Abstract
Huntington disease and its related autosomal-dominant polyglutamine (pQ) neurodegenerative diseases are characterized by intraneuronal accumulation of protein aggregates. Studies on protein aggregates have revealed the importance of the ubiquitin-proteasome system as the front line of protein quality control (PQC) machinery against aberrant proteins. Recently, we have shown that the autophagy-lysosomal system is also involved in cytoplasmic aggregate degradation, but the nucleus lacked this activity. Consequently, the nucleus relies entirely on the ubiquitin-proteasome system for PQC. According to previous studies, nuclear aggregates possess a higher cellular toxicity than do their cytoplasmic counterparts, however degradation kinetics of nuclear aggregates have been poorly understood. Here we show that nuclear ubiquitin ligases San1p and UHRF-2 each enhance nuclear pQ aggregate degradation and rescued pQ-induced cytotoxicity in cultured cells and primary neurons. Moreover, UHRF-2 is associated with nuclear inclusion bodies in vitro and in vivo. Our data suggest that UHRF-2 is an essential molecule for nuclear pQ degradation as a component of nuclear PQC machinery in mammalian cells.
Huntington disease (HD)2 and related polyglutamine (pQ) diseases are caused by the expansion of trinucleotide repeats encoding pQ within the mutant gene product (1). The pQ length dependence of disease onset and severity in pQ diseases correlates strongly with the tendency of expanded pQ proteins to aggregate in disease models (2, 3). Because of this pQ chain, the gene product assembles into oligomers, further aggregates to form microscopically visible inclusion bodies (IBs), and shows pathological extensity in diseased brains (4).
Aggregated forms of pQ-expanded huntingtin (Htt) can disrupt cellular function in a variety of ways, including inactivation of transcription factors (5, 6) and impairment of the ubiquitin proteasome system (UPS) (7). The precise mechanism of UPS inhibition remains to be solved, but could be of particular interest, because it has been shown to occur in in vivo disease models (8). UPS impairment triggers aggresome formation (9), an active cellular mechanism to enrich cytoplasmic aggregates and autophagy-lysosomal components to the microtubule-organizing center by retrograde transport, which in turn enhances the efficiency and selectivity of autophagic degradation of cytoplasmic aggregates supporting the UPS as an alternative protein quality control (PQC) system (10, 11). However, autophagy is ineffective in clearing nuclear aggregates (12), and there is no known nuclear PQC mechanism other than the UPS. Therefore, the nucleus is a relatively protected environment for aggregates than the cytoplasm.
The majority of the pQ proteins are functional in the nucleus, and the strong correlation between aggregate toxicity and nuclear translocation of pQ proteins has led to the hypothesis that the nucleus is the primary center for action of these proteotoxins (13). This was shown by redirecting nuclear ataxin-1 or androgen receptor to the cytoplasm, resulting in a drastic reduction in neuronal toxicity and aggregate formation in mice models (14, 15).
Therefore, to gain a better understanding of the disease pathomechanism, it is essential to study the nuclear PQC system. Because the autophagy-lysosome system is incapable of this task, we attempted to investigate the role of the nuclear UPS in aggregate degradation. Among the three enzymatic components of the UPS, ubiquitin ligases (E3s) possess substrate recognition capability and are of the greatest number and diversity (16), making them attractive candidates for the degradation of nuclear pQ aggregates. In yeast cells, an E3 San1p has specificity for aberrant proteins and provides PQC against deleterious accumulation of nuclear protein aggregates (17). However, a mammalian functional ortholog has yet to be identified. In this study, we identified UHRF-2 as a mammalian nuclear E3 promoting nuclear pQ degradation. Our findings not only provide a new therapeutic target for pQ diseases but also demonstrate that UHRF-2 is a component of novel nuclear PQC machinery in mammalian cells.
EXPERIMENTAL PROCEDURES
Expression Constructs—At the carboxyl terminus of Htt exon-1 fused with enhanced GFP (Htt-GFP), a nuclear export signal (NES: LALKLAGLDIN) or a triple nuclear localization signal (NLS: DPKKKRKVDPKKKRKVDPKKKRKV) was attached. For yeast expression, Htt-GFP was inserted into pMK vector and San1p was inserted into pGBT9 vector (Clontech, Palo Alto ,CA). For mammalian expression, Htt-GFPs were inserted into pCMV vector (Invitrogen), and San1p, UHRF-1, and UHRF-2 were inserted into p3×FLAG14 vector (Sigma). Full-length ataxin-1 cDNA was inserted into pcDNA4 mycHis vector, and full-length dentato-rubro-pallido-luysian atrophy (DRPLA) cDNA was tagged with the amino terminus FLAG sequence and subcloned into pcDNA 3.1 vector (Invitrogen). The sequences of all constructs used were confirmed by direct sequencing of both strands.
Antibodies—Anti-GFP antibody was purchased from Roche Applied Science; anti-V5-HRP antibody was from Invitrogen; anti-FLAG M2-HRP, anti-HA-HRP, anti-His-HRP, and anti-MAP2 antibodies were from Sigma; anti-β-actin and anti-ubiquitin antibodies were from Chemicon (Billerica, MA); and anti-UHRF-2 antibody was from Proteogenix (Oberhausbergen, France). Anti-UHRF-2 antibody was raised in rabbits by injection of the peptide RPDPDHLPGTSTQIEAK, corresponding to amino acids 74–90 of UHRF-2.
Yeast Experiments—Yeast W303 cell line was maintained in selection media containing 2% glucose. G-418 cassette transformation from pFA6aKanMX6 to yeast cells was used to generate san1Δ cells. Htt-GFP expression was measured by a Fluoroskan II fluorescence plate reader (Labsystems, Waltham, MA) with excitation at 488 nm and emission at 500 nm. Each arbitrary fluorescent value was divided by the optical density at 660 nm (A660) for normalization.
