Abstract
The goal of this investigation was to study the expression and regulation of β1,3-Glucuronosyltransferase-I (GlcAT-I), a key enzyme regulating GAG synthesis in cells of the intervertebral disc. There was a robust expression of GlcAT-I in the nucleus pulposus in vivo. Treatment with the calcium ionophore ionomycin resulted in increased GlcAT-I expression, whereas GlcAT-I promoter constructs lacking TonE site or a mutant TonE were unresponsive to the ionophore. Experiments using TonEBP and DN-TonEBP constructs showed that TonEBP positively regulated GlcAT-I promoter activity. ChIP analysis confirmed binding of TonEBP to the promoter. We further validated the role of TonEBP in controlling GlcAT-I expression using mouse embryo fibroblasts from TonEBP null mice. GlcAT-I promoter activity in null cells was significantly lower than the wild type cells. In contrast to wild type cells, treatment with ionomycin failed to increase GlcAT-I promoter activity in null cells. We then investigated if calcineurin (Cn)-NFAT signaling played a regulatory role in GlcAT-I expression. Inhibition of Cn following ionomycin treatment did not block GlcAT-I and tauT, a TonEBP-responsive reporter activity. GlcAT-I promoter activity was suppressed by co-expression of Cn, NFAT2, NFAT3, and NFAT4. Moreover, following ionomycin treatment, fibroblasts from CnAα and CnAβ null mice exhibited robust induction in GlcAT-I promoter activity compared with wild type cells. Results of these studies demonstrate that calcium regulates GlcAT-I expression in cells of the nucleus pulposus through a signaling network comprising both activator and suppressor molecules. The results suggest that by controlling both GAG and aggrecan synthesis, disc cells can autoregulate their osmotic environment and accommodate mechanical loading.
The intervertebral disc is a specialized structure that permits rotation as well as flexure and extension of the human spine. It consists of an outer ligament, the annulus fibrosus, that encloses a gel-like tissue, the nucleus pulposus. Although sparse, cells in the nucleus pulposus secrete a complex extracellular matrix that contains fibrillar collagens and the proteoglycan aggrecan. Glycosoaminoglycan (GAG)2 components of the aggrecan molecule provide a robust hydrodynamic system that serves to accommodate applied biomechanical forces (1, 2). Surprisingly, although the importance of aggrecan secretion and function has been discussed by many investigators, mechanisms of control of GAG synthesis are poorly understood.
In the nucleus pulposus, the principle GAG is chondroitin sulfate. Structurally, this molecule is a heteropolysaccharide containing repeating units of N-acetylgalactosamine linked to glucuronic acid. In its fully sulfated form, the molecule exhibits a high charge density, and when hydrated, it assumes a linear configuration. Bound to the aggrecan core protein and associated with hyaluronic acid, the chondroitin sulfate chains form a giant polydispersed supramolecular structure. The high osmotic pressure of the aggregate contains the forces applied to the spine (3).
In an earlier study, we showed that nucleus pulposus cells responded to changes in osmotic pressure by up-regulating the transcription factor, TonEBP (tonicity enhancer-binding protein) (4). When expressed, this protein modulated aggrecan expression; whether it can regulate chondroitin chain synthesis has not been determined. It is noteworthy that other workers have shown that galactose-β1,3-glucuronysltransferase-1 (GlcAT-I) activity was required for GAG chain synthesis (5); hence, there is the possibility that this enzyme serves as the rate-limiting step in GAG synthesis for chondrocytes and possibly other cell types (6, 7). Related to this point, it is now known that IL-1β suppressed GAG biosynthesis by down-regulating GlcAT-I expression and activity (8). A second factor regulating aggrecan synthesis is the intracellular calcium concentration (9, 10). In smooth muscle and Sertoli cells, calcium channel blockers have been shown to alter proteoglycan synthesis (11, 12). Furthermore, using p-nitrophenyl-β-d-xyloside as an exogenous primer for chain initiation, it was demonstrated that GAG synthesis was inhibited by calcium blocking agents (12). Since these agents affected both chondroitin sulfate and heparin sulfate, the results suggested that calcium ions controlled a common early step in the GAG biosynthetic pathway. In a recent study, calcium was shown to control GlcAT-I expression through Sp1 transcription factor (13).
