Abstract
The bacterial proton-translocating NADH:quinone oxidoreductase (NDH-1) consists of two domains, a peripheral arm and a membrane arm. NuoH is a counterpart of ND1, which is one of seven mitochondrially encoded hydrophobic subunits, and is considered to be involved in quinone/inhibitor binding. Sequence comparison in a wide range of species showed that NuoH is comprehensively conserved, particularly with charged residues in the cytoplasmic side loops. We have constructed 40 mutants of 27 conserved residues predicted to be in the cytoplasmic side loops of Escherichia coli NuoH by utilizing the chromosomal DNA manipulation technique and investigated roles of these residues. Mutants of Arg37, Arg46, Asp63, Gly134, Gly145, Arg148, Glu220, and Glu228 showed low deamino-NADH-K3Fe(CN)6 reductase activity, undetectable NDH-1 in Blue Native gels, low contents of peripheral subunits (especially NuoB and NuoCD) bound to the membranes, and a significant loss of the membrane potential and proton-pumping function coupled to deamino-NADH oxidation. The results indicated that these conserved residues located in the cytoplasmic side loops are essential for the assembly of the peripheral subunits with the membrane arm. Implications for the involvement of NuoH (ND1) in maintaining the structure and function of NDH-1 are discussed.
The H+-translocating NADH:quinone (Q)3 oxidoreductase (EC 1.6.5.3) is a complex membrane-bound enzyme that catalyzes electron transfer from NADH to Q coupled with proton pumping across the inner mitochondrial membrane (complex I) or the bacterial cytoplasmic membrane (NDH-1) (1, 2). The Escherichia coli NDH-1 is composed of 13 subunits and all 13 nuo-encoded subunits from E. coli (NuoA-N) have their homologues present in the mitochondrial enzyme that contains 45 subunits (3–5). A chromosomal deletion of all nuo genes has been achieved earlier by homologous recombination in E. coli (6). Thus NDH-1 serves as a model system to elucidate the structure and function of complex I due to its structural simplicity and ease of gene manipulation (7–10). Electron microscopic analyses have established that NDH-1/complex I has a characteristic L-shaped structure consisting of two domains, a peripheral arm protruding into the cytoplasm (or the matrix) and a membrane domain embedded within the cytoplasmic membrane (or the inner mitochondrial membrane) (11–13). The NADH binding site and all known redox cofactors of complex I are located in the peripheral domain (14, 15). This was also confirmed by the crystal structure of the peripheral arm from Thermus thermophilus (16). Unlike the peripheral arm, the membrane arm does not contain any prosthetic group identified so far. Information about the structural and functional roles of these membrane domain subunits are rather limited despite recent progress in the field of structural biology. Based on the projection structure of the membrane domain and detergent-based fractionation study that led to the disruption of the membrane arm into fragments containing NuoL/M, NuoA/K/N, and NuoH/J subunits, a speculative arrangement of the membrane segment of E. coli NDH-1 has been proposed, wherein subunits NuoA, NuoK, NuoN, NuoJ, and NuoH are present in the vicinity of the peripheral arm, whereas the NuoL and NuoM subunits are distantly located from the peripheral segment (17–20). It is believed that these membrane-bound subunits play roles in proton translocation (8–10) and quinone/inhibitor binding (21, 22) and their alteration is linked to many human diseases (23).
The ND1 subunit is one of seven mitochondrial-encoded hydrophobic subunits (ND1–6 and 4L) of the membrane arm of complex I. The ND1 subunit is a mutational hot spot for mitochondrial diseases like LHON (24) and MELAS (25), underscoring its physiological role. Subunit NuoH in E. coli (a homolog of the mitochondrial ND1 subunit) is one of the most conserved subunits of the membrane domain. Mutations in the NuoH (Nqo8) subunit from Paracoccus denitrificans were previously reported to alter the affinity of complex I for ubi-Q (26). Various inhibitors were reported to label the ND1/NuoH subunit (21, 22, 27, 28). Subunit NuoH/ND1 is believed to be involved in Q/inhibitor binding (29). The essential role of ND1 on the activity and assembly of complex I was implied earlier in Chlamydomonas reinhardtii, in which deletion of the ND1 subunit prevented the assembly of complex I (30). Subunit NuoH/ND1 was thus assumed to play roles in assembly/communication between the peripheral and membrane domains (31). However, experimental evidence is lacking. Sequence comparison among various species shows that charged residues in all four cytoplasmic side loops of the NuoH subunit are comprehensively conserved. There are numerous implications for short-range electrostatic forces involved in maintaining subunit-subunit interactions as reported for V-ATPase (32). Thus, it is likely that the NuoH subunit interacts electrostatically with neighboring subunits by means of these charged amino acids. To investigate the role of the residues that could be associated with subunit-subunit interaction, we have systematically generated a set of point mutations of highly conserved amino acid residues of the NuoH subunit from E. coli, and thus attempted to understand the structural and functional importance of this subunit. The results indicate that NuoH, through its conserved residues in the cytoplasmic side loops, is involved in connecting the membrane arm with the peripheral arm, presumably with the NuoB and/or NuoCD subunit of NDH-1.
EXPERIMENTAL PROCEDURES
Materials—The pGEM-T Easy Vector was from Promega (Madison, WI). The QuikChange® II XL site-directed mutagenesis kit and Herculase®-enhanced DNA polymerase were obtained from Stratagene (Cedar Creek, TX). The endonucleases were from New England Biolabs (Beverly, MA). Materials for PCR product purification, gel extraction, and plasmid preparation were from Qiagen (Valencia, CA). The gene replacement vector, pKO3, was a generous gift from Dr. George M. Church (Harvard Medical School, Boston, MA). Oxonol VI and ACMA were from Molecular Probes (Eugene, OR). Capsaicin-40 was generously provided by Dr. Hideto Miyoshi (Kyoto University, Kyoto, Japan). The BCA protein assay kit and SuperSignal West Pico chemiluminescent substrate were from Pierce (Rockford, IL). dNADH, NADH, DB, chloramphenicol, and spectinomycin (Spc) were from Sigma. p-Nitro blue tetrazolium was from EMD Biosciences (La Jolla, CA). Goat anti-rabbit IgG horseradish peroxidase conjugate was from GE Healthcare (Piscataway, NJ). Oligonucleotides were synthesized by IDT (Coralville, IA). All other materials were reagent grade and obtained from commercial sources.
