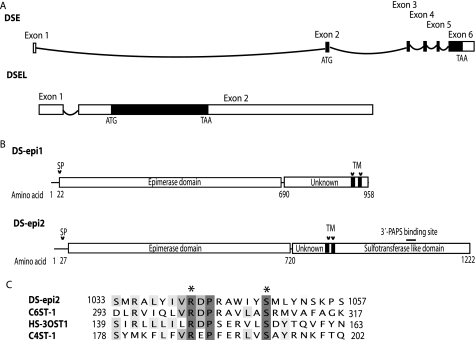
Genomic and Domain Structures of DS-epi1 and DS-epi2—Using the amino acid sequence of DS-epi1, a putative second epimerase (DS-epi2), encoded by the DSEL gene located at 18q22, was also identified. The DS-epi1 gene has six exons, and the open reading frame spans five exons and encodes a protein of 958 amino acids (Fig. 1A). The DS-epi2 gene has two exons with an uninterrupted open reading frame in exon 2 encoding a 1,222-amino acid-long protein. Alignment of the DS-epi1 and DS-epi2 sequences showed that they share 51% amino acid identity between amino acid positions 62 and 692 (corresponding to the human sequence of DS-epi2; Fig. 1B). This covers the domain in DS-epi1 that contains the active site (16). In addition, amino acids implicated in the catalytic activity of DS-epi1 are also conserved in DS-epi2 (16). The C-terminal domain of DS-epi2 has significant similarity in primary structure with members of the HS/CS 6-O-sulfotransferase family. Thus, alignment of the amino acid sequence of C6ST-1 with that of DS-epi2 showed 16% identity and 31% similarity in a region between positions 841 and 1,222. Furthermore, DS-epi2 contains a sequence that is highly similar to the 3′-PAPS-binding site in O-sulfotransferases (Fig. 1C). In summary, DS-epi2 has an N-terminal epimerase domain in common with DS-epi1 and an additional C-terminal O-sulfotransferase domain.
FIGURE 1.
Gene and domain structure of the DS epimerases. A, schematic view of the DSE and DSEL genomic structure. Exons are shown with rectangles, and introns with curved lines. Coding regions are shown as filled boxes. B, domain structure of the two epimerases, showing a common N-terminal epimerase domain followed by two distinct domains of different length (of unknown function) and, in the case of DS-epi2, by an O-sulfotransferase-like domain. The locations of signal peptides (SP) and predicted transmembrane regions (TM) are also shown. A potential 3′-PAPS-binding site is depicted with a straight line. C, amino acid conservation of the 3′-PAPS-binding site is shown by alignment (generated with ClustalW) to the protein sequences of human C6ST-1, HS-3OST1, and C4ST-1. Catalytic amino acids are marked with asterisks.
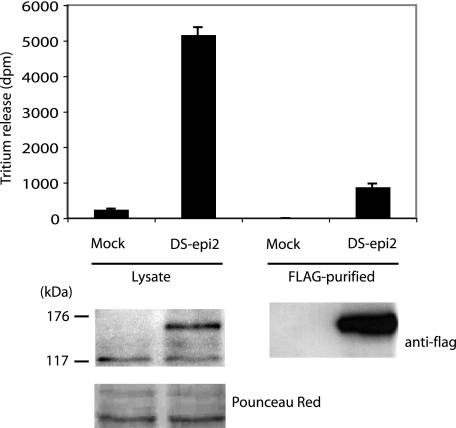
DS-epi2 Has Epimerase Activity—DS-epimerase converts glucuronic acid to iduronic acid at the polymer level. Activity is assayed by incubating enzyme preparations with C-5-3H-labeled chondroitin, resulting in the release of tritium, measured as tritiated water after distillation. Transient overexpression of a FLAG-tagged DS-epi2 in 293HEK cells increased the epimerase activity in cellular lysates 21-fold over the detectable endogenous activity (Fig. 2). Nine percent of the activity was recovered as anti-FLAG gel-bound material, whereas no endogenous epimerase activity was unspecifically bound to the gel during the immunopurification procedure. The overexpressed DS-epi2 migrated on SDS-PAGE as a 140–150-kDa band, compatible with being the uncleaved protein of the expected size. The specific activity relative to the 140–150-kDa band was lower in the immunopurified material than in the lysate, possibly because of inactivation and/or nonoptimal assay of the gel-bound enzyme. The activity measured in the immunopurified fraction could not be ascribed to endogenous DS-epi1 that had been co-purified with DS-epi2. In fact, no DS-epi1 signal could be detected in the FLAG-purified fraction by Western blot (data not shown), and the activity measured in the FLAG-purified fraction was higher than the total activity of the control lysate.