The major objective of the investigation was to examine the regulation of GlcAT-I expression by nucleus pulposus cells of the intervertebral disc. We show for the first time that TonEBP regulates GlcAT-I expression and that regulation is dependent on intracellular calcium ions. We also demonstrate that calcium-dependent calcineurin (Cn)-NFAT signaling serves as a negative regulator of GlcAT-I expression in these cells. From this perspective, by controlling GAG as well as aggrecan synthesis, TonEBP permits nucleus pulposus cells to autoregulate the osmotic environment of the disc.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents—Rabbit polyclonal TonEBP antibody was a kind gift from Dr. H. Moo Kwon (University of Maryland). TonEBP/NFAT5 wild type and null MEFs (originally from Dr. Steffan N. Ho), were provided by Dr. Feng Chen (Washington University, St. Louis, MO). Primary kidney medullary fibroblasts derived from CnAα null, CnAβ null, and CnA wild type mice were provided by Dr. Jennifer Gooch (Emory University). Plasmids were kindly provided by Dr. Takashi Ito (Osaka University, Japan) (tauT (taurine transporter) wild type (WT) and mutant (MT) reporter) (14) and Dr. Ben C. Ko (University of Hong Kong) (FLAG-DN-TonEBP, FLAG-TonEBP, and FLAG-CMV2) (15). DN-TonEBP contains amino acids 157–581 of human TonEBP (from clone KIAA0827). Dr. Gerald Crabtree (Stanford University) provided catalytic subunit (CnA) and regulatory subunit (CnB). Dr. Jeffery Molkentin (Cincinnati Children's Hospital Medical Center) supplied NFAT4, pECE, and Dr. Gerald Thiel (University of Saarland Medical Center, Germany) supplied DN-Sp1. Plasmids for NFAT1 (catalogue number 11100) and constitutively active NFAT2 (CA-NFAT2) (catalog number 11102) with key serine residues changed to alanine to prevent phosphorylation developed by Dr. Anjana Rao (16), plasmid for NFAT3 (catalog number 10961) developed by Dr. Toren Finkel (17), and plasmid for 3×NFAT-Luc (catalog number 17870) developed by Dr. Gerald Crabtree (18) were obtained from Addgene. 3×NFAT-Luc contains three NFAT binding sites upstream of the minimal interleukin-2 promoter and used to measure calcineurin-dependent NFAT activation. As an internal transfection control, vector pRL-TK (Promega) containing the Renilla reniformis luciferase gene was used. The amount of transfected plasmid, the pretransfection period after seeding, and the post-transfection period before harvesting have been optimized for rat nucleus pulposus cells using pSV β-galactosidase plasmid (Promega) (19).
Immunohistological Studies—Freshly isolated discs were immediately fixed in 4% paraformaldehyde in PBS and then embedded in paraffin. Transverse and coronal sections, 6–8 μm in thickness, were deparaffinized in xylene, rehydrated through graded ethanol, and stained with Alcian blue, eosin, and hematoxylin. For localizing GlcAT-I, sections were incubated with the anti-GlcAT-I antibody (Novus) in 2% bovine serum albumin in PBS at a dilution of 1:100 at 4 °C overnight. After thoroughly washing the sections, the bound primary antibody was incubated with biotinylated universal secondary antibody, at a dilution of 1:20 (Vector Laboratories) for 10 min at room temperature. Sections were incubated with a streptavidin-peroxidase complex for 5 min and washed with PBS, and color was developed using 3′,3-diaminobenzidine (Vecta Stain Universal Quick Kit; Vector Laboratories).
Isolation of Nucleus Pulposus Cells and Treatments of Cells—Rat nucleus pulposus cells were isolated using a method reported earlier by Risbud et al. (19). Nucleus pulposus cells and MEFs were maintained in Dulbecco's modified Eagle's medium and 10% fetal bovine serum supplemented with antibiotics. In some experiments, cells were treated with 1 μm ionomycin and PMA (100 ng/ml) with or without FK505 (10 ng/ml) and cyclosporine A (1 μg/ml) or BAPTA-AM (10 μm) or bisanthracycline (WP631; 50–100 nm).