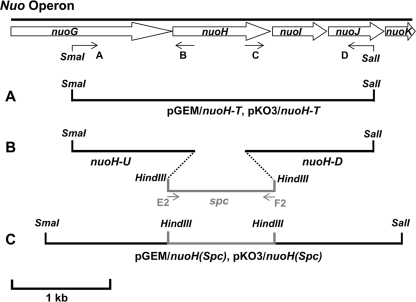
Cloning and Mutagenesis of the E. coli nuoH Gene—The strategies used for cloning and mutagenesis for the E. coli nuoH gene were in principle similar to those we reported for nuoA, nuoJ, nuoK, and nuoM genes (7–10) and illustrated in Fig. 1. Sequences of primers used for this purpose are listed in supplemental Table S1. A DNA fragment, which includes the nuoH gene, its upstream 1-kb DNA segment, and its downstream 1-kb DNA segment, was amplified from E. coli DH5α by PCR using primers A and D. The amplified DNA fragment containing the nuoH gene was then cloned into pGEM-T-Easy vector following the manufacturer's protocol, and its sequence was verified by DNA sequencing. The fragment bearing the nuoH gene was then subcloned into integration vector pKO3. The resulting plasmids were designated pGEM/nuoH-T and pKO3/nuoH-T, respectively (Fig. 1A). Later, a 1-kb DNA upstream segment including the 5′-sequence of the nuoH gene was cloned by PCR using primers A and B, generating fragment nuoH-U. Similarly, a 1-kb DNA downstream segment including the 3′ sequence of the nuoH gene was cloned by PCR using primers C and D, generating fragment nuoH-D. Both fragments were cloned in pGEM-T Easy Vector. The spectinomycin encoding gene from transposon Tn554 of Staphylococcus aureus (33) was obtained by PCR amplification using primers E2 and F2, both containing the HindIII restriction site. The spc fragment obtained was cloned in pGEM-T Easy Vector. Furthermore, the spc gene was inserted between nuoH-U and nuoH-D fragments with the help of the HindIII site generating the nuoH(spc) fragment (Fig. 1B) and then assembled in pGEM generating the pGEM/nuoH(spc) plasmid (Fig. 1C). The fragment harboring the spc cassette was finally subcloned in pKO3 with the help of SmaI and SalI restriction sites generating the pKO3/nuoH(spc) plasmid (Fig. 1C). To prepare pKO3 vectors carrying mutated nuoH genes, pGEM/nuoH-T was first mutagenized individually with the synthetic oligonucleotides listed in supplemental Table S1 using the QuikChange II XL site-directed mutagenesis kit according to the manufacturer's instructions to introduce nuoH point mutations. The generation of the desired mutations was confirmed by direct DNA sequencing. The pGEM constructs were then digested with SmaI and SalI, and DNA fragments containing the desired nuoH point mutations were finally transferred to integration plasmid pKO3 at the SmaI/SalI sites. The resulting plasmids were referred to as pKO3/nuoH (mutant). To evaluate possible effects of the entire process of gene manipulation in generation of point mutations on the E. coli cells, we also constructed a control pKO3 vector, using the same procedure except that the wild-type nuoH gene derived from pGEM(nuoH-T) without any mutation was inserted into the SmaI and SalI sites of pKO3.
FIGURE 1.
Schematic representation of the strategy of nuoH cloning, insertion of a Spc cassette in the E. coli nuoH gene, and construction of site-specific nuoH mutants. Arrows (A–D, E2, and F2) indicate the primers used in this study.
Preparation of nuoH Knock-out and Mutant Cells—E. coli strain MC4100 (F–, araD139, Δ(arg F-lac)U169, ptsF25, relA1, flb5301, rpsL 150.λ–) was used to generate knock-out and site-specific mutations of nuoH employing the pKO3 system. The process was carried out according to the method of Link et al. (34) with a minor modification as described previously by Kao et al. (9). E. coli nuoH knock-out (NuoH-KO mutant) was constructed by replacement of the nuoH gene in the nuo operon for the nuoH(spc) fragment inserted in the pKO3 plasmid. The NuoH-KO mutant obtained was verified by PCR sequencing using specific oligonucleotides and stored in glycerol at –80 °C. In subsequent steps the pKO3/nuoH (mutant) plasmids were used for construction of site-directed nuoH mutants. The control mutant (KO-rev) was generated in the same way except that, instead of using pKO3/nuoH (mutant), the pKO3 (nuoH-T) carrying the wild-type nuoH gene was employed in the recombination process. The correct introduction of point mutations in the chromosome was finally confirmed by direct DNA sequencing.
Bacterial Growth and Membrane Preparation—The E. coli membrane preparation was done according to Kao et al. (8). In brief, cells were first grown in LB medium from freshly streaked plates at 37 °C for 10 h and used to inoculate 250 ml of terrific broth and cells were cultivated at 37 °C with shaking until A600 was ∼2. The cells were then harvested and resuspended at 10% (w/v) in a buffer containing 10 mm Tris-HCl (pH 7.0), 1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 15% (w/v) glycerol. 0.5 mg/ml of lysozyme was added to the cell suspension followed by incubation on ice for 30 min and sonicated twice with a 15-s pulse. The cell suspension was then passed twice through the “French pressure cell press” at 25,000 p.s.i. After cell debris was removed, the supernatant was ultracentrifuged at 50,000 × g for 35 min in a Beckman Spinco 60Ti rotor. The collected pellet was resuspended in the same buffer, and the resulting membrane suspension was either used immediately for enzyme assays or stored in small aliquots at –80 °C for later use.
Gel Electrophoresis and Immunoblotting Analysis—10 μg of protein from each membrane suspension was first subjected to SDS-PAGE using the discontinuous system of Laemmli (35). The expression of the NDH-1 subunits was then determined by using antibodies specific to NuoB, NuoCD, NuoE, NuoF, NuoG, NuoI, and NuoM in Western blot experiments. The assembly of NDH-1 was evaluated by using BN-PAGE according to Schägger and von Jagow (36) with minor modifications (10). For NADH dehydrogenase activity staining of NDH-1 gels were incubated in 2 mm Tris-HCl (pH 7.5) containing 2.5 mg/ml of p-nitro blue tetrazolium and 150 μm NADH for 2 h at 37 °C. The reaction was stopped with 7% acetic acid.
Enzymatic Assay—In addition to NDH-1, E. coli also contains an alternative NADH-Q oxidoreductase, NDH-2. The E. coli NDH-2 is a small, FAD-dependent enzyme that is insensitive to capsaicin (37, 38) and cannot oxidize dNADH (39). Thus, to measure the activities derived solely from NDH-1, we used dNADH as the substrate in enzymatic assays throughout this study. All measurements of enzymatic activity were carried out at 30 °C using a Olis-SLM DW-2000 spectrophotometer. The dNADH oxidase activity was assayed at 340 nm as described previously (10). The dNADH-DB reductase activity measurements were conducted in the presence of 10 mm KCN, 0.15 mm dNADH, and 0.1 mm DB. The dNADH-K3Fe(CN)6 reductase activity was measured at 420 nm in the presence of 10 mm KCN, 0.15 mm dNADH, and 1 mm K3Fe(CN)6. The ε340 = 6.22 mm–1 cm–1 for dNADH and ε420 = 1.00 mm–1 cm–1 for K3Fe(CN)6 were used for analyses.
The generation of membrane potential by NuoH mutants was monitored optically with indicator dye oxonol VI as described previously (8–10). The reaction was started by addition of 0.2 mm dNADH and absorbance changes at 630 minus 603 nm were monitored. Proton-pumping activity of NDH-1 was estimated from ACMA fluorescence quenching as described by Amarneh and Vik (40). The reaction was initiated by adding 0.2 mm dNADH.
Other Analytical Procedures—Protein concentrations were determined by the BCA protein assay kit (Pierce) using bovine serum albumin as the standard according to the manufacturer's instructions. Any variations from the procedures and other details are described in the figure legends.
RESULTS
Sequence Analysis of the NuoH Subunit—The E. coli NuoH subunit is a polypeptide of 325 amino acid residues. A topology of the E. coli NuoH subunit was proposed based on the most reasonable predictions offered by a composite of computer programs including TopPredII (41, 42), PHDhtm (43), TMHMM (44), SOSUI (45–47), Tmpred (48), and HMMTOP (49). The E. coli NuoH subunit is predicted to contain eight transmembrane segments (designated TM1–8 from the N to C terminus), with both the N- and C-terminal regions oriented toward the periplasmic side of the membrane (Fig. 2). The final model also accommodated the experimentally determined topology of the Rhodobacter capsulatus NuoH subunit (31).