Cell Culture and Expression in Mammalian Cells—Stable cells were generated by using 400 μg/ml G-418 selection followed by sorting with a Fluorescence-activated cell sorting (FACS Aria, BD Biosciences, San Jose, CA). Cell viability was measured using the colorimetric CellTiter 96 Aqueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI) with a Model 450 plate reader at A490 (Bio-Rad). Immunoblots were visualized with ECL Western blotting system (Amersham Biosciences) and analyzed by LAS-3000 mini (Fujifilm, Japan). Statistical analysis was performed by using StatView 5.0 (SAS Institute, Cary, NC) or Instat software (GraphPad, San Diego, CA). Significance was tested by the Bonferroni/Dunn post hoc test. Mouse primary cortical neurons were prepared from E16 or E17 ICR mice. Transfection was performed by Nucleofector with a mouse neuron kit (VPG-1001, AMAXA Biosystems, Gaithersburg, MD).
Pulse-chase Experiment—Cells grown in 6-well plates were incubated in methionine- and cysteine-free medium for 30 min and pulse-labeled with 9.25 MBq (250 μCi) EasyTag EXPRESS protein labeling mix 35S (PerkinElmer Life Sciences). After SDS-PAGE, the polyacrylamide gels were dried and subjected to imaging with a Cyclone phosphorimaging device (Packard BioScience, Meriden, CT). Images were quantitatively analyzed by using ImageJ (National Institutes of Health).
In Vitro Ubiquitylation Reaction—A Bac-to-Bac baculovirus expression system (Invitrogen) was applied to express recombinant proteins in sf-9 cells cultured in SF-900-II medium (Invitrogen). In vitro ubiquitylation was performed with 100 ng of E1 (Sigma), E2 (ubcH5a, Biomol, Plymouth Meeting, PA), E3, and 100 ng of FLAG-ubiquitin (Sigma) in 50 mm Tris (pH 7.5), 5 mm MgCl2, 1 mm ATP, and 1 mm dithiothreitol for 1 h at 30 °C.
Microscopic Images—After 4% paraformaldehyde fixation, the primary antibody was incubated at 4 °C overnight, followed by 1-h incubation at room temperature with Alexa 488 or 546 labeled secondary antibodies (Molecular Probes, Eugene, OR) and 50 μg/ml bisbenzimide (Sigma) or 0.5 μm SYTOX orange (Molecular Probes) for nuclear staining. Microscopic images were obtained by an LSM-510 confocal microscope system (Carl Zeiss, Oberkochen, Germany).
Quantitative RT-PCR—Total RNA was purified with TRIzol (Invitrogen), reverse transcription was carried out with Rever-Tra Ace (Toyobo), quantitative PCR was performed with the HT-7900 system (Applied Biosystems, Foster City, CA) using the Assay ID Hs00287648_m1 probe set for human UHRF-2 and HuGAPDH (Applied Biosystems) as a control probe set for human analysis and Mm00520047_m1 and MsACTB probe sets for mouse analysis.
RNA Interference—Control siRNA (AM4611) was purchased from Ambion (Austin, TX), and siRNA for UHRF-2 was purchased from Invitrogen (HSS133012) or from Ambion (110239). Mouse short hairpin RNA plasmid set was purchased from Superarray (Frederick, MD, KM35540N). Lipofectamine 2000 was used for transfection.
RESULTS
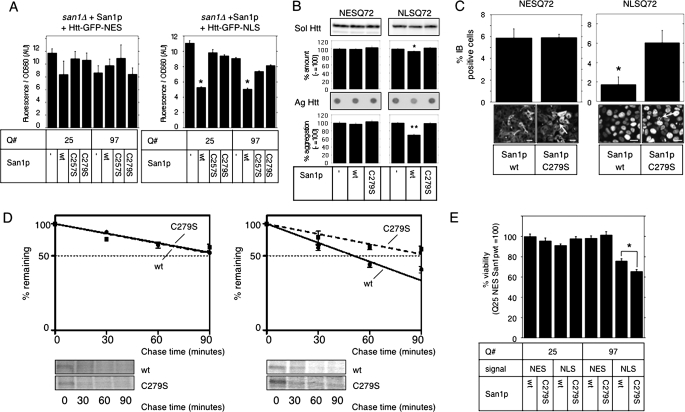
San1p Degrades Nuclear Huntingtin in Yeast and Mammalian Cells—First, we tested whether nuclear ubiquitin ligase San1p is capable of accelerating nuclear pQ degradation in yeast cells. For this purpose, Htt-GFP carrying an NLS or an NES was coexpressed with San1p and its mutants in san1Δ cells. Immunoblot analysis showed no GFP cleavage from Htt; therefore, we estimated the expression level of Htt-GFP by measuring the fluorescence intensity to precisely evaluate the total amount of expressed Htt regardless of the solubility. Expression of wild-type (wt) San1p rescued function, resulting in successful reduction of nuclear Htt-GFP, whereas it had no effect on cytoplasmic Htt-GFP. This reduction of nuclear Htt-GFP depended upon the ubiquitin ligase activity of San1p, because the C257S and C279S mutants, deficient in ligase activity, had no effect on the expression of either Htt-GFP (Fig. 1A). San1p seemed to degrade both short and long pQ proteins, probably due to very high expression under GAL4 promoter producing aberrant protein even with short glutamines. Therefore, we concluded that San1p had the ability to promote nuclear Htt degradation in yeast cells.
FIGURE 1.
San1p accelerates nuclear Htt degradation. A, rescue expression of wt San1p in san1Δ yeast cells reduces Htt-NLS. ADH1 promoter-driven wild-type San1p or its RING mutants (C257S and C279S) were cotransformed into san1Δ cells with GAL4-driven Htt-GFP-NES (left) or NLS (right) containing 25 or 97 glutamines (Q#). Normalized Htt-GFP expression was measured after 2% galactose induction for 4 h. Bars indicate ±S.E. *, p < 0.0001 versus cotransformation with empty vector (–), C257S, or C279S. B, San1p reduces NLS-Q72 aggregates in HeLa cells. FLAG-tagged San1p was expressed in NESQ72 or NLSQ72 cells. After 48 h, the amount of soluble GFP-Htt (Sol Htt) was measured by immunoblot (upper bands and graphs) and those of insoluble aggregates (Ag Htt) were measured by filter trap analysis (lower dots and graphs). The intensity (relative to empty vector (–)) of bands or filter-retained spots was quantified and plotted. Bars indicate ±S.E. *, p = 0.0016; **, p < 0.001 versus empty vector. C, effect of San1p overexpression on NESQ72 (left) or NLSQ72 (right) aggregation was assessed by IB frequency. Cells with IBs were counted (n > 100 from three independent trials) and quantified (top graphs). Black bars indicate ±S.E. Representative images from each experiment are shown at the bottom. Arrows indicate IBs; scale bar = 20 μm. *, p = 0.0108 versus C279S. D, San1p accelerates NLSQ46 degradation. wt or C279S San1p was overexpressed in NESQ46 or NLSQ46 cells for 48 h, and a pulse-chase experiment was performed. Band intensities from SDS-PAGE were normalized by the ratio to those at time = 0 and plotted onto semi-log charts below with fitted lines (wt: straight lines; C279S: broken lines). Bars indicate ±S.E. E, San1p overexpression rescues acute pQ toxicity. NES- or NLS-attached Htt-GFP (signal) with 25 or 97 glutamines (Q#) were transiently coexpressed with wt San1p or C279S mutant (San1p) in HeLa cells for 72 h. Cell viability was measured by colorimetric assay (relative to NESQ25 plus San1pwt; n = 3). Bars indicate ±S.E. *, p = 0.005511.