FIGURE 2.
DS-epi2 has epimerase activity. Epithelial 293HEK cells were transiently transfected with empty pCS2 expression vector or with the vector containing DS-epi2 in-frame with an N-terminal FLAG tag. Top, after 48 h, cell lysates were prepared, and 280 μg of desalted proteins was assayed. Alternatively, 2 mg of lysate was immunopurified by an anti-FLAG M2-agarose gel, and one-fifth of the resuspended gel-bound material was used in each assay. Data represent mean ± 2 S.D. of triplicates. Repeated transfections (n = 12) ranged from 12- to 35-fold overexpression of epimerase activity, compared with mock controls. B, 20 μg of lysates or one-fifth of the immunopurified gel-bound material was subjected to Western blot and probed with peroxidase-conjugated anti-FLAG antibody.
The activity of immunopurified DS-epi2 was measured in different buffers to establish the best assay conditions for this new enzyme. The optimal pH was 5.5, and the divalent cation manganese activated its activity, whereas the monovalent NaCl inhibited its activity (IC50 25 mm) (data not shown). The optimal assay conditions were identical to those of DS-epi1. These results demonstrate conclusively that DS-epi2 is an active epimerase.
Analysis of O-Sulfotransferase Activity of DS-epi2—The presence of a putative O-sulfotransferase domain in DS-epi2 prompted us to assay this enzymatic activity upon overexpression of the protein. The acceptor substrates tested were chondroitin, dermatan, two partially desulfated CS-4, and two partially desulfated CS-6. None of the substrates tested were sulfated by either the full-length protein (FLAG-31–1,222/pCS2) or the portion of the protein that was devoid of the epimerase domain (FLAG-722–1,222/pCS2), when overexpressed in 293HEK cells and FLAG-purified (data not shown). Both proteins were abundantly expressed and were detected as bands of the expected molecular size in Western blots (data not shown).
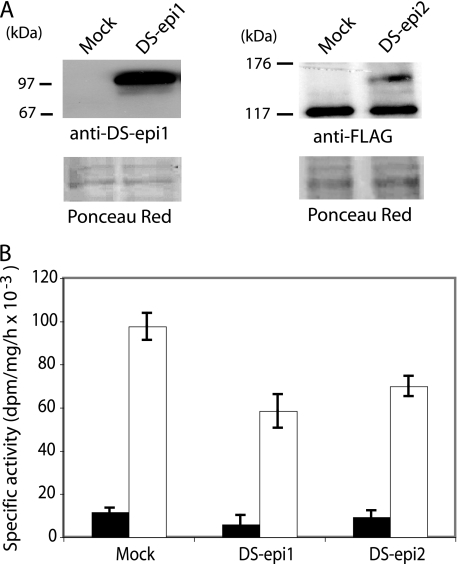
To test the possibility that only DS-epi2 correctly localized to the Golgi compartment would be an active O-sulfotransferase, microsomes from 293HEK cells overexpressing the full-length enzyme were assayed. We hypothesized that detergent would disrupt interactions of DS-epi2 with other protein(s), which could be required for an active DS-epi2 O-sulfotransferase. Thus, enzymatic assays were performed with or without detergent. Epimerase activity was also monitored as a control of expression. Microsomes from cells overexpressing DS-epi2 or DS-epi1 contained 3- and 29-fold more epimerase activity than the control, respectively, when assayed without detergent (data not shown). Addition of detergent led to more DS-epi2 activity, increasing it to 10 times more than that of the mock sample, whereas no further DS-epi1 activity could be gained. Western blot analysis of the microsomes showed that DS-epi2 is an unprocessed protein of 140–150 kDa, identical to the one found in total lysates (Fig. 3A). DS-epi1, however, runs as a 97-kDa band, incompatible with the molecular mass of ∼120 kDa of a glycanated full-size protein (16). Based on observed molecular weight of the overexpressed proteins and on amino acid sequencing of endogenous DS-epi1 (5), the difference in detergent extractability of DS-epi1 and DS-epi2 could be explained by the fact that DS-epi1 has lost the C-terminal portion containing two transmembrane domains (amino acids 901–952), whereas overexpressed DS-epi2 still retains two transmembrane domains (amino acids 774–833). O-Sulfotransferase activity was not significantly different in microsomes prepared from mock-, DS-epi1-, and DS-epi2-transfected cells, when assayed without detergent (Fig. 3B). Solubilization with detergent considerably increased O-sulfotransferase activity in all three samples. Overexpression of DS-epi1 and DS-epi2 resulted in approximately a 40% decrease in O-sulfotransferase activity, compared with the control. It is possible that overexpression of the epimerases disrupts biosynthetic complexes, leading to decreased presence of O-sulfotransferase(s). Furthermore, O-sulfotransferase activity in DS-epi2-transfected microsomes was not significantly higher than in DS-epi1-transfected microsomes.