FIGURE 2.
Speculative topology of the E. coli NuoH subunit. The prediction has been performed on the basis of a composite of computer programs, including the reported topology of its Rhodobacter counterpart (31). The eight transmembrane segments of the E. coli NuoH subunit from the N terminus to the C terminus are designated TM1–8. The four cytoplasmic side loops are marked as C1–C4, whereas P1–P3 represent three periplasmic side loops. The N and C terminus of the subunit are exposed to the periplasmic side of the membrane. Amino acid residues mutated in this study are displayed as black squares, and some are marked by their E. coli numbering.
According to the sequence alignment, many residues were found to be extensively conserved particularly in all four cytoplasmic side loops (C1–4) (supplemental Fig. S1). These loops, especially C1 and C3, are highly enriched in charged residues, indicative of the electrostatic interaction with neighboring subunits. Our data base search revealed that in C1, Arg37, Arg46, Pro49, and Asp63 are almost perfectly conserved among all species examined. Residues Glu36, Gln44, Lys70, and Glu71 are highly conserved except that, in some species, they are replaced by some other similar residues. Similarly, C3 contains Asp213, Glu220, and Glu228 as almost invariant residues, whereas Arg209, Glu216, and Glu218 are highly conserved. It should be noted that most of these residues are also conserved in the EchB, HycD, and HyfC subunits of multisubunit membrane-bound energy-converting [Ni-Fe] Ech hydrogenase, hydrogenase 3, and hydrogenase 4 complexes, respectively.
Subunit ND1 is described to be a mutational hot spot for LHON (24) and MELAS (25). A good number of disease-related mutations in the ND1 subunit are reported to occur in residues that are located in highly conserved domains of extramembrane loops facing the matrix side (24, 50). LHON/MELAS mutation E24K (51) (E. coli numbering E36), MELAS mutations R25Q (E. coli numbering Arg37) (52), G131S (53) (E. coli numbering Gly145), and E214K (25) (E. coli numbering Glu228) are examples of four residues associated with these pathogenic mutations present in the matrix side extramembrane loops of subunit ND1. These residues are highly conserved between the human ND1 subunit and homologues of many species. Residues associated with LHON mutation E143K (24) (E. coli numbering Glu157) are almost completely conserved.
To clarify the structural and functional roles of the conserved residues as well as residues corresponding to those linked to human diseases, we have generated a set of 40 point mutations covering 27 different residues of the E. coli NuoH subunit using homologous recombination targeted directly to the chromosome (9) (see Table 1 for the complete list of mutants).
TABLE 1.
Enzyme activities of the membrane-bound NDH-1 of E. coli wild-type and various NuoH mutants
a Activity in nanomole of dNADH/mg of protein/min.
b Activity in nanomole of K3Fe(CN)6/mg of protein/min.
Effects of NuoH Mutation on the dNADH-K3Fe(CN)6 Reductase Activity of NDH-1—Table 1 includes dNADH-K3Fe(CN)6 reductase activity of all mutants together with wild-type. This activity derives from the NADH dehydrogenase segment of complex I/NDH-1 and, therefore, could be generally used as an estimate for the amount of the active peripheral domain associated with the membrane. The NuoH-KO mutant retained very little activity of the dNADH-K3Fe(CN)6 reductase (∼11% of parent membranes), which was similar to most of the other hydrophobic subunit knock-out mutants, except for NuoM-KO (∼35%) (10). Correspondingly, the NuoH-KO mutant exhibited almost null activity of the energy-coupled NDH-1 activities (dNADH oxidase and dNADH-DB reductase; see below) comparable with deletion mutants of other hydrophobic subunits studied to date (NuoA, J, K, and M) (7–10). This in fact agrees with the commonly accepted geometry of the membrane domain that places NuoJ, NuoH, NuoA, NuoK, and NuoN near the peripheral arm and NuoM and NuoL in a distant location (13, 19, 54, 55).
As shown in Table 1, the KO-rev mutant displayed properties indistinguishable from the wild-type strain in all enzymatic activities measured. The complete restoration of the activity to the level of the parental MC4100 strain demonstrates that the homologous recombination procedure adopted here worked out exactly as expected. Consequently, the KO-rev mutant could serve as an appropriate reference strain in addition to the original MC4100 wild-type strain. In good agreement with previous assembly experiments (7–10), membranes of all point mutants of various transmembrane segments (V206G, E241A, and E241Q) displayed the dNADH-K3Fe(CN)6 reductase activity comparable with that of the wild-type. In contrast, point mutants of all C1–4 displayed various degrees of inhibition of the dNADH-K3Fe(CN)6 reductase activity. The most drastic decrease in the dNADH-K3Fe(CN)6 reductase activity was seen for mutation of invariantly conserved residues, particularly those with charged residues in C1 and C3. Mutations of invariantly conserved Arg residues located in C1 (R37A and R46A) resulted in a ∼80 and ∼53% reduction of activities, respectively. Replacement of the same Arg residues by other basic Lys (R37K and R46K) only partially restored the activity by ∼15–20%, highlighting the importance of Arg residues at this location for the structure. Replacing the highly conserved residue Asp63 with either a corresponding amide Asn or a nonpolar Ala resulted in loss of activity of ∼76 or ∼82%, respectively. In contrast to this seemingly drastic effect, replacing Asp63 with an acidic Glu brought about a complete restoration of ferricyanide activity. Less drastic but still significant diminution of the activities occurred with mutations at Pro49 and Glu71 positions. Two mutants of Glu36, E36D and E36A, exhibited small but measurable inhibition (15–25%) in the dNADH-K3Fe(CN)6 reductase activity. The negative charge at this position does not seem to be essential for the activity.
In C2, mutation of the invariantly conserved Arg148 resulted in ∼50% reduction in the activity. The disease-related mutants, G134V and G134L, exhibited a more significant reduction (∼50%) of dNADH-K3Fe(CN)6 reductase activity than the G134A mutant (∼30%). A similar result was obtained with mutation of Gly145, the residue associated with the human ND1 mutation, G3697A (human numbering G131S). The G145A mutant had activities close to those from the wild-type strain, whereas G145V had detectably lower activities (∼50%). Mutation of the other three residues Ser155, Tyr156, and Glu157 to Ala did not have any appreciable effects on the activities. Glu157 is well conserved, and the corresponding position in human ND1 subunit is associated with LHON. Interestingly, mutation (E157K) mimicking the LHON (human numbering E143K) in this position augmented the activity to 160% (see Table 1 and supplemental Fig. S1). A similar result has been reported earlier by Valentino et al. (24), where they attributed the higher activity for the LHON mutation (G3733A, human numbering E143K) to a higher mitochondrial proliferation. Similarly, mutation of the highly conserved residues carrying a carboxyl group (D213A, E220A, and E228A) in C3 resulted in 50–60% reduction in the activities. A small activity loss was observed with mutations of R209A. Similar trends were reported by recent studies of MELAS mutations (human numbering E214K) using the Paracoccus and E. coli systems (50). In the case of mutations related to C4, replacing the conserved Asp295 residue with Ala resulted in ∼42% loss of the activity, whereas mutation of D295E resulted in the complete restoration of activity. Smaller but still considerable reduction of the activities occurred with mutations at the Arg286 and Lys303 positions, whereas mutation of another conserved charged residue, Arg291, did not affect the activity.