Real Time RT-PCR Analysis—Following treatment, total RNA was extracted from nucleus pulposus cells using RNAeasy minicolumns (Qiagen). Before elution from the column, RNA was treated with RNase-free DNase I. 100 ng of total RNA was used as template for real time PCR analysis. Reactions were set up in microcapillary tubes using 1 μl of RNA with 9 μl of a LightCycler FastStart DNA Master SYBR Green I mix (Roche Applied Science) to which gene-specific forward and reverse PCR primers were added (GlcAT-I (NCBI number NM_001128184), forward (5′-atgcccagtttgatgctactgcac-3′) and reverse (5′-tgttcctcctgcttcatcttcggt-3′)). Each set of samples included a template-free control. PCRs were performed in a LightCycler (Roche Applied Science) according to the manufacturer's instructions. All the primers used were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Immunofluorescence Microscopy—Cells were plated in flat bottom 96-well plates (5 × 103/well) and treated with ionomycin for 6–24 h. After incubation, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS for 10 min, blocked with PBS containing 5% fetal bovine serum, and incubated with antibodies against GlcAT-I (1:200) (Novus), TonEBP (1:200) (Calbiochem), NFAT-2 (1:200; Abcam), NFAT-3 (1:100; Cell Signaling), or NFAT-4 (1:200; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 4 °C overnight. As a negative control, cells were reacted with isotype IgG under similar conditions. After washing, the cells were incubated with Alexa Fluor-488-conjugated anti-mouse secondary antibody (Invitrogen) at a dilution of 1:50 and 10 μm propidium iodide for 1 h at room temperature. Cells were imaged using a laser-scanning confocal microscope (Olympus Fluoview).
Nuclear Protein Extraction and Western Blotting—Cells were placed on ice immediately following treatment and washed with ice-cold Hanks' balanced saline solution. Nuclear and cytosolic proteins were prepared using the CellLytic NuCLEAR extraction kit (Sigma). All of the wash buffers and final resuspension buffer included 1× protease inhibitor mixture (Pierce), NaF (5 mm), and Na3VO4 (200 μm). Nuclear or total cell proteins were resolved on 8–12% SDS-polyacrylamide gels and transferred by electroblotting to nitrocellulose membranes (Bio-Rad). The membranes were blocked with 5% nonfat dry milk in TBST (50 mm Tris, pH 7.6, 150 mm NaCl, 0.1% Tween 20) and incubated overnight at 4 °C in 3% nonfat dry milk in TBST with the anti-GlcAT-I (1:500; Novus) or anti-TonEBP antibody (1:3,000; from Dr. Kwon). Immunolabeling was detected using the ECL reagent (Amersham Biosciences).
Generation of GlcAT-I Reporter and Deletion Constructs—PCR amplification using genomic DNA of a 1,231-bp fragment containing 1,170 bp of the upstream promoter sequence linked to 61 bp of exon 1 (i.e. –1170 to +61) of the human GlcAT-I gene was performed using the following primers (forward, 5′ CTAGCTAGCGCGCCCAGCTGCTAACAGCA-3′ (NheI site underlined); reverse, 5′-CCCAAGCTTGCGAGAAACACGTTCTTCAGC-3′ (HindIII site underlined)) with the addition of GC buffer 1 and LA Taq polymerase (Takara Mirus Bio). Similarly, to generate successive 5′ deletions, promoter fragments of 335 bp (–274 to +61) and 184 bp (–123 to +61) were PCR-amplified using specific forward primers and the same reverse primer. The resulting DNA fragments were subcloned into pCR2.1 TA vector (Invitrogen), isolated by restriction digestion with NheI and HindIII, and ligated into the luciferase basic expression vector, pGL3 (Promega). The identity of each GlcAT-I promoter sequence was confirmed by sequencing.
Site-directed Mutagenesis of TonE—GlcAT-I-D reporter plasmid was used to mutate the TonE site (TTTCCA to TAAAAA). Mutants were generated using the QuikChange II XL site-directed mutagenesis kit (Stratagene), using forward and reverse primer pair containing the desired mutation, following the manufacturer's instructions. The mutation was verified by sequencing.
Chromatin Immunoprecipitation (ChIP) Assay—COS7 cells were transfected with GlcAT-I-D along with either FLAG-TonEBP (increasing amounts) or FLAG-CMV2 vector and cultured for 48 h. ChIP analysis was performed as described before (20). Cross-linked and fragmented lysates were immunoprecipitated with monoclonal anti-FLAG M2 antibody (Sigma). PCR analysis to identify the coprecipitated TonE fragment in GlcAT-I promoter was performed using the following primer sequences: forward, 5′-GGAAAAATGAGAAGGATGGCTTG-3′; reverse, 5′-TGGACACCCGATTCCAGCATCTCTA-3′.