Because our goal was to elucidate the role of the mammalian UPS in nuclear aggregate degradation, we decided to examine the effect of yeast San1p on Htt-GFP expression in mammalian cells. To evaluate the effect of various interventions on aggregate formation in mammalian cells, we generated HeLa cell lines stably expressing Htt-GFP in selected cellular compartments (supplemental Fig. S1A). Although the amount of soluble Htt-GFP seemed to decrease with pQ length (supplemental Fig. S1B), filter retardation assay showed pQ length-dependent aggregate formation (supplemental Fig. S1C). To test whether San1p suppresses Htt-GFP aggregation in these cells, wt or C279S-mutant San1p was overexpressed in Htt-GFP-NES-Q72 (NESQ72) or Htt-GFP-NLS-Q72 (NLSQ72) cells. Steady-state levels of aggregated Htt were assessed by filter retardation assay (Fig. 1B) or by the frequency of IBs (Fig. 1C), decreased in NLSQ72 cells upon wt San1p expression. We next performed pulse-chase experiments to determine the degradation half-life of Htt-GFP upon San1p overexpression. The half-life of NLSQ46 was significantly shorter when wt San1p was expressed (51.1 ± 3.8 min in wt San1p versus 99.2 ± 19.0 min in C279S mutant; p = 0.034), and the half-life of NESQ46 was not affected (99.3 ± 10.1 min in wt San1p versus 101.3 ± 6.8 min in C279S mutant (Fig. 1D)). We also confirmed that San1p ubiquitylated nuclear pQs (supplemental Fig. S1D). These results confirmed that San1p specifically accelerated nuclear Htt-GFP degradation by ubiquitylation. Because accumulation of aggregated proteins especially in the nucleus leads to cellular dysfunction and cell death, we assessed the effect of San1p on pQ-induced cytotoxicity. Expression of wt San1p significantly restored cellular viability impaired by transiently expressed nuclear pQ97 but not cytoplasmic pQ97 (Fig. 1E). Therefore, we concluded that San1p was capable of accelerating nuclear Htt degradation in mammalian cells and could thereby protect cells from toxicity. We also confirmed that mammalian cells have nuclear PQC machinery driven by the UPS that is responsible for aggregate clearance.
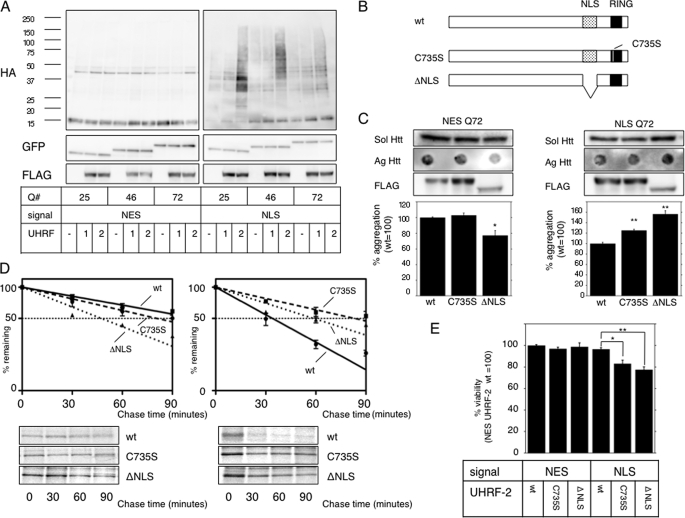
UHRF-2 Ubiquitylates Nuclear Htt-GFP and Promotes Its Degradation—We next attempted to search for mammalian nuclear ubiquitin ligase capable of degrading nuclear pQs. Because San1p does not have apparent homologous proteins in mammals, we searched through a sequence database under criteria of possessing ubiquitin ligase-specific domain and nuclear-localizing potentials, and selected two candidates, UHRF-1 and UHRF-2, because they have RING domains possessing proteins previously reported to show nuclear localization. To test whether UHRF-1 or UHRF-2 ubiquitylates nuclear pQ, we coexpressed hemagglutinin (HA)-tagged ubiquitin with UHRF-1 or UHRF-2 in NESQ or NLSQ cells, and Htt-GFP was immunoprecipitated. An immunoblot for tagged ubiquitin clearly revealed enhancement of the polyubiquitylated smear above the molecular weight of monomeric Htt-GFP only in the presence of nuclear pQ and UHRF-2 (Fig. 2A).
FIGURE 2.