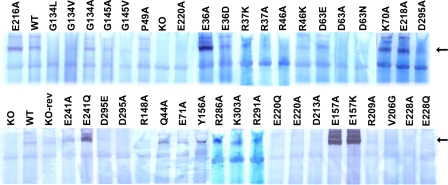
Subunit Assembly of NDH-1 of NuoH Mutants—We used BN-PAGE and NADH dehydrogenase activity staining to directly verify the presence of assembled NDH-1 in the constructed mutants. As illustrated in Fig. 3, no intact NDH-1 was observed in the NuoH-KO mutant. In contrast, membranes isolated from the wild-type and NuoH KO-rev mutants contained similar amounts of fully assembled NDH-1, once again validating that there was no problem in the process of chromosomal manipulation. As expected, mutants that had low ferricyanide activities displayed either no assembly (R37A, R46A, D63A, D63N, G134L, E220A, E220Q, E228A, and E228Q) or a significantly diminished level of the assembled complex (R37K, R46K, G134V, G145V, R148A, D213A, and D295A). The point mutants that had ferricyanide activity similar to wild-type (K70A, Y156A, E157A, E157K, E241Q, and E241A) were akin to the wild-type with regard to their assembly.
FIGURE 3.
NADH dehydrogenase activity staining of BN-PAGE gels of membrane preparations from E. coli. Membranes from the wild-type (WT), NuoH knock-out (KO), NuoH knock-out revertant (KO-rev), and site-specific NuoH mutants were compared. The arrow shows location of the NDH-1 band. The concentration of dodecyl β-maltoside used was 1%. Electrophoresis was performed as described under “Experimental Procedures.” After electrophoresis the gels were incubated with 2.5 mg/ml p-nitro blue tetrazolium and 150 μm NADH for 2 h at 37 °C. The reaction was stopped by 7% acetic acid.
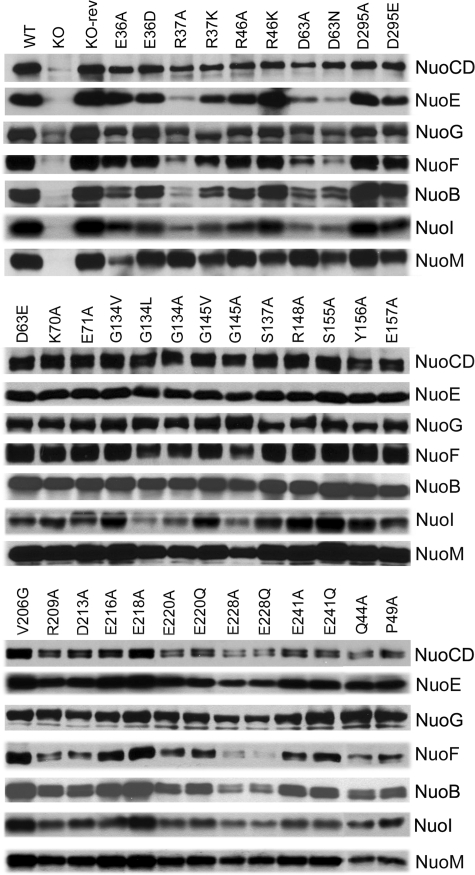
Subunit Contents of NDH-1 of NuoH Mutants—Immunochemical analyses were performed to systematically examine the subunit contents of the NDH-1 in the cytoplasmic membranes from all NuoH mutants. We selected a total of seven subunit-specific antibodies for detection; six antibodies are against all subunits present in the peripheral domain (NuoB, NuoCD, NuoE, NuoF, NuoG, and NuoI) and one antibody against a subunit located in the membrane domain (NuoM) (Fig. 4). Interestingly, divergent results were observed in the subunits that make up the peripheral domain of E. coli NDH-1. As seen in Fig. 4, peripheral subunits NuoCD, NuoE, NuoF, NuoI, and NuoB and the hydrophobic subunit NuoM were almost entirely missing in membranes of the NuoH KO cells. Membranes of the NuoH KO cells contained very small amounts of NuoG subunit. This observation further validates that NuoH is required for efficient assembly of the NDH-1 complex. The amounts of the peripheral subunits of point mutants such as K70A, R148A, Y156A, E157A, and E241A were identical to those of the wild-type. It appears that NDH-1 of mutant R148A is inactively assembled and very fragile as judged from the results of BN-PAGE and dNADH-K3Fe(CN)6 reductase. As expected, for many of the constructed point mutants of different cytoplasmic loops, significant and divergent quantitative differences were observed for the seven NDH-1 subunits tested. Most interestingly, the membranes of many of the loop mutants that had reasonably lower ferricyanide activity (less than ∼50%) (Table 1) contained very small amounts of peripheral subunits of NuoB and NuoCD. In C1, membranes from the mutants of invariantly conserved residues Arg37, Arg46, and Asp63 contained fairly low amounts of NuoE, NuoF, NuoB, NuoI, and NuoCD subunits. Lower levels of NuoE and NuoB subunits were observed for mutants of Glu36. Similarly, the mutants of invariantly charged residues Glu220 and Glu228 in C3 had rather low amounts of NuoB, NuoE, NuoF, NuoG, and NuoCD subunits (Fig. 4).
FIGURE 4.
Immunoblotting of membrane preparations from wild-type (WT), NuoH knock-out (KO), NuoH knock-out revertant (KO-rev), and site-specific NuoH mutants. E. coli membranes (10 μg of protein per lane) were loaded on a 15% Laemmli SDS-polyacrylamide gel. After electrophoresis, the proteins were transferred onto nitrocellulose membranes and Western blotting was carried out using the SuperSignal West Pico system. Antibodies specific to NuoB, NuoCD, NuoE, NuoF, NuoG, NuoI, and NuoM were used. Goat anti-rabbit IgG horseradish peroxidase conjugate was used as secondary antibody for the detection of the bands. Note that mutants E157K, R286A, K303A, and R291A are not shown as they were the same as the wild-type.
Taken together, it is quite clear that many of the site-specific NuoH mutations of various cytoplasmic loops analyzed in this study drastically affected the assembly of NDH-1. Invariantly conserved residues Arg37, Arg46, Asp63, Gly134, Gly145, Glu220, and Glu228 are essential for the assembly of peripheral subunits to the membrane arm and, thus activity of NDH-1.
Effects of NuoH Mutation on the Energy-transducing NDH-1 Activity—In this work, we also introduced site-directed mutations in some of the charged amino acids that are conserved and predicted to be in the TMs of the NuoH subunit. The point mutants analyzed displayed varied degrees of inhibition of dNADH oxidase and dNADH-DB reductase activity. Mutation of the highly conserved Glu36, which is present at the interface of TM1, significantly reduced the energy-transducing NDH-1 activity (E36A, ∼80% inhibition). The well conserved Glu157, which is present at the interface of TM3, has the corresponding position in human ND1 subunit associated with LHON. Mutation E157A diminished the activity by 71% but scarcely affected the dNADH dehydrogenase activity, although, interestingly, mutation E157K had energy-coupled NDH-1 activity comparable with the wild-type. Although neither Glu36 nor Glu157 are likely to be directly involved in proton translocation because the mutants of these residues did not inhibit completely the energy-transducing activities, it seems possible that Glu36 and Glu157 might act as catalysts for turnover (conformational change) of the energy-coupled NDH-1 reaction. Less drastic but still significant diminution of the energy-transducing activity occurred with mutations at the Gln44, Glu218, Glu241, and Asp295 positions (∼40–60% inhibition).