Transfections and Dual Luciferase Assay—Cells were transferred to 24-well plates at a density of 6 × 104 cells/well 1 day before transfection. To investigate the effect of TonEBP overexpression on GlcAT-I promoter activity, cells were cotransfected with 100–500 ng of pTonEBP or DN-TonEBP or backbone vector pcDNA3.1 with 250 ng of GlcAT-I reporter and 250 ng of pRL-TK plasmid. In some experiments, cells were transfected with 500 ng of GlcAT-I reporter plasmids with 500 ng of pRL-TK plasmid. Lipofectamine 2000 (Invitrogen) was used as a transfection reagent. For each transfection, plasmids were premixed with the transfection reagent. For measuring the effect of ionomycin on GlcAT-I reporter activity, 24 h after transfection, the cells in some wells were ionomycin-treated with or without inhibitors FK506 and CsA or BAPTA-AM. The next day, the cells were harvested, and a Dual-Luciferase™ reporter assay system (Promega) was used for sequential measurements of firefly and Renilla luciferase activities. Transfection efficiency for rat nucleus pulposus cells was about 50–60%, whereas for fibroblasts, it was close to 90%. Quantification of luciferase activities and calculation of relative ratios were carried out using a luminometer (TD-20/20; Turner Designs, CA). At least three independent transfections were performed, and all analyses were carried out in triplicate.
Statistical Analysis—All measurements were performed in triplicate; data are presented as mean ± S.D. Differences between groups were analyzed by Student's t test. *, p < 0.05.
RESULTS
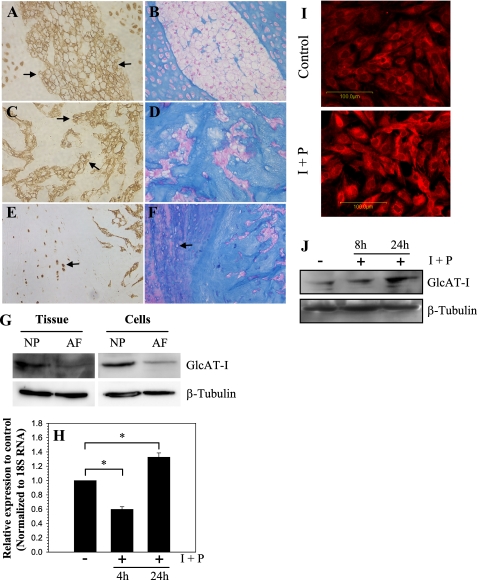
Saggital sections of the neonatal rat (Fig. 1, A and B) and skeletally mature rat discs (Fig. 1, C–F) were stained with an antibody to GlcAT-I (Fig. 1, A, C, and E) or counterstained with hematoxylin and eosin and Alcian blue for morphology assessment (Fig. 1, B, D, and F). GlcAT-I is expressed by cells of the nucleus pulposus, annulus fibrosus, and cartilaginous end plate in rat disc (Fig. 1, A, C, and E). In all cases, staining is localized to the cytosol (Fig. 1, A and C). Expression of GlcAT-I in native disc tissues and cultured cells was studied using Western blot analysis. Fig. 1G indicates that nucleus pulposus tissue expresses a prominent 43-kDa GlcAT-I band. Moreover, the expression level of GlcAT-I in nucleus pulposus tissue is higher than in the annulus fibrosus (Fig. 1G). Similar to native tissue, cultured rat disc cells exhibit a similar pattern of expression (Fig. 1G). To explore the premise that intracellular calcium regulated GlcAT-I expression, nucleus pulposus cells were treated with ionomycin, a calcium ionophore, along with PMA, and expression of GlcAT-I was analyzed. Fig. 1H shows that treatment with ionomycin for 24 h results in increased GlcAT-I mRNA levels in nucleus pulposus cells. In addition, we studied the expression of GlcAT-I in nucleus pulposus cells using immunofluorescence microscopy and Western blot analysis. Ionomycin treatment results in increased GlcAT-I protein expression (Fig. 1, I and J); the increase is pronounced 24 h after treatment (Fig. 1J).
FIGURE 1.