UHRF-2 shows specific E3 activity for nuclear pQ. A, UHRF-2 ubiquitylates nuclear Htt in NES- or NLS-stable HeLa cells. FLAG-tagged UHRF-1 (1), UHRF-2 (2), or empty vector (–) was cotransfected with HA-tagged ubiquitin to NES or NLS HeLa cells for 48 h. After an additional 4 h of 5 μm MG-132 treatment, cell lysates were immunoprecipitated by anti-GFP antibody and subjected to immunoblots by anti-HA antibody (upper panels). This was reblotted by anti-GFP antibody to show immunoprecipitated monomeric Htt-GFP (middle panels). Lower panels show UHRF-1 or UHRF-2 expression in pre-immunoprecipitated lysates. B, schematic diagram showing the wt and mutants of UHRF-2. The RING domain (black box) is at its carboxyl terminus, and one of the critical cysteine residues is at amino acid 735, which was mutated to disrupt ligase activity (C735S). ΔNLS is a nuclear localization signal-defective mutant, from which amino acids 602–693 (dotted box) were deleted. C, wt UHRF-2 decreases aggregate burden. NESQ72 (left) or NLSQ72 (right) cells were transfected with wt, C735S, or ΔNLS mutants of UHRF-2 and evaluated for filter-trapped Htt-GFP. The middle panels show expression of the UHRF-2s detected by anti-FLAG antibody (FLAG). Bars indicate ±S.E. *, p = 0.0149; **, p < 0.0001 versus wt. D, UHRF-2 accelerates Htt-GFP-Q72 degradation. wt, RING domain mutant (C735S), or ΔNLS UHRF-2 was overexpressed in NESQ72 or NLSQ72 cells and a pulse-chase experiment was performed after 48 h. Relative band intensities from SDS-PAGE were normalized to 100% at time = 0 and plotted onto semi-log charts with fitted lines (wt: straight line; C735S: broken line; ΔNLS: dotted line). Bars indicate ±S.E. E, UHRF-2 rescues pQ toxicity. Htt-GFP-NES or NLS with 25Q or 97Q was transiently coexpressed with wt or C735S UHRF-2 in 293T cells. After 48 h, cell viability was measured by colorimetric assay (relative to NES plus wt UHRF-2; n = 12). Bars indicate ±S.E.; *, p = <0.01; **, p < 0.001.
UHRF-2 has a RING domain at its carboxyl terminus (Fig. 2B). To confirm ubiquitin ligase activity dependence on this domain, we generated a cysteine-to-serine mutation at residue 735 (C735S), because mutation of the conserved zinc (corresponding to residue 279 of San1p) binding cysteine residue results in loss of activity in other E3s (18, 19). This UHRF-2 “C735S” did not show any difference in cellular localization (supplemental Fig. S1E, center panels) but lacked in vitro ligase activity (supplemental Fig. S2A). Therefore, we concluded that the RING domain was essential for ligase activity. Because UHRF-2 lacks a canonical NLS sequence, we generated a series of deletion mutants to locate a region critical for its nuclear localization and identified this region between amino acids 602 and 693. The NLS-deficient mutant (ΔNLS) exhibited almost exclusive cytoplasmic localization (supplemental Fig. S1E, right panels) while maintaining in vitro E3 activity (supplemental Fig. S2B). These two mutants served as controls for subsequent assays of UHRF-2 on pQ degradation. To determine the effect of UHRF-2 on Htt-GFP aggregation, we overexpressed UHRF-2 (wt and the mutants) in NESQ72 or NLSQ72 cells. The ΔNLS mutant reduced cytoplasmic aggregates in NESQ72 cells, suggesting that ΔNLS acts as a cytoplasmic E3 (Fig. 2C, left). When we transfected UHRF-2 to NLSQ72 cells, only wt UHRF-2 had a positive effect on aggregate reduction, with no effect on soluble Htt-GFP (Fig. 2C, right). UHRF-2 seemed to accelerate the degradation of aberrant form of Htt, because the soluble amount of Htt was not affected by its overexpression (supplemental Fig. S2C). We then performed pulse-chase experiments to determine the half-life of Htt-GFP-Q46 upon UHRF-2 expression. Half-lives of NESQ46 were calculated as 83.4 ± 13.9 min, 106.2 ± 11.0 min, and 47.4 ± 4.7 min in the presence of wt UHRF-2, C735S, and ΔNLS, respectively. The half-lives of NLSQ46 were 33.8 ± 1.3 min (p = 0.0015 versus C735S, p = 0.0041 versus ΔNLS), 87.5 ± 17.1 min, and 58.8 ± 4.0 min with wt UHRF-2, C735S, and ΔNLS, respectively (Fig. 2D). We therefore concluded that UHRF-2 specifically promoted nuclear Htt degradation. To assess the effect of UHRF-2 on pQ cytotoxicity, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxy-methonyphenol)-2-(4-sulfophenyl)-2H-tetrazolium assay was performed on cells expressing Htt-GFP-Q97 and UHRF-2. Only wt UHRF-2 rescued nuclear pQ toxicity (Fig. 2E). Thus, we concluded that UHRF-2 accelerated nuclear Htt degradation by promoting its ubiquitylation, leading to suppression of cellular toxicity.
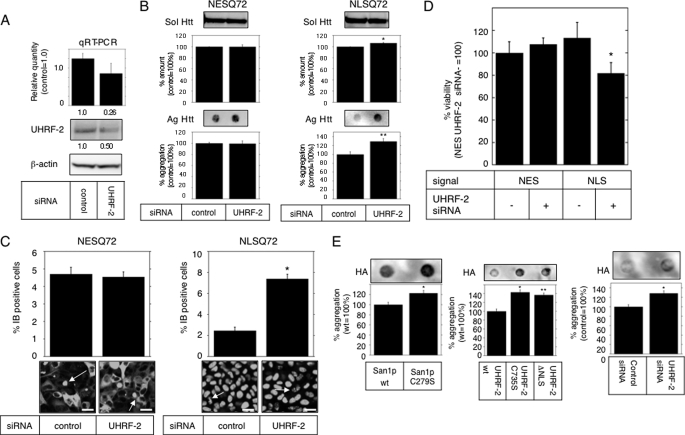
UHRF-2 Contributes to Nuclear Htt Degradation—To test the importance of UHRF-2 in nuclear pQ degradation, we decided to reduce the endogenous UHRF-2 level by RNAi. To detect endogenous UHRF-2, we generated an antibody that specifically recognized UHRF-2 over UHRF-1 (supplemental Fig. S2D) and was able to detect endogenous human UHRF-2 (supplemental Fig. S2E). To knock down UHRF-2, we tested 14 independent siRNAs (data not shown) but found only two that had relatively high efficiency (Fig. 3A). To assess the effect of reduced UHRF-2 levels on the aggregate burden, this siRNA was transfected to NESQ72 or NLSQ72 cells to show the increase in the amount of nuclear aggregates measured by filter trap assay (Fig. 3B) or by the frequency of IBs (Fig. 3C). We next tested the effect of UHRF-2 knockdown on acute pQ toxicity: UHRF-2 knockdown resulted in a significant reduction in cell viability only in the presence of nuclear Htt-Q97 (Fig. 3D). To test whether UHRF-2 is able to accelerate non-Htt pQ degradation, we used a full-length DRPLA protein with 80 glutamines (DRPLAQ80) as another model pQ protein. Overexpression of San1p and UHRF-2 reduced the amount of filter-trapped DRPLAQ80 aggregates; conversely, UHRF-2 knockdown increased the aggregate burden (Fig. 3E), consistent with previous experiments. The pulse-chase experiment resulted in accelerated degradation (half-life of 110.1 ± 1.9 min with wt UHRF-2 and 167.0 ± 7.0 min with C735S) of DRPLA only by wt UHRF-2 (supplemental Fig. S2F). The results were confirmed by two different siRNAs to eliminate the off-target effect (data not shown). From these data, we concluded that UHRF-2 contributed deeply to nuclear pQ degradation.