We determined kinetic parameters for DB to investigate whether the residues in C1–4 play some roles in Q binding. An apparent Km of 2.8 μm for DB was observed for the wild-type strain. Regarding the mutants (Km in parentheses), C1 mutant E36D (1.1 μm) showed a somewhat increased affinity for DB, whereas C4 mutant D295E (2.5 μm) had about the same value as that of the wild-type. Mutants Q44A (6.2 μm), D63E (4.9 μm), E71A (5.0 μm), E157K (6.1 μm), E216A (5.9 μm), and K303A (6.5 μm) all exhibited a moderately lower affinity for DB. Overall, no drastic change in Km value for DB was observed among the mutants. We also investigated the effects of the NuoH mutation on inhibitory characteristics of capsaicin-40. The IC50 value of capsaicin-40 of most of these mutants was determined to be in the range of 0.11–0.21 μm, which is almost the same as that of wild-type (0.14–0.17 μm). The data suggest that neither Q binding nor capsaicin-40 binding are significantly modified by these point mutations.
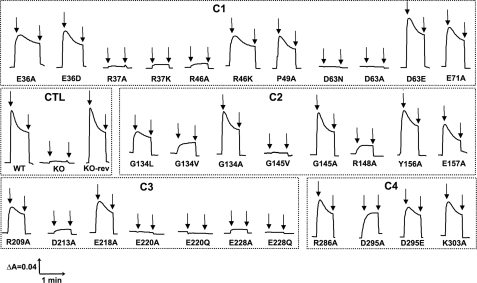
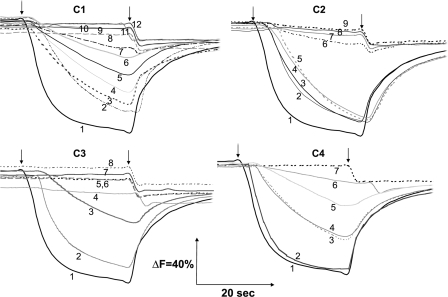
Measurements of Electrochemical Potential and Proton Translocation of NuoH Mutants—Because electron transfer activities of the mutants varied from normal to almost null with varying effects on the assembly of the enzyme, it was of interest to examine the generation of membrane potential and proton gradient. Fig. 5 shows representative traces of oxonol VI response, organized according to cytoplasmic loops. As expected, no ΔΨ was observed in membrane vesicles of the knock-out mutant, and the membrane vesicles of KO-rev mutant generated a signal comparable with that of the wild-type strain. Similarly, negligible ΔΨ was observed for the membrane vesicles of the mutants that had no assembly (R37A, R37K, R46A, D63N, D63A (C1); G145V (C2); E220A, E220Q, E228A, E228Q (C3)) as per our observation. The mutant R46K generated a small membrane potential. E36A and E36D mutations had moderate effects (C1). Mutation of another highly conserved Gly134, either to bulky Val or Leu caused a considerable reduction in ΔΨ, whereas substituting Ala had an insignificant effect (C2). Likewise, in case of invariantly conserved Gly145, replacing this small group with a bulky group Val resulted in an almost complete loss of the ability of the enzyme to generate ΔΨ, whereas Ala substitution brought back the normal membrane potential (C2). The same nearly total diminution of ΔΨ was seen with the mutant of invariant residue Arg148 (C2). Interestingly, mutation D295A had a moderate decrease in membrane potential, and its (moderately) mutated twin, D295E (C4), as well as E71A (C1) and E218A (C3) had a slightly higher absorbance change but was still much lower than that of the wild-type strain.
FIGURE 5.
Detection of the membrane potential generated by dNADH oxidation in E. coli NuoH mutants. Membrane vesicles were prepared from each of the constructed mutants, and the membrane potential changes were monitored by the absorbance changes of oxonol VI at 630–603 nm at 37 °C. The assay mixture typically contained 50 mm MOPS (pH 7.3), 10 mm MgCl2, 50 mm KCl, 2 μm oxonol VI, and E. coli membrane samples (330 μg of protein/ml). The first arrow indicates addition of 0.2 mm dNADH and the second arrow indicates addition of 2 μm carbonyl cyanide p-trifluoromethoxyphenylhydrazone to the assay mixture. The traces for the mutants are depicted on the basis of their presence in the cytoplasmic loops (C1–C4) along with the controls (CTL). It should be noted that Y156A and E157A are kept along with C2 mutants as they were at the interface. The mutants having the membrane potential similar to the wild-type are not shown.
The formation of the proton gradient was monitored using ACMA (Fig. 6). Wild-type and the KO-rev mutant exhibited an almost identical response. The signal was completely dissipated by carbonyl cyanide p-trifluoromethoxyphenylhydrazone. No proton translocation was observed in the NuoH-KO mutant. The results for nearly all of the mutants are in good accord with the membrane potential or with data obtained from the assay of dNADH oxidase and dNADH-DB reductase activities. Identical to their inability to generate membrane potential, the mutants (R37A, R37K, R46A, D63N, D63A, (C1); G134V, G145V, R148A, (C2); D213A, E220A, E220Q, E228A, E228Q, (C3)) did not show any sign of proton translocation. The mutants R46K (C1), G134L (C2), and D295A (C4) displayed significantly slower proton translocation and did not reach stationary phase within the experimental time. E36A, E36D, E71A (C1), and D295E (C4) showed a slightly higher proton pumping ability but were still much lower than that of the wild-type strain. Other mutants exhibited no significant effects on proton pumping activity.
FIGURE 6.
Generation of a pH gradient coupled to dNADH oxidation in E. coli NuoH mutants. Membrane vesicles were prepared from each of the constructed mutants, and the extent of proton translocation was measured by quenching the fluorescence of ACMA at room temperature with an excitation wavelength of 410 nm and emission wavelength of 480 nm. At the time indicated by the arrows, 0.2 mm dNADH (first arrow) or 10 μm carbonyl cyanide p-trifluoromethoxyphenylhydrazone (second arrow) was added to the assay mixture containing 50 mm MOPS (pH7.3), 10 mm MgCl2, 50 mm KCl, 2 μm ACMA, and E. coli membrane samples (150 μg of protein/ml). C1 mutants: 1, WT; 2, D63E; 3, P49A; 4, E36D; 5, E36A; 6, E71A; 7, R46K; 8, R37A; 9, R37K; 10, R46A; 11, D63A; 12, D63N. C2 mutants: 1, WT; 2, E157A; 3, G134A; 4, Y156A; 5, G145A; 6, G134L; 7, G134V; 8, G145V; 9, R148A. C3 mutants: 1, WT; 2, E218A; 3, R209A; 4, E220A; 5, E228A; 6, D213A; 7, E220Q; 8, E228Q. C4 mutants: 1, WT; 2, KO-rev; 3, R286A; 4, K303A; 5, D295E; 6, D295A; 7, NuoH KO. Controls (i.e. WT, KO, and KO-rev mutants) are displayed along with the mutants of cytoplasmic loop 4 (C4). As shown are Y156A and E157A in group C2. The mutants that exhibited proton pumping activity akin to the wild-type are not shown.