FIGURE 3.
Analysis for O-sulfotransferase activity of DS-epi2. Microsomes were prepared from 293HEK cells overexpressing full-length DS-epi1 or DS-epi2 and resuspended in dialysis buffer without detergent. A, Western blot of 25 μg of microsomal preparations. B, O-sulfotransferase activity was assayed (mean ± 2 S.D. of triplicates) without detergent (black bars) or with detergent (empty bars). The O-sulfotransferase assay shown was performed at pH 6.8 using chondroitin as substrate. Twenty-five μg of microsomes from mock cells incorporated 2,700 dpm, when assayed with detergent.
To investigate whether the putative O-sulfotransferase activity of DS-epi2 might be responsible for the biosynthesis of the less prominent disulfated moieties in CS/DS, the disaccharides, obtained after complete chondroitinase ABC digestion of CS/DS chains that had been released into the medium by primary human lung fibroblasts (HFL-1), were analyzed. Control cells were compared with cells overexpressing (pcDNA3.1/DS-epi2) or cells in which DS-epi2 was down-regulated (DS-epi2 siRNA). Compared with mock-transfected sample, the amounts of the three types of disulfated disaccharides (recovered as ΔB, ΔD, and ΔE structures after the analysis; where ΔB is Δ4,5-unsaturated hexuronic acid-2-sulfated-N-acetylgalactosamine-4-sulfated; ΔD is Δ4,5-unsaturated hexuronic acid-2-sulfated-N-acetylgalactosamine-6-sulfated; and ΔE is Δ 4,5-unsaturated hexuronic acid-N-acetylgalactosamine-4,6-disulfated) remained unchanged. Only slight changes in the monosulfated disaccharides were noted. This supports the conclusion that the putative O-sulfotransferase domain of DS-epi2 is also inactive in intact cells. In summary, no O-sulfotransferase activity could be ascribed to DS-epi2.
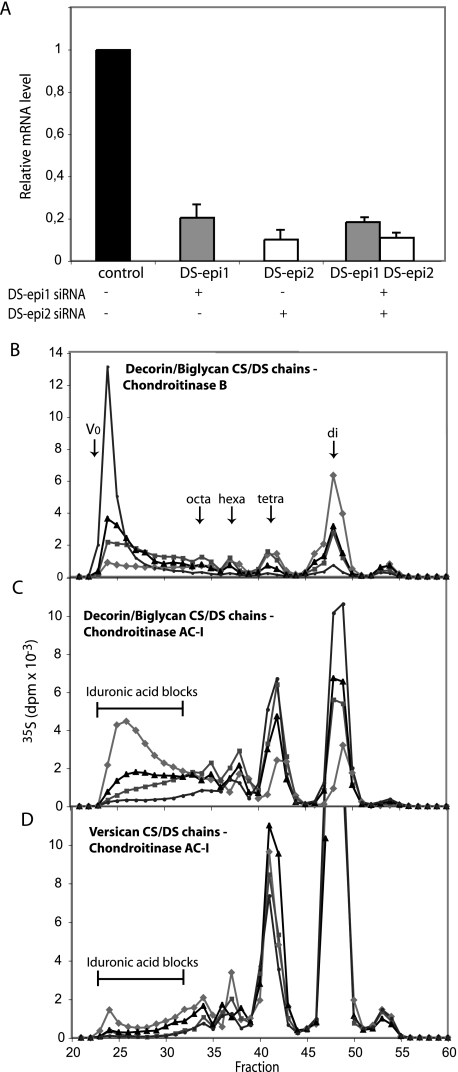
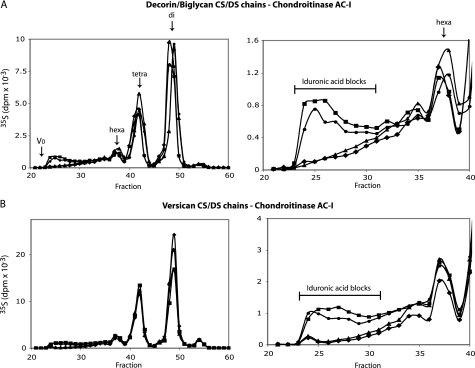
Role of Endogenous DS-epi1 and DS-epi2 in Biosynthesis of the Iduronic Acid Blocks in Fibroblasts—To determine whether the iduronic acid moieties synthesized by the two endogenous DS-epimerases are differently arranged along the DS chain, primary human lung fibroblasts (HFL-1) were transfected with DS-epi1 and DS-epi2 siRNAs. The transfections resulted in 5- and 10-fold reduction of expression of DS-epi1 and DS-epi2 mRNAs, respectively, as revealed by qRT-PCR (Fig. 4A). To study the effect on DS structure, labeled proteoglycans released into the medium were purified and fractionated on a Superose 6 size-permeation column into versican and a mixture of decorin and biglycan. Subsequent size-permeation chromatograms on Superose 6 of decorin/biglycan- or versican-derived CS/DS chains