FIGURE 3.
UHRF-2 contributes to nuclear Htt degradation. A, effect of UHRF-2 RNAi on HeLa cells was evaluated by quantitative RT-PCR and immunoblot. The quantitative RT-PCR results showed 74% knockdown of mRNA (upper graph: relative quantity shown below the graph), resulting in a 50% protein reduction (lower blot: measured intensity shown below the blot). Bars indicate 95% confidence level. B, UHRF-2 knockdown increases nuclear aggregates in HeLa stable cell lines. After 72 h of transfection, the amount of soluble GFP-Htt (Sol Htt) was measured by immunoblot analysis (upper bands and graphs), and those of insoluble aggregates (Ag Htt) were measured by filter trap analysis (lower dots and graphs). Bars indicate ±S.E.; *, p = 0.0289; **, p = 0.0174 versus control. C, UHRF-2 knockdown increases IB frequency in HeLa stable cells. Effect of UHRF-2 on NESQ72 (left) or NLSQ72 (right) aggregation was assessed by IB frequency after 72 h of transfection. Cells with IBs (representative field, GFP) were counted (n > 100 from three independent trials) and quantified (top). Black bars indicate ±S.E.; *, p < 0.0001 versus control siRNA. Representative images are shown at the bottom. Arrows indicate IBs; scale bars = 20 μm. D, UHRF-2 rescues nuclear pQ toxicity. NES- or NLS-attached Htt-GFP 97Q was cotransfected with UHRF-2 siRNA (upper panel) or short hairpin RNA plasmid (lower panel) to 293T cells for 48 h. The effect of UHRF-2 RNAi on pQ toxicity was measured by colorimetric assay (relative to NES plus control; n = 13). Bars indicate ±S.E. *, p < 0.001. E, nuclear E3s degrade DRPLA aggregates. San1p (left panel,*, p = 0.0341 versus wt), UHRF-2 (middle panel,*, p = 0.001; ** p = 0.0023 versus wt) or siRNA against UHRF-2 (right panel,*, p = 0.0144 versus control) were coexpressed with HA-tagged DRPLA proteins in HeLa cells. After 48 h of transfection, the aggregate burden was analyzed with filter trap analysis. Intensities (relative to wt or control) of filter-retained spots were quantified by image analysis (upper corresponding dots) and plotted (lower graphs). Bars indicate ±S.E.
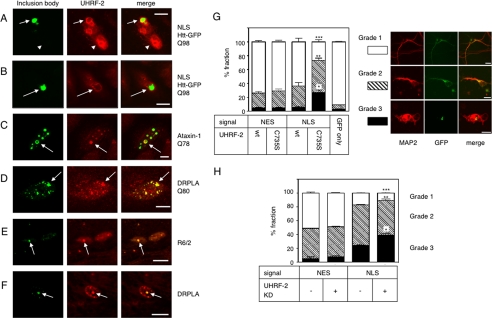
UHRF-2 Associates with IBs in Vitro and in Vivo—To evaluate the physiological role of UHRF-2 in HD pathogenesis, immunocytochemical studies were performed. First, we determined whether overexpressed UHRF-2 associates with IBs (Fig. 4A) and found positive results in almost all coexpressing cells. We then tested whether endogenous UHRF-2 associates with Htt-NLS IBs (Fig. 4B). The UHRF-2 signal seemed to surround Htt IBs without exact colocalization, due to the structural tightness of Htt IBs not allowing most of the antibodies to penetrate inside the IBs and, therefore, decorated the rim of the IBs. We also examined the colocalization of endogenous UHRF-2 with nuclear IBs formed by ataxin-1 (Fig. 4C) and DRPLA (Fig. 4D), which also produced positive results. Given our results demonstrating the in vitro association of UHRF-2 to IBs, we next tested whether UHRF-2 associates with in vivo IBs formed in R6/2 mice. Our UHRF-2 antibody recognized mouse UHRF-2, although mouse UHRF-2 does not have the exact same sequence as the immunogen we used (supplemental Fig. S2G). Indirect immunohistochemical staining of the mouse sections revealed that ∼50% of the ubiquitin-positive IBs formed in 6-week-old R6/2 mice cortex were positive for UHRF-2 (Fig. 4E), suggesting a physiological role in disease pathogenesis. Moreover, nuclear ubiquitin-positive inclusion in a postmortem section from a DRPLA patient was positive for UHRF-2 (Fig. 4F).
FIGURE 4.