DISCUSSION
The membrane domain of NDH-1 contains seven subunits, all of which are homologues of mammalian mitochondrially encoded subunits. Systematic mutagenesis of hydrophobic domain subunits NuoA (ND3), NuoJ (ND6), NuoK (ND4L), NuoM (ND4), and NuoN (ND2) has not only helped us to characterize the essential carboxyl residues necessary for energy-coupled activities but also established E. coli NDH-1 as a useful model system for studying complex I (7–10, 40, 56–58). Detergent-based fractionation studies showed that the ND1 subunit (mitochondrial counterpart of NuoH) is present in subcomplex Iα, which comprises the peripheral domain and part of the membrane domain (19, 59). This leads to a suggestion that ND1 may be located near the peripheral domain. Upon sequence comparison the NuoH subunit was found to be extensively conserved among various species, particularly in all four loops facing the cytoplasmic side (supplemental Fig. S1). Interestingly, this is the only subunit among all seven membrane domain subunits, wherein residues in the cytoplasmic side loops are so vastly conserved. These cytoplasmic side loops are relatively large in comparison to a majority of other membrane domain subunits. Furthermore, these loops are highly enriched in charged residues (Fig. 2 and supplemental Fig. S1), particularly C1 and C3, and thus appear to be involved in binding with peripheral subunits.
The dNADH-K3Fe(CN)6 reductase activity that evaluates the effects on the stability of the mutations (Table 1) showed that, in a majority of point mutants of these cytoplasmic side loops, the activity was significantly lower compared with that of the wild-type (20–60%). Further analyses with BN-PAGE (Fig. 3) and immunological methods (Fig. 4) corroborated the dNADH-K3Fe(CN)6 reductase activity results, and the assembly of NDH-1 was found to be completely or partially disturbed in the loop mutants studied. Similarly, the membrane potential (Fig. 5) and proton translocation ability (Fig. 6) of nearly all of these mutants were in good agreement with the above result. Remarkably, nearly all of these residues associated with disturbing the assembly of NDH-1 (Arg37, Arg46, Asp63, Gly134, Gly145, Arg148, Glu220, or Glu228) are almost universally conserved among a wide variety of species (see supplemental Fig. S1). Furthermore, these NuoH mutants exhibit diminished peripheral subunits (Fig. 4), due to incomplete assembly of the whole complex. The most prominent observation was that some of the mutants showed quite low amounts of NuoB and NuoCD. It is interesting to note that Nqo4 (NuoD) and Nqo6 (NuoB) also contain an unusually high proportion of charged amino acids (see, for example, Ref. 20). From the crystal structure it is quite comprehensible that NuoD/Nqo4/49K and NuoB/Nqo6/PSST have many charged residues present in the loops that face the interface between the membrane domain and the Q-cavity. According to Sazanov (20), these charged residues are likely to be involved in interaction with hydrophobic subunits. Subunit NuoB/Nqo6/PSST is the central subunit housing the terminal iron-sulfur cluster N2 that acts as an immediate electron donor to Q (60, 61). There may therefore be a role for short-range electrostatics in maintaining NuoH/NuoB and/or NuoH/NuoCD interactions (possibly maintaining the Q-cavity/NuoH interaction) (29). The proposed interaction of NuoH and NuoB is in good agreement with the fact that ND1 (NuoH) and PSST (NuoB) were earlier shown to be functionally coupled in a photoaffinity labeling experiment (62). Additional support is from the energy-converting hydrogenase (Ech) family (63). Ech is composed of 6 different subunits (EchA–F). These subunits are homologues of NuoM(or L), NuoH, NuoB, NuoC, NuoD, and NuoI, respectively. The membrane segment of Ech is composed of two subunits: EchA (NuoM or L) and EchB (NuoH) (64). Considering that the peripheral arm of Ech is made up of four homologues of NDH-1 and that NuoM or NuoL of NDH-1 does not interact with the peripheral arm of NDH-1, we can speculate that EchB (NuoH) is involved in interaction with the peripheral arm. In addition, as described above, the eight essential residues in cytoplasmic loops of NuoH are conserved in EchB (see supplemental Fig. S1).
According to a recent report (65), formation of fully assembled complex I as a result of the union of various pre-assembled subcomplexes, occurs in a semi-sequential manner; the membrane part is formed at an early stage of assembly, wherein ND1 (NuoH), ND6 (NuoJ), and PSST (NuoB) subunits are added to the B17 subunit containing subcomplex. This further binds to a hydrophilic subcomplex containing the 30- and 49-kDa subunits (fused together as NuoCD in E. coli) and subsequently more subcomplexes and subunits are added leading to the formation of holo-complex I. Given the observation that the NuoH subunit is incorporated into the NDH-1 structure at an early stage of its assembly (66), it is conceivable that the reduced levels of fully assembled complex I observed in this work might be due to an impaired assembly of complex I with consequent degradation of unincorporated subunits.
Piericidin and capsaicin structurally resemble Q and are demonstrated to act as a competitive inhibitor for Q in complex I (67, 68) as well as in the E. coli membrane-bound glucose dehydrogenase (69, 70). Therefore, these inhibitors and catalytic Q most likely share the same binding site. Our present results showed that the Km value for DB and the IC50 value of capsaicin-40 were hardly affected by mutating residues of the cytoplasmic side loops of the NuoH subunit, suggesting that they apparently do not directly participate in Q/inhibitor binding. It should be noted that the kinetics assays could not be performed on mutants that have significantly lost NDH-1 activities. Similar studies from other laboratories reported alteration of Km values for short chain Q in certain mutants of the Paracoccus Nqo8 subunit (a homologue of NuoH) (26, 71). In their case, mutated residues, Glu158, Glu212, and Glu247 (Glu157, Val206, and Glu241 in E. coli NuoH) are all predicted to be located in the transmembrane segments. They also reported that the Paracoccus mutation (A65T) mimicking LHON affected the Km value for DB. This Ala65 is not conserved and is replaced by Met64 in E. coli.
Mutants of the eight essential residues in NuoH exhibited almost no activities of the dNADH oxidase and dNADH-DB reductase, whereas their dNADH-K3Fe(CN)6 reductase activities were moderately inhibited. These results are interpreted in at least two ways. It is known that dNADH-K3Fe(CN)6 reductase activity requires only a two-subunit assembly (NuoE and F) (72), whereas the dNADH-DB reductase activity needs a delicate architecture of NDH-1. Therefore, one explanation is that those essential residues are located near the catalytic Q-binding site and thus affect only DB reduction. Another explanation has to do with possible structural alterations. Given the finding that modification of certain residues in the cytoplasmic loops causes dislodging of the peripheral arm, it would not be surprising to see, in other mutations, imperfect association between the peripheral and membrane domain subunits that leads to little or no Q reduction.
In conclusion, the present study highlights various aspects of the NuoH/ND1 subunit in NDH-1 assembly and activity. Our data suggest that invariantly conserved residues Arg37, Arg46, Asp63, Gly134, Gly145, Arg148, Glu220, and Glu228 are all essential for stability and, thus activity of complex I. The results clearly indicate that NuoH, through its conserved residues in the cytoplasmic side loops, is involved in connecting the membrane arm with the peripheral arm. Based on our results we hypothesize that various conserved residues in all four cytoplasmic loops connect the NuoH subunit of the membrane arm with the NuoB and/or NuoCD subunit of the peripheral arm of NDH-1, placing it in a region that is hypothesized to be critical for the catalytic Q binding. The results from the disease-mimicking mutation using E. coli NDH-1 demonstrates that the bacterial system is a useful model of studies of mitochondrial complex I diseases.