were superimposable for control, DS-epi1, DS-epi2, and concomitant DS-epi1 and DS-epi2 siRNAs samples (data not shown). Furthermore, the 35S-labeled CS/DS-proteoglycan production of the different samples was similar (data not shown). Therefore, down-regulation of either epimerase did not result in any changes of the CS/DS production and chain length. Decorin/biglycan CS/DS chains were digested with chondroitinase B, which cleaves only GalNAc∼IdoUA linkages, and the split products were analyzed by gel filtration. The proportion of iduronic acid in the total hexuronic acid in the chains, calculated from the chromatograms, amounted to 60% in the control cells. The single down-regulation of DS-epi1 or DS-epi2 reduced the iduronic acid content to 32%, which was reduced further to 9% by the combined treatment. The disaccharide peaks in particular, derived from contiguous (IdoUA-GalNAc)n stretches in the native chain, were affected. They amounted to 49% of the total radioactivity of the split products in the control cells, 19% in DS-epi1 down-regulated cells, 24% in DS-epi2 down-regulated cells, and 6% in DS-epi1 and DS-epi2 down-regulated cells (Fig. 4B). The distribution of iduronic acid was confirmed by digestion with chondroitinase AC-I, which cleaves only GalNAc∼GlcUA linkages. The iduronic acid blocks, with the structure (IdoUA-GalNAc)n ≥ 4, representing 51% of the total split products in the control chains, were reduced to 15% in the DS-epi1 down-regulated samples and to 24% in the DS-epi2 down-regulated samples and further decreased to 5% with combined treatment (Fig. 4C).
FIGURE 4.
Endogenous DS-epi1 and DS-epi2 are both active in the biosynthesis of iduronic acid blocks, as revealed by siRNA-mediated down-regulation in human lung fibroblasts. DS-epi1 and DS-epi2 were down-regulated by siRNA treatment, either individually or in combination. A, relative qRT-PCR quantification of DS-epi1 mRNA (grey bars) or DS-epi2 mRNA (empty bars) extracted from the cells after the labeling period. Data represent expression relative to that in the control cells and show mean ± 2 S.D. of triplicates. B–D, labeled proteoglycans were size-fractionated into versican and a mixture of decorin and biglycan. Purified CS/DS chains were cleaved with chondroitinase B or chondroitinase AC-I, as indicated. Split products were separated on the size-permeation Superdex Peptide column. Elution positions of di-, tetra-, hexa-, and octasaccharides are indicated by arrows. AC-I-resistant iduronic acid blocks are shown. Diamonds, control; squares, DS-epi1 siRNA; triangles, DS-epi2 siRNA; circles, DS-epi1 and DS-epi2 siRNAs.