Physiological role of UHRF-2 in pQ pathogenesis. A, fluorescence micrograph of Htt-NLS-Q97 IB surrounded by coexpressed FLAG-tagged UHRF-2 in HeLa cells. IBs are visualized by GFP fluorescence; UHRF-2 was visualized by anti-FLAG and Alexa 546 secondary antibody. Scale bar = 10 μm. Arrows indicate IB; arrowheads indicate neighboring cell nuclei expressing UHRF-2 alone. B, Htt-NLS-Q97 IB transiently expressed in HeLa cells is surrounded by endogenous UHRF-2. Scale bar = 10 μm. Arrows indicate IBs. C, ataxin-1-Q78 IBs transiently formed in HeLa cells colocalize with UHRF-2. IB was labeled with anti-Myc antibody and Alexa 488 secondary antibody. Scale bar = 2 μm. Arrows indicate IBs. D, DRPLAQ80 IBs transiently formed in HeLa cells are positive for UHRF-2. IBs are labeled with anti-FLAG antibody and Alexa 488 secondary antibody. Scale bar = 2 μm. Arrows indicate IBs. E, Htt inclusion is positive for UHRF-2 in vivo. Section from a 6-week-old R6/2 mouse was stained with anti-UHRF-2 antibody. Anti-UHRF-2 antibody stained the IBs. IBs are labeled with anti-ubiquitin antibody. Scale bar = 10 μm. F, DRPLA inclusion is positive for UHRF-2 in vivo. Section from a postmortem DRPLA patient was stained with anti-UHRF-2 antibody. Anti-UHRF-2 antibody stained the IB labeled with anti-ubiquitin antibody. Scale bar = 10 μm. G, UHRF-2 overexpression rescues pQ toxicity in primary cultured mouse neurons. Mouse primary cortical neurons were cotransfected with Htt-Q97 and UHRF-2. After 96 h, coverslips were stained with anti-MAP2 antibody to identify neurons. The shape of the neurons determined by neurite outgrowth was classified into three grades: healthy, fully neurite-grown cells were grouped as grade 1 (a representative cell is shown in the right upper panels); cells with moderately grown neurites were grade 2 (middle panels); cells with unhealthy appearance were grade 3 (lower panels). At least 100 cells from four different coverslips were counted for each transfection. Scale bars = 10 μm. Black bars indicate ±S.E.; *, p = 0.0020; **, p = 0.0407; ***, p = 0.0009 versus wt NLS. H, UHRF-2 knockdown enhances pQ toxicity in primary cultured mouse neurons. Mouse primary cortical neurons were cotransfected with Htt-Q97 and mUHRF-2 or control short hairpin RNA plasmid. After 96 h, coverslips were stained with anti-MAP2 antibody to identify neurons. Evaluation was carried out by the same method as in the overexpression experiment. At least 100 cells from three different coverslips were counted for each transfection. Black bars indicate S.E.; *, p = 0.0049; **, p = 0.0182; ***, p = 0.007 versus NLS RNAi (–).
UHRF-2 Is Essential for Neuroprotection against Nuclear pQs—Our final goal was to test the significance of UHRF-2-mediated nuclear aggregate degradation in neurons. To do so, we first tested if overexpressed UHRF-2 could rescue neurotoxicity of nuclear pQ aggregates by overexpressing UHRF-2 in mouse primary cultured cortical neurons. The effect of pQ neurotoxicity was evaluated by the appearance of MAP2-positive cells observed under microscopy. As a transfection control, GFP showed minimal damage to neurons. By counting neurons classified by the appearances into three grades (Fig. 4G, right panels), expression of NESQ97 showed some degree of toxicity, which was not altered by UHRF-2 coexpression. NLSQ97 expressed with UHRF-2 C735S showed significant neurotoxicity that was rescued by wt UHRF-2 (Fig. 4G, left chart). To test whether UHRF-2 is critical for neuroprotection, an RNAi experiment was performed. Clearly, knockdown of mouse UHRF-2 resulted in enhanced neurotoxity only in the presence of NLSQ97 (Fig. 4H). Consequently, we concluded that UHRF-2 was an essential molecule for the protection of nuclear pQ toxicity in neurons.
DISCUSSION
Understanding the Nuclear Aggregate Degradation Machinery—Accumulation of protein aggregates in intracellular IBs has long been recognized as a pathognomonic feature of pQ diseases, and research on potential therapeutics has been focused on their removal. Indeed, reduction of aggregated proteins by accelerating their degradation leads to improved phenotypes (20–22); these findings have also contributed to the basic understanding of the cellular PQC system. To date, studies on pQ aggregates have focused mostly on the cytoplasmic degradation system, such as molecular chaperones, the UPS, and autophagy, because details about these systems had been well characterized. Previously, we have shown that autophagy activated under proteasomal impairment contributes to partial degradation of cytoplasmic protein aggregates through an aggresome formation pathway, which however has no effect on nuclear aggregates (12). Therefore, the study indicated that the nucleus has relatively poor PQC machinery compared with the cytoplasm, and nuclear translocated pQs could be resistant to degradation. A common pathological feature linking pQ diseases is the presence of intranuclear IBs rather than cytoplasmic IBs, which are indirect markers of the presence of more toxic oligomeric aggregates in the cellular compartment. Each time a cell divides, the nuclear envelope breaks up to mix its nuclear and cytoplasmic components. This gives cells the opportunity to capture nuclear aggregates with cytoplasmic machinery (e.g. autophagy) and to promote degradation, which could partially explain some of our data presented in this study that nuclear E3s seemed to have some activity on aggregate degradation across the cellular compartment (Fig. 2C, NES versus C735S). However, as neurons are terminally differentiated cells that do not undergo further division, any aggregates with diameters larger than the nuclear pore will be unable to exit the nucleus and will therefore never be captured by the autophagylysosome system in diseased brain unless there is an active nuclear export system for aggregates, which has not been discovered so far. Data provided in this study establish the role of nuclear E3s in the intranuclear PQC system and demonstrate its effectiveness in reducing nuclear aggregates.
San1p and Nuclear pQ Aggregate Degradation—To date, the machinery responsible for the nuclear PQC system has not been well characterized. Although a number of chaperones have been implicated in protein refolding and disaggregation in the nucleus (23, 24), virtually no components of the degradation mechanism were found in the nucleus until San1p was reported to function in this role in yeast (17), and such a system has never been reported for mammalian cells. We were able to show that San1p can degrade nuclear Htt in yeast and also in a mammalian system, clearly demonstrating that the mammalian nucleus is endowed with UPS components capable of pQ aggregate degradation. San1p expression in mammalian cells seems to be regulated by UPS activity, because its expression level was minimal without UPS inhibition. This may have been due to its auto-ubiquitylation activity, because the C279S mutant showed excellent expression even without proteasome inhibition. The identification of San1p as a nuclear pQ degradation enhancer encouraged us to seek similar mechanisms in mammalian cells.