Supplementary Material
Acknowledgments
We thank Dr. Hideto Miyoshi (Kyoto University, Kyoto, Japan), for kindly providing capsaicin-40, Dr. George M. Church (Harvard medical School, Boston, MA) for allowing us to use the pKO3 plasmid, and Drs. Byoung Boo Seo, Mathieu Marella, and Jennifer Barber-Singh (TSRI, La Jolla) for discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM033712 (to T. Y. and A. M.-Y.). This is Publication 19803-MEM from The Scripps Research Institute, La Jolla, CA.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
Footnotes
The abbreviations used are: Q, quinone(s); complex I, mitochondrial proton-translocating NADH-quinone oxidoreductase; NDH-1, bacterial proton-translocating NADH-quinone oxidoreductase; DB, 2,3-dimethoxy-5-methyl-6-decyl-1,4-benzoquinone; dNADH, reduced nicotinamide hypoxanthine dinucleotide (deamino-NADH); Spc, spectinomycin; LHON, Leber's hereditary optic neuropathy; MELAS, mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes; oxonol VI, bis-(3-propyl-5-oxoisoxazol-4-yl)pentamethine oxonol; ACMA, 9-amino-6-chloro-2-methoxyacridine; TM, transmembrane segment(s); BN, blue native; MOPS, 4-morpholinepropanesulfonic acid.
References
- 1.Hirst, J. (2005) Biochem. Soc. Trans. 33 525–529 [DOI] [PubMed] [Google Scholar]
- 2.Galkin, A. S., Grivennikova, V. G., and Vinogradov, A. D. (1999) FEBS Lett. 451 157–161 [DOI] [PubMed] [Google Scholar]
- 3.Carroll, J., Fearnley, I. M., Skehel, J. M., Shannon, R. J., Hirst, J., and Walker, J. E. (2006) J. Biol. Chem. 281 32724–32727 [DOI] [PubMed] [Google Scholar]
- 4.Yagi, T., Yano, T., and Matsuno-Yagi, A. (1993) J. Bioenerg. Biomembr. 25 339–345 [DOI] [PubMed] [Google Scholar]
- 5.Yagi, T., Yano, T., Di Bernardo, S., and Matsuno-Yagi, A. (1998) Biochim. Biophys. Acta 1364 125–133 [DOI] [PubMed] [Google Scholar]
- 6.Amarneh, B., De Leon-Rangel, J., and Vik, S. B. (2006) Biochim. Biophys. Acta 1757 1557–1560 [DOI] [PubMed] [Google Scholar]
- 7.Kao, M. C., Di Bernardo, S., Nakamaru-Ogiso, E., Miyoshi, H., Matsuno-Yagi, A., and Yagi, T. (2005) Biochemistry 44 3562–3571 [DOI] [PubMed] [Google Scholar]
- 8.Kao, M. C., Nakamaru-Ogiso, E., Matsuno-Yagi, A., and Yagi, T. (2005) Biochemistry 44 9545–9554 [DOI] [PubMed] [Google Scholar]
- 9.Kao, M. C., Di Bernardo, S., Perego, M., Nakamaru-Ogiso, E., Matsuno-Yagi, A., and Yagi, T. (2004) J. Biol. Chem. 279 32360–32366 [DOI] [PubMed] [Google Scholar]
- 10.Torres-Bacete, J., Nakamaru-Ogiso, E., Matsuno-Yagi, A., and Yagi, T. (2007) J. Biol. Chem. 282 36914–36922 [DOI] [PubMed] [Google Scholar]
- 11.Guénebaut, V., Schlitt, A., Weiss, H., Leonard, K., and Friedrich, T. (1998) J. Mol. Biol. 276 105–112 [DOI] [PubMed] [Google Scholar]
- 12.Clason, T., Zickermann, V., Ruiz, T., Brandt, U., and Radermacher, M. (2007) J. Struct. Biol. 159 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranova, E. A., Holt, P. J., and Sazanov, L. A. (2007) J. Mol. Biol. 366 140–154 [DOI] [PubMed] [Google Scholar]
- 14.Yagi, T., and Dinh, T. M. (1990) Biochemistry 29 5515–5520 [DOI] [PubMed] [Google Scholar]
- 15.Yano, T., and Yagi, T. (1999) J. Biol. Chem. 274 28606–28611 [DOI] [PubMed] [Google Scholar]
- 16.Sazanov, L. A., and Hinchliffe, P. (2006) Science 311 1430–1436 [DOI] [PubMed] [Google Scholar]
- 17.Di Bernardo, S., and Yagi, T. (2001) FEBS Lett. 508 385–388 [DOI] [PubMed] [Google Scholar]
- 18.Kao, M. C., Matsuno-Yagi, A., and Yagi, T. (2004) Biochemistry 43 3750–3755 [DOI] [PubMed] [Google Scholar]
- 19.Sazanov, L. A., Peak-Chew, S. Y., Fearnley, I. M., and Walker, J. E. (2000) Biochemistry 39 7229–7235 [DOI] [PubMed] [Google Scholar]
- 20.Sazanov, L. A. (2007) Biochemistry 46 2275–2288 [DOI] [PubMed] [Google Scholar]
- 21.Murai, M., Ishihara, A., Nishioka, T., Yagi, T., and Miyoshi, H. (2007) Biochemistry 46 6409–6416 [DOI] [PubMed] [Google Scholar]
- 22.Yagi, T., and Hatefi, Y. (1988) J. Biol. Chem. 263 16150–16155 [PubMed] [Google Scholar]
- 23.Mitchell, A. L., Elson, J. L., Howell, N., Taylor, R. W., and Turnbull, D. M. (2006) J. Med. Genet. 43 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valentino, M. L., Barboni, P., Ghelli, A., Bucchi, L., Rengo, C., Achilli, A., Torroni, A., Lugaresi, A., Lodi, R., Barbiroli, B., Dotti, M., Federico, A., Baruzzi, A., and Carelli, V. (2004) Ann. Neurol. 56 631–641 [DOI] [PubMed] [Google Scholar]
- 25.Kirby, D. M., McFarland, R., Ohtake, A., Dunning, C., Ryan, M. T., Wilson, C., Ketteridge, D., Turnbull, D. M., Thorburn, D. R., and Taylor, R. W. (2004) J. Med. Genet. 41 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurki, S., Zickermann, V., Kervinen, M., Hassinen, I., and Finel, M. (2000) Biochemistry 39 13496–13502 [DOI] [PubMed] [Google Scholar]
- 27.Earley, F. G. P., Patel, S. D., Ragan, C. I., and Attardi, G. (1987) FEBS Lett. 219 108–113 [DOI] [PubMed] [Google Scholar]
- 28.Vgenopoulou, I., Gemperli, A. C., and Steuber, J. (2006) J. Bacteriol. 188 3264–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yagi, T., and Matsuno-Yagi, A. (2003) Biochemistry 42 2266–2274 [DOI] [PubMed] [Google Scholar]
- 30.Cardol, P., Matagne, R. F., and Remacle, C. (2002) J. Mol. Biol. 319 1211–1221 [DOI] [PubMed] [Google Scholar]