The CS/DS chains of versican were analyzed in a similar way. The total iduronic acid content, calculated from the chromatograms after chondroitinase B treatment (not shown), was 6.8% in the control versican chains, decreased to 2.0% in the DS-epi1 down-regulated samples, to 4.3% in the DS-epi2 down-regulated samples, and to 1.3% in the DS-epi1 and DS-epi2 down-regulated samples. Chromatograms after chondroitinase AC-I digestion showed that 3.1% of the total radioactivity was located in iduronic acid blocks in the control sample, with corresponding values of 0.7, 1, and 0.3% in the DS-epi1, DS-epi2, and combined DS-epi1 and DS-epi2 down-regulated samples (Fig. 4D). In summary, these data indicate that both epimerases are involved in the biosynthesis of the iduronic acid blocks carried by decorin/biglycan and by versican.
Effects of Overexpression of DS-epi1 and DS-epi2 on Formation of Iduronic Acid Domains in 293HEK Cells—DS-epi1 (pcDNA3.1-DS-epi1/myc-His) or DS-epi2 (pCS2-FLAG-DS-epi2) overexpression in 293HEK cells resulted in 37- and 21-fold increased epimerase activity in the total lysates, respectively. Overexpression of DS-epi1 led to the appearance of iduronic acid blocks in both decorin/biglycan chains (Fig. 5A) and versican chains (Fig. 5B). Twenty one-fold expression of DS-epi1 resulted as well in the same increase of block structures (5). Overexpression of DS-epi2 did not result in block formation in either proteoglycan but produced increased amounts of hybrid structures, recovered as tetra-octasaccharides after chondroitinase AC-I treatment (Fig. 5, A and B). Concomitant overexpression of both enzymes did not increase further the biosynthesis of the blocks, compared with overexpression of DS-epi1 alone.
FIGURE 5.
Differences in epimerization pattern obtained after overexpression of DS-epi1 or DS-epi2 in epithelial 293HEK cells. Full-length DS-epi1 or DS-epi2 was overexpressed in 293HEK cells. Labeled CS/DS chains were obtained from purified versican or decorin/biglycan proteoglycans. Purified CS/DS chains were cleaved with chondroitinase AC-I, and split products were separated as in Fig. 4. On the left, the full chromatograms are shown, and on the right a 6× zoom of the fractions equal or larger than hexasaccharides is presented. Diamonds, control; squares, DS-epi1; triangles, DS-epi2; circles, DS-epi1 and DS-epi2.
DISCUSSION
In this study, we have identified the protein encoded by the DSEL (DSE-like) gene. This protein is composed of an epimerase domain followed by an O-sulfotransferase domain and is indeed an active epimerase. In contrast, no O-sulfotransferase activity was detected. Thus, we propose naming the two DS-epimerases DS-epi1, encoded by the DSE gene, and DS-epi2, encoded by the DSEL gene.
DS-epi1 and DS-epi2 are present in the genome of a wide range of animals such as Xenopus tropicalis, Danio rerio, mice, and humans but not in Caenorhabditis elegans or in Drosophila melanogaster. DS-epi1 and DS-epi2 are ubiquitously expressed in humans (6, 17). The two proteins have an N-terminal epimerase domain (amino acids 22–690 of DS-epi1 and amino acids 27–720 of DS-epi2), which is very similar in amino acid sequence and in structural models (16). As predicted, we found that DS-epi2 overexpressed in 293HEK cells is an active epimerase (Fig. 2). During CD/DS biosynthesis, formation of iduronic acid is the first polymer modification reaction, which results in creation of structurally variable domains within the chain. The distribution of N-sulfated-glucosamine in heparan sulfate is a similar process (18). The presence of two DS-epimerases and four HS N-deacetylase/N-sulfotransferases probably reflects the need to fine-tune these processes. On the other hand, in HS the location of iduronic acids, synthesized by a single HS epimerase, is mostly dictated by the close vicinity to N-sulfated glucosamine residues (19).