Ubiquitin Ligase UHRF-2 as a Nuclear pQ Aggregate Degrader—To explore the nuclear aggregate degradation system in human cells, we searched through a sequence data base for human nuclear E3 and found UHRFs. UHRFs (ubiquitin plant homeodomain RING fingers) are a family of proteins initially found in cancer cells that are reportedly associated with S-phase progression and cell cycle regulation (25–27). They have a ubiquitin-like domain, a plant homeodomain, and a RING domain (28). The family includes two homologous proteins, UHRF-1 and UHRF-2. UHRF-1 is better characterized than UHRF-2 and is reportedly associated with cell proliferation (29), ubiquitylation of histone H3 (30), DNA damage (30), and DNA methylation promotion (31). Less is known about UHRF-2; it shares 52.6% sequence homology with UHRF-1 (28) and is up-regulated upon carcinogen exposure (32). It is reportedly well expressed in the brain (33). Unlike San1p, UHRFs have been reported to harbor their NLS at the N terminus, where charged amino acids are relatively abundant (29, 30, 34). We therefore constructed an N terminus deletion mutant, which failed to show its cytoplasmic localization. Attempts to generate an NLS-defective mutant were thus carried out by generating various deletion mutants. We were able to identify an NLS responsible for localization, but surprisingly it was more proximal to the carboxyl terminus. The dependence of E3 activity at its RING domain, which has been predicted, was confirmed by generating cysteine residue mutants disrupting the cross-brace architecture characterizing RING domains. These data strongly supported our conclusion that UHRF-2 is also a nuclear E3.
UHRF-2 as a Key Molecule in pQ Pathogenesis—Although UHRF-2 promoted nuclear Htt-GFP ubiquitylation, it had no or little effect on cytoplasmic Htt or nuclear GFP itself. The effect of UHRF-2 knockdown on the aggregate amount was not that prominent compared with overexpression in the Htt-GFP filter retardation assay. Two explanations for this observation are possible. One is that the effect of knockdown was not sufficient, because the reduction of the protein level was 50% even by the best siRNA tested. This possibility could be explained by an experiment using another siRNA, which had a 13% reduction in the protein amount without any change in the amounts of aggregates (data not shown). The second possibility is that there could be contributions of other nuclear E3s possessing functional redundancy with UHRF-2. The results obtained in cellular viability assays upon overexpression or RNAi of UHRF-2 showed significant change. To further confirm this, we overexpressed UHRF-2 in primary cultured neurons to show significant effect on rescuing pQ-induced neurotoxicity. RNAi was more effective in mouse cells than in human cells to achieve almost 80% knockdown. This system was further applied to mouse primary cultured neurons to show a quite dramatic effect of UHRF-2 knock down resulting in enhanced neurotoxicity.
Therefore, we conclude that nuclear Htt is a specific substrate of UHRF-2. Data presented with overexpression and knockdown of UHRF-2 clearly show that UHRF-2 is involved in nuclear pQ degradation and is essential for neuroprotection against nuclear pQ aggregates. Moreover, immunohistochemical data showing an association of UHRF-2 with in vitro and in vivo Htt-IBs and IBs of other pQ molecules, ataxin-1 (35) and DRPLA (36), support the hypothesis that it is important in pQ disease pathogenesis. Lastly, UHRF-2 is effective not only for degrading aggregates of Htt but also for DRPLA, suggesting a more general role as a global degradation component for pQ. Taken together, these data implicate UHRF-2 as a key molecule for the degradation of nuclear pQ. In addition to demonstrating that mammalian cells are endowed with nuclear PQC machinery, our findings shed new light on the understanding of pQ diseases and offer a novel and viable therapeutic target for intervention into these debilitating conditions.
Supplementary Material
Acknowledgments
We thank Masaru Kurosawa for technical assistance, John Christianson for critical readings and manuscript revision, and Akira Yamashita for critical readings.
This work was supported by a grant-in aid from the Japan Society for the Promotion of Science and by research funding from the Takeda science foundation (Osaka, Japan), the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Sankyo Foundation for Life Science, and Janssen Pharmaceutical K.K. (Tokyo, Japan).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: HD, Huntington disease; pQ, polyglutamine; IB, inclusion body; Htt, huntingtin; UPS, ubiquitin-proteasome system; PQC, protein quality control; E3, ubiquitin-protein isopeptide ligase; GFP, green fluorescent protein; NES, nuclear export signal; NLS, nuclear localization signal; CMV, cytomegalovirus; DRPLA, dentato-rubro-pallido-luysian atrophy; HRP, horseradish peroxidase; RNAi, RNA interference; wt, wild type.
References
- 1.Zoghbi, H. Y., and Orr, H. T. (2000) Annu. Rev. Neurosci. 23 217–247 [DOI] [PubMed] [Google Scholar]
- 2.Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R., Bates, G. P., Davies, S. W., Lehrach, H., and Wanker, E. E. (1997) Cell 90 549–558 [DOI] [PubMed] [Google Scholar]
- 3.Jackson, G. R., Salecker, I., Dong, X., Yao, X., Arnheim, N., Faber, P. W., MacDonald, M. E., and Zipursky, S. L. (1998) Neuron 21 633–642 [DOI] [PubMed] [Google Scholar]