- 31.Roth, R., and Hagerhall, C. (2001) Biochim. Biophys. Acta 1504 352–362 [DOI] [PubMed] [Google Scholar]
- 32.Jones, R. P., Durose, L. J., Findlay, J. B., and Harrison, M. A. (2005) Biochemistry 44 3933–3941 [DOI] [PubMed] [Google Scholar]
- 33.Murphy, E., Huwyler, L., and Freire Bastos, M. C. (1985) EMBO J. 4 3357–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Link, A. J., Phillips, D., and Church, G. M. (1997) J. Bacteriol. 179 6228–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. (1970) Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- 36.Schagger, H., and Von Jagow, G. (1991) Anal. Biochem. 199 223–231 [DOI] [PubMed] [Google Scholar]
- 37.Satoh, T., Miyoshi, H., Sakamoto, K., and Iwamura, H. (1996) Biochim. Biophys. Acta 1273 21–30 [DOI] [PubMed] [Google Scholar]
- 38.Yagi, T. (1990) Arch. Biochem. Biophys. 281 305–311 [DOI] [PubMed] [Google Scholar]
- 39.Matsushita, K., Ohnishi, T., and Kaback, H. R. (1987) Biochemistry 26 7732–7737 [DOI] [PubMed] [Google Scholar]
- 40.Amarneh, B., and Vik, S. B. (2003) Biochemistry 42 4800–4808 [DOI] [PubMed] [Google Scholar]
- 41.Claros, M. G., and Von Heijne, G. (1994) Comput. Appl. Biosci. 10 685–686 [DOI] [PubMed] [Google Scholar]
- 42.Von Heijne, G. (1992) J. Mol. Biol. 225 487–494 [DOI] [PubMed] [Google Scholar]
- 43.Rost, B., Fariselli, P., and Casadio, R. (1996) Protein Sci. 5 1704–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. L. (2001) J. Mol. Biol. 305 567–580 [DOI] [PubMed] [Google Scholar]
- 45.Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998) Bioinformatics 14 378–379 [DOI] [PubMed] [Google Scholar]
- 46.Mitaku, S., Hirokawa, T., and Tsuji, T. (2002) Bioinformatics 18 608–616 [DOI] [PubMed] [Google Scholar]
- 47.Mitaku, S., and Hirokawa, T. (1999) Protein Eng. 12 953–957 [DOI] [PubMed] [Google Scholar]
- 48.Hofmann, K., and Stoffel, W. (1993) Biol.-Chem. Hoppe Seyler 374 166. [DOI] [PubMed] [Google Scholar]
- 49.Tusnády, G. E., and Simon, I. (2001) Bioinformatics 17 849–850 [DOI] [PubMed] [Google Scholar]
- 50.Kervinen, M., Hinttala, R., Helander, H. M., Kurki, S., Uusimaa, J., Finel, M., Majamaa, K., and Hassinen, I. E. (2006) Hum. Mol. Genet. 15 2543–2552 [DOI] [PubMed] [Google Scholar]
- 51.Blakely, E. L., de Silva, R., King, A., Schwarzer, V., Harrower, T., Dawidek, G., Turnbull, D. M., and Taylor, R. W. (2005) Eur. J. Hum. Genet. 13 623–627 [DOI] [PubMed] [Google Scholar]
- 52.Horvath, R., Reilmann, R., Holinski-Feder, E., Ringelstein, E. B., and Klopstock, T. (2008) Neuromuscul. Disord. 18 553–556 [DOI] [PubMed] [Google Scholar]
- 53.Spruijt, L., Smeets, H. J., Hendrickx, A., Bettink-Remeijer, M. W., Maat-Kievit, A., Schoonderwoerd, K. C., Sluiter, W., de Coo, I. F., and Hintzen, R. Q. (2007) Arch. Neurol. 64 890–893 [DOI] [PubMed] [Google Scholar]
- 54.Baranova, E. A., Morgan, D. J., and Sazanov, L. A. (2007) J. Struct. Biol. 159 238–242 [DOI] [PubMed] [Google Scholar]
- 55.Sazanov, L. A., and Walker, J. E. (2000) J. Mol. Biol. 302 455–464 [DOI] [PubMed] [Google Scholar]
- 56.Kervinen, M., Patsi, J., Finel, M., and Hassinen, I. E. (2004) Biochemistry 43 773–781 [DOI] [PubMed] [Google Scholar]
- 57.Patsi, J., Kervinen, M., Finel, M., and Hassinen, I. E. (2008) Biochem. J. 409 129–137 [DOI] [PubMed] [Google Scholar]
- 58.Euro, L., Belevich, G., Verkhovsky, M. I., Wikstrom, M., and Verkhovskaya, M. (2008) Biochim. Biophys. Acta 1777 1166–1172 [DOI] [PubMed] [Google Scholar]
- 59.Hirst, J., Carroll, J., Fearnley, I. M., Shannon, R. J., and Walker, J. E. (2003) Biochim. Biophys. Acta 1604 135–150 [DOI] [PubMed] [Google Scholar]
- 60.Brandt, U. (2006) Annu. Rev. Biochem. 75 69–92 [DOI] [PubMed] [Google Scholar]
- 61.Ohnishi, T., and Salerno, J. C. (2005) FEBS Lett. 579 4555–4561 [DOI] [PubMed] [Google Scholar]
- 62.Schuler, F., and Casida, J. E. (2001) Biochim. Biophys. Acta 1506 79–87 [DOI] [PubMed] [Google Scholar]
- 63.Hedderich, R., and Forzi, L. (2005) J. Mol. Microbiol. Biotechnol. 10 92–104 [DOI] [PubMed] [Google Scholar]
- 64.Kunkel, A., Vorholt, J. A., Thauer, R. K., and Hedderich, R. (1998) Eur. J. Biochem. 252 467–476 [DOI] [PubMed] [Google Scholar]
- 65.Ugalde, C., Vogel, R., Huijbens, R., van den Heuvel, B., Smeitink, J., and Nijtmans, L. (2004) Hum. Mol. Genet. 13 2461–2472 [DOI] [PubMed] [Google Scholar]
- 66.Antonicka, H., Ogilvie, I., Taivassalo, T., Anitori, R. P., Haller, R. G., Vissing, J., Kennaway, N. G., and Shoubridge, E. A. (2003) J. Biol. Chem. 278 43081–43088 [DOI] [PubMed] [Google Scholar]
- 67.Miyoshi, H. (2001) J. Bioenerg. Biomembr. 33 223–231 [DOI] [PubMed] [Google Scholar]
- 68.Degli Esposti, M. (1998) Biochim. Biophys. Acta 1364 222–235 [DOI] [PubMed] [Google Scholar]
- 69.Elias, M. D., Nakamura, S., Migita, C. T., Miyoshi, H., Toyama, H., Matsushita, K., Adachi, O., and Yamada, M. (2004) J. Biol. Chem. 279 3078–3083 [DOI] [PubMed] [Google Scholar]
- 70.Kobayashi, K., Mustafa, G., Tagawa, S., and Yamada, M. (2005) Biochemistry 44 13567–13572 [DOI] [PubMed] [Google Scholar]
- 71.Zickermann, V., Barquera, B., Wikström, M., and Finel, M. (1998) Biochemistry 37 11792–11796 [DOI] [PubMed] [Google Scholar]
- 72.Yano, T., Sled, V. D., Ohnishi, T., and Yagi, T. (1996) J. Biol. Chem. 271 5907–5913 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.