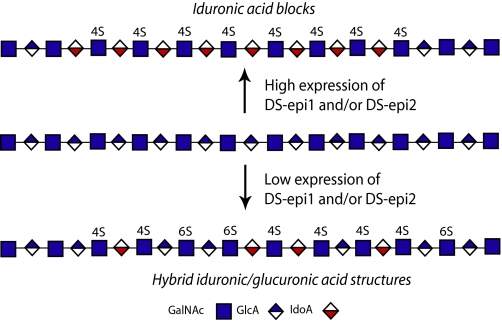
Iduronic acid moieties in dermatan sulfate chains can be clustered together to form long blocks or interspersed among the unmodified glucuronic acid, forming hybrid iduronic-glucuronic structures (Fig. 6). The distribution of iduronic acid determines some of the subsequent O-sulfation reactions. For instance, the long (IdoUA-GalNAc)n stretches are never found with 6-O-sulfates on GalNAc, which instead are mostly 4-O-sulfated. The above structures are substrates for the 2-O-sulfation reaction, giving rise to (IdoUA2OS-GalNAc4OS)n sequences.
FIGURE 6.
Formation of different iduronic acid domains by DS-epi1 and DS-epi2.
DS-epi2 has the capacity to form both hybrid structures and blocks. In fact, an increase in hybrid structures is seen after overexpression of DS-epi2 (Fig. 5). In support of this, in vivo CS/DS chains derived either from the whole body or from skin decorin/biglycan of DS-epi1 knock-out mice still retain hybrid structures in a similar amount to that found in wild-type CS/DS.3 The capacity of DS-epi2 to form blocks is revealed by siRNA-mediated down-regulation of the endogenous enzyme in fibroblasts (Fig. 4). DS-epi1 is also capable of forming the blocks, apparently with higher efficiency than DS-epi2. In fact, down-regulation of DS-epi1 in fibroblasts leads to a decrease in iduronic acid block formation (Fig. 4), and overexpression in epithelial cells leads to block formation (Fig. 5), which is different from DS-epi2 overexpression. The capability of DS-epi1 to form hybrid structures in vivo may be revealed by the DS-epi2 knock-out model, which is underway in our laboratory. To summarize, reduced expression of either epimerase affects block formation (Fig. 6), as noted previously after treating fibroblasts with transforming growth factor-β1, epidermal growth factor, and platelet-derived growth factor (20). It is possible that the relative abundance of the two enzymes, which could not been determined in this study, might also be a regulatory mechanism for the iduronic acid distribution. In an analogous manner, it has been proposed that the relative proportion of NDST1 and EXT2 determines the outcome of the heparan sulfate structure (21).
Enzymes for GAG biosynthesis are believed to work in complexes, to facilitate the transfer of the polysaccharide substrate from one enzyme to the following one (22), even though the evidence to date has only been for enzymes interacting in pairs (21, 23–25). These hypothetical complexes, generally called GAGosomes, which would differ in the composition of the individual enzymes, could be the major factor responsible for the generation of CS/DS chains with different structures. Another mechanism could be the co-localization in different Golgi microcompartments of individual GAGosomes and specific core proteins, thus explaining the core protein-specific CS/DS structure (26). We hypothesized that DS-epi1 and DS-epi2 can interact. However, we could not detect any DS-epi1 bound to overexpressed DS-epi2. Other proteins, either with enzymatic properties or with other roles, such as scaffolding or targeting functions, could be part of such complex(es). The overexpression of DS-epi1 in epithelial 293HEK cells results in a modest amount of iduronic acid blocks (Fig. 5), suggesting that the implicated GAGosome could have other cell-specific limiting components, in addition to the two epimerases.
DS-epi2 has a C-terminal domain that is similar in primary sequence to several 6-O-sulfotransferases, including the sequence of a 3′-PAPS-binding site common to all HS/CS O-sulfotransferases. However, we were unable to demonstrate any O-sulfotransferase activity in an immunopurified overexpressed protein or to trace this activity in the structure of CS/DS chains synthesized by cells in which DS-epi2 was overexpressed or down-regulated (Fig. 3). It can be speculated that an enzymatically defective O-sulfotransferase domain in DS-epi2 could have allosteric properties that would affect the epimerase reaction upon binding to the substrate(s). The idea of a possible link between epimerization and the C-terminal domain is supported by the observation that we did not detect any epimerase activity on overexpression of the isolated epimerase domain (amino acids 31–734) in 293HEK, despite the fact that an abundant protein of the expected size was expressed (data not shown). Interestingly, it has been shown that substrate binding to heparan sulfate 3-O-sulfotransferase-1 induces a conformational change (27).