- 4.Ross, C. A., and Poirier, M. A. (2004) Nat. Med. 10 Suppl. S10–S17 [DOI] [PubMed] [Google Scholar]
- 5.Shimohata, T., Nakajima, T., Yamada, M., Uchida, C., Onodera, O., Naruse, S., Kimura, T., Koide, R., Nozaki, K., Sano, Y., Ishiguro, H., Sakoe, K., Ooshima, T., Sato, A., Ikeuchi, T., Oyake, M., Sato, T., Aoyagi, Y., Hozumi, I., Nagatsu, T., Takiyama, Y., Nishizawa, M., Goto, J., Kanazawa, I., Davidson, I., Tanese, N., Takahashi, H., and Tsuji, S. (2000) Nat. Genet. 26 29–36 [DOI] [PubMed] [Google Scholar]
- 6.Yamanaka, T., Miyazaki, H., Oyama, F., Kurosawa, M., Washizu, C., Doi, H., and Nukina, N. (2008) EMBO J. 27 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bence, N. F., Sampat, R. M., and Kopito, R. R. (2001) Science 292 1552–1555 [DOI] [PubMed] [Google Scholar]
- 8.Bennett, E. J., Shaler, T. A., Woodman, B., Ryu, K. Y., Zaitseva, T. S., Becker, C. H., Bates, G. P., Schulman, H., and Kopito, R. R. (2007) Nature 448 704–708 [DOI] [PubMed] [Google Scholar]
- 9.Johnston, J. A., Ward, C. L., and Kopito, R. R. (1998) J. Cell Biol. 143 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopito, R. R. (2000) Trends Cell Biol. 10 524–530 [DOI] [PubMed] [Google Scholar]
- 11.Iwata, A., Riley, B. E., Johnston, J. A., and Kopito, R. R. (2005) J. Biol. Chem. 280 40282–40292 [DOI] [PubMed] [Google Scholar]
- 12.Iwata, A., Christianson, J. C., Bucci, M., Ellerby, L. M., Nukina, N., Forno, L. S., and Kopito, R. R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13135–13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lunkes, A., Lindenberg, K. S., Ben-Haiem, L., Weber, C., Devys, D., Landwehrmeyer, G. B., Mandel, J. L., and Trottier, Y. (2002) Mol. Cell 10 259–269 [DOI] [PubMed] [Google Scholar]
- 14.Katsuno, M., Adachi, H., Kume, A., Li, M., Nakagomi, Y., Niwa, H., Sang, C., Kobayashi, Y., Doyu, M., and Sobue, G. (2002) Neuron 35 843–854 [DOI] [PubMed] [Google Scholar]
- 15.Klement, I. A., Skinner, P. J., Kaytor, M. D., Yi, H., Hersch, S. M., Clark, H. B., Zoghbi, H. Y., and Orr, H. T. (1998) Cell 95 41–53 [DOI] [PubMed] [Google Scholar]
- 16.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425–479 [DOI] [PubMed] [Google Scholar]
- 17.Gardner, R. G., Nelson, Z. W., and Gottschling, D. E. (2005) Cell 120 803–815 [DOI] [PubMed] [Google Scholar]
- 18.Joazeiro, C. A., Wing, S. S., Huang, H., Leverson, J. D., Hunter, T., and Liu, Y. C. (1999) Science 286 309–312 [DOI] [PubMed] [Google Scholar]
- 19.Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S., and Weissman, A. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warrick, J. M., Chan, H. Y., Gray-Board, G. L., Chai, Y., Paulson, H. L., and Bonini, N. M. (1999) Nat. Genet. 23 425–428 [DOI] [PubMed] [Google Scholar]
- 21.Cummings, C. J., Mancini, M. A., Antalffy, B., DeFranco, D. B., Orr, H. T., and Zoghbi, H. Y. (1998) Nat. Genet. 19 148–154 [DOI] [PubMed] [Google Scholar]
- 22.Waza, M., Adachi, H., Katsuno, M., Minamiyama, M., Sang, C., Tanaka, F., Inukai, A., Doyu, M., and Sobue, G. (2005) Nat. Med. 11 1088–1095 [DOI] [PubMed] [Google Scholar]
- 23.Rossi, J. M., and Lindquist, S. (1989) J. Cell Biol. 108 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsell, D. A., Kowal, A. S., Singer, M. A., and Lindquist, S. (1994) Nature 372 475–478 [DOI] [PubMed] [Google Scholar]
- 25.Uemura, T., Kubo, E., Kanari, Y., Ikemura, T., Tatsumi, K., and Muto, M. (2000) Cell Struct. Funct. 25 149–159 [DOI] [PubMed] [Google Scholar]
- 26.Mousli, M., Hopfner, R., Abbady, A. Q., Monte, D., Jeanblanc, M., Oudet, P., Louis, B., and Bronner, C. (2003) Br. J. Cancer 89 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori, T., Li, Y., Hata, H., and Kochi, H. (2004) FEBS Lett. 557 209–214 [DOI] [PubMed] [Google Scholar]
- 28.Bronner, C., Achour, M., Arima, Y., Chataigneau, T., Saya, H., and Schini-Kerth, V. B. (2007) Pharmacol. Ther. 115 419–434 [DOI] [PubMed] [Google Scholar]
- 29.Fujimori, A., Matsuda, Y., Takemoto, Y., Hashimoto, Y., Kubo, E., Araki, R., Fukumura, R., Mita, K., Tatsumi, K., and Muto, M. (1998) Mamm. Genome 9 1032–1035 [DOI] [PubMed] [Google Scholar]
- 30.Citterio, E., Papait, R., Nicassio, F., Vecchi, M., Gomiero, P., Mantovani, R., Di Fiore, P. P., and Bonapace, I. M. (2004) Mol. Cell. Biol. 24 2526–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bostick, M., Kim, J. K., Esteve, P. O., Clark, A., Pradhan, S., and Jacobsen, S. E. (2007) Science 317 1760–1764 [DOI] [PubMed] [Google Scholar]
- 32.Sakai, A., Kikuchi, Y., Muroi, M., Masui, T., Furihata, C., Uchida, E., Takatori, K., and Tanamoto, K. (2003) Biol. Pharm. Bull. 26 347–351 [DOI] [PubMed] [Google Scholar]
- 33.Muto, M., Fujimori, A., Nenoi, M., Daino, K., Matsuda, Y., Kuroiwa, A., Kubo, E., Kanari, Y., Utsuno, M., Tsuji, H., Ukai, H., Mita, K., Takahagi, M., and Tatsumi, K. (2006) Radiat. Res. 166 723–733 [DOI] [PubMed] [Google Scholar]
- 34.Bonapace, I. M., Latella, L., Papait, R., Nicassio, F., Sacco, A., Muto, M., Crescenzi, M., and Di Fiore, P. P. (2002) J. Cell Biol. 157 909–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orr, H. T., Chung, M. Y., Banfi, S., Kwiatkowski, T. J., Jr., Servadio, A., Beaudet, A. L., McCall, A. E., Duvick, L. A., Ranum, L. P., and Zoghbi, H. Y. (1993) Nat. Genet. 4 221–226 [DOI] [PubMed] [Google Scholar]
- 36.Koide, R., Ikeuchi, T., Onodera, O., Tanaka, H., Igarashi, S., Endo, K., Takahashi, H., Kondo, R., Ishikawa, A., Hayashi, T., Saito, M., Miike, T., Naito, H., IKuta, F., and Tsuji, S. (1994) Nat. Genet. 6 9–13 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.