A structural function of iduronic acid is to increase the flexibility of DS chains, in turn facilitating the interaction with proteins (28). The binding of CS/DS to most growth factors requires the presence of iduronic acid, in as yet incompletely described sequences (29). For instance (IdoUA-GalNAc4OS)n, which represents the major structural element in the iduronic acid blocks, is important in FGF-2 binding (30). Moreover, three disulfated disaccharide units have been found to be part of the sites that interact with several growth factors, many of which are expressed in brain, such as pleiotrophin, midkine, FGF-10, and FGF-18 (1). Many of the structures containing disulfated disaccharides have been found to be IdoUA/GlcUA hybrid structures (31). The gene encoding DS-epi2 has been genetically linked to bipolar disorder (6), and interestingly, brain is the tissue with the highest DS-epi2 expression and the lowest DS-epi1 expression (6, 17). In the central nervous system, the interaction between DS and growth factors affects axonal regeneration, neuronal plasticity, and neurite outgrowth (1). Any involvement of functional DS in these neurobiological processes might explain the linkage between human bipolar disorder and alteration of DS-epi2.
Finally, the present results indicate that modulation of the expression of DS-epi1 and DS-epi2 could reveal the biological significance of specific DS domains. DS-epi1 and DS-epi2 gene knock-out models are clearly an approach to this end.
Acknowledgments
We are grateful to Shirui Hou and Edgar Pera for advice concerning the pCS2 expression vector and to Olivia Rizescu for skillful technical assistance.
This work was supported by grants from the Swedish Science Research Council, the Medical Faculty of Lund University, the AlbertÖsterlund Foundation, the Greta and Johan Kock Foundation, Polysackaridforskning AB, and the Tissue in Motion Medical Faculty Program.
Footnotes
The abbreviations used are: CS/DS, chondroitin sulfate/dermatan sulfate; DS-epi1, DS-epimerase 1; DS-epi2, DS-epimerase 2; GlcUA, glucuronic acid; GalNAc, N-acetylgalactosamine; HS, heparan sulfate; IdoUA, iduronic acid; MES, 4-morpholineethanesulfonic acid; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; ST, sulfotransferase; siRNA, short interfering RNA; HEK, human embryo kidney; BSA, bovine serum albumin; GAG, glycosaminoglycan; qRT, quantitative reverse transcription.
M. Maccarana, S. Kalamajski, A. Oldberg, and A. Malmstrom, manuscript in preparation.
References
- 1.Sugahara, K., and Mikami, T. (2007) Curr. Opin. Struct. Biol. 17 536–545 [DOI] [PubMed] [Google Scholar]
- 2.Trowbridge, J. M., and Gallo, R. L. (2002) Glycobiology 12 117R–125R [DOI] [PubMed] [Google Scholar]
- 3.Prabhakar, V., and Sasisekharan, R. (2006) Adv. Pharmacol. 53 69–115 [DOI] [PubMed] [Google Scholar]
- 4.Malmstrom, A. (1984) J. Biol. Chem. 259 161–165 [PubMed] [Google Scholar]
- 5.Maccarana, M., Olander, B., Malmstrom, J., Tiedemann, K., Aebersold, R., Lindahl, U., Li, J. P., and Malmstrom, A. (2006) J. Biol. Chem. 281 11560–11568 [DOI] [PubMed] [Google Scholar]
- 6.Goossens, D., Van Gestel, S., Claes, S., De Rijk, P., Souery, D., Massat, I., van den Bossche, D., Backhovens, H., Mendlewicz, J., Van Broeckhoven, C., and Del Favero, J. (2003) Mol. Psychiatry 8 83–89 [DOI] [PubMed] [Google Scholar]
- 7.Hou, S., Maccarana, M., Min, T. H., Strate, I., and Pera, E. M. (2007) Dev. Cell 13 226–241 [DOI] [PubMed] [Google Scholar]
- 8.Hannesson, H. H., Hagner-McWhirter, A., Tiedemann, K., Lindahl, U., and Malmstrom, A. (1996) Biochem. J. 313 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikami, T., Mizumoto, S., Kago, N., Kitagawa, H., and Sugahara, K. (2003) J. Biol. Chem. 278 36115–36127 [DOI] [PubMed] [Google Scholar]
- 10.Kantor, T., and Scubert, M. (1957) J. Am. Chem. Soc. 79 152–153 [Google Scholar]
- 11.Nagasawa, K., Inoue, Y., and Tokuyasu, T. (1979) J. Biochem. (Tokyo) 86 1323–1329 [DOI] [PubMed] [Google Scholar]
- 12.Wlad, H., Maccarana, M., Eriksson, I., Kjellen, L., and Lindahl, U. (1994) J. Biol. Chem. 269 24538–24541 [PubMed] [Google Scholar]
- 13.Tufvesson, E., Malmstrom, J., Marko-Varga, G., and Westergren-Thorsson, G. (2002) Eur. J. Biochem. 269 3688–3696 [DOI] [PubMed] [Google Scholar]
- 14.Shively, J. E., and Conrad, H. E. (1976) Biochemistry 15 3932–3942 [DOI] [PubMed] [Google Scholar]
- 15.Midura, R. J., Salustri, A., Calabro, A., Yanagishita, M., and Hascall, V. C. (1994) Glycobiology 4 333–342 [DOI] [PubMed] [Google Scholar]
- 16.Pacheco, B., Maccarana, M., Goodlett, D. R., Malmstrom, A., and Malmstrom, L. (2008) J. Biol. Chem. 284 1741–1747 [DOI] [PubMed] [Google Scholar]
- 17.Nakao, M., Shichijo, S., Imaizumi, T., Inoue, Y., Matsunaga, K., Yamada, A., Kikuchi, M., Tsuda, N., Ohta, K., Takamori, S., Yamana, H., Fujita, H., and Itoh, K. (2000) J. Immunol. 164 2565–2574 [DOI] [PubMed] [Google Scholar]
- 18.Kjellen, L. (2003) Biochem. Soc. Trans. 31 340–342 [DOI] [PubMed] [Google Scholar]
- 19.Maccarana, M., Sakura, Y., Tawada, A., Yoshida, K., and Lindahl, U. (1996) J. Biol. Chem. 271 17804–17810 [DOI] [PubMed] [Google Scholar]
- 20.Tiedemann, K., Olander, B., Eklund, E., Todorova, L., Bengtsson, M., Maccarana, M., Westergren-Thorsson, G., and Malmstrom, A. (2005) Glycobiology 15 1277–1285 [DOI] [PubMed] [Google Scholar]
- 21.Presto, J., Thuveson, M., Carlsson, P., Busse, M., Wilen, M., Eriksson, I., Kusche-Gullberg, M., and Kjellen, L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esko, J. D., and Selleck, S. B. (2002) Annu. Rev. Biochem. 71 435–471 [DOI] [PubMed] [Google Scholar]
- 23.Pinhal, M. A., Smith, B., Olson, S., Aikawa, J., Kimata, K., and Esko, J. D. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormick, C., Duncan, G., Goutsos, K. T., and Tufaro, F. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izumikawa, T., Koike, T., Shiozawa, S., Sugahara, K., Tamura, J., and Kitagawa, H. (2008) J. Biol. Chem. 283 11396–11406 [DOI] [PubMed] [Google Scholar]
- 26.Seidler, D. G., Breuer, E., Grande-Allen, K. J., Hascall, V. C., and Kresse, H. (2002) J. Biol. Chem. 277 42409–42416 [DOI] [PubMed] [Google Scholar]
- 27.Munoz, E., Xu, D., Kemp, M., Zhang, F., Liu, J., and Linhardt, R. J. (2006) Biochemistry 45 5122–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferro, D. R., Provasoli, A., Ragazzi, M., Casu, B., Torri, G., Bossennec, V., Perly, B., Sinay, P., Petitou, M., and Choay, J. (1990) Carbohydr. Res. 195 157–167 [DOI] [PubMed] [Google Scholar]
- 29.Nandini, C. D., and Sugahara, K. (2006) Adv. Pharmacol. 53 253–279 [DOI] [PubMed] [Google Scholar]
- 30.Taylor, K. R., Rudisill, J. A., and Gallo, R. L. (2005) J. Biol. Chem. 280 5300–5306 [DOI] [PubMed] [Google Scholar]
- 31.Bao, X., Muramatsu, T., and Sugahara, K. (2005) J. Biol. Chem. 280 35318–35328 [DOI] [PubMed] [Google Scholar]