Saggital and coronal sections of disc tissue from neonatal (A) and mature (C and E) rat spines stained with an antibody against GlcAT-I or stained with hematoxylin and eosin and Alcian blue (B, D, and F). Note that nucleus pulposus cells in the neonatal (A) as well as skeletally mature disc cells (C and E) express GlcAT-I protein; much of the staining is localized to the cytosol and plasma membrane (A and C; arrows). Furthermore, inner annulus fibrosus cells localized in amorphous Alcian blue-positive matrix (F; arrow) express GlcAT-I protein (E; arrow). Isotype and secondary antibody controls were negative (not shown). Magnification was ×20. G, Western blot analysis of GlcAT-I expression by nucleus pulposus (NP) and annulus fibrosus (AF) tissue and cultured cells. Note, the expression of the 43-kDa GlcAT-I band in tissue extracts. The native nucleus pulposus tissue and cells in culture expressed higher GlcAT-I protein levels than the annulus tissue and cells. H, real time reverse transcription-PCR analysis of GlcAT-I expression by cells treated with ionomycin (1 μm) and PMA (100 ng) (I + P) for 4–24 h. There was a time-dependent change in mRNA expression following treatment. At 4 h, GlcAT-I expression was suppressed; at 24 h, there was increased expression of the gene. I, immunofluorescent analysis of nucleus pulposus cells treated with ionomycin and PMA. Cells showed increased GlcAT-I expression 24 h after the treatment. J, Western blot of nucleus pulposus cells treated with ionomycin and PMA. Note the increased GlcAT-I expression 24 h after treatment.
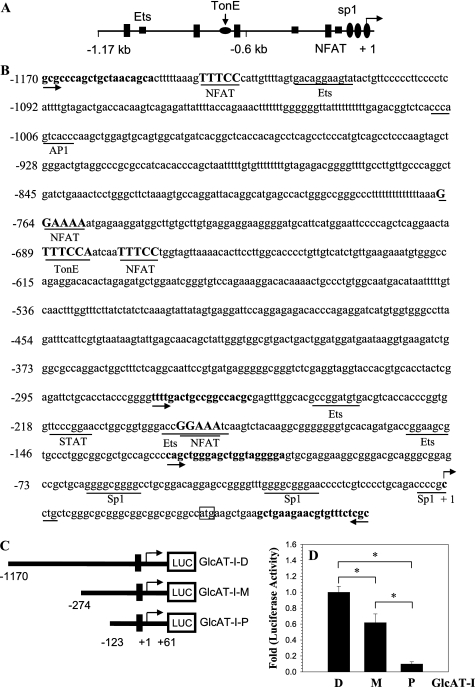
To investigate the regulation of expression, we analyzed the 1.17-kb promoter sequence of human GlcAT-I and measured the activity of different size promoter fragments. Analysis revealed that the GlcAT-I promoter contains four NFAT (GGGAA or TTTCC) as well as a conserved TonE (TTTCCA) motif at –684 bp (Fig. 2, A and B). To analyze promoter function, we generated luciferase reporter constructs containing a –1170/+61 bp (pGlcAT-I-D), a –274/+61 bp (pGlcAT-I-M), and a –123/+61 bp (pGlcAT-I-P) fragment of the human GlcAT-I promoter (Fig. 2C). We measured the basal activity of all three fragments in nucleus pulposus cells. Fig. 2D shows that the –1170/+61 bp fragment has maximal basal activity, whereas the –123/+61 bp fragment exhibits the least activity.
FIGURE 2.
GlcAT-I promoter contains TonEBP and NFAT binding motifs. A, promoter organization of the human GlcAT-I gene. The transcription start site is marked as +1. The TonE site is shown as a flattened circle, Spls are indicted as ovals, and the NFAT binding motifs are shown as rectangles. B, DNA sequence of the promoter region of the human GlcAT-I gene. TonE (TTTCCA) and NFAT (TTTCC or GGAAA) consensus sequences are marked in boldface type and underlined. The arrows indicate the starting location of the primers used to generate promoter constructs. The transcription start site is marked as +1; ATG marks the translation start site. Spl sites are underlined and lie within first 100 bases. C, schematic diagram showing a map of successive PCR-generated 5′ deletion constructs of the human GlcAT-I promoter. The GlcAT-I-D construct consists of a 1,231-bp fragment containing 1,170 bp of the upstream GlcAT-I promoter sequence linked to 61 bp of exon 1 (i.e. –1170/+61), whereas GlcAT-I-M and GlcAT-I-P constructs contain a 335-bp fragment (–274/+61) and a 184-bp fragment (–123/+61), respectively. D, basal activities of GlcAT-I promoter constructs relative to full-length construct GlcAT-I-D in nucleus pulposus cells. Cells showed maximal luciferase activity for the GlcAT-I-D construct, whereas the shortest construct, GlcAT-I-P, showed minimal activity. Values shown are mean ± S.D. of three independent experiments. *, p < 0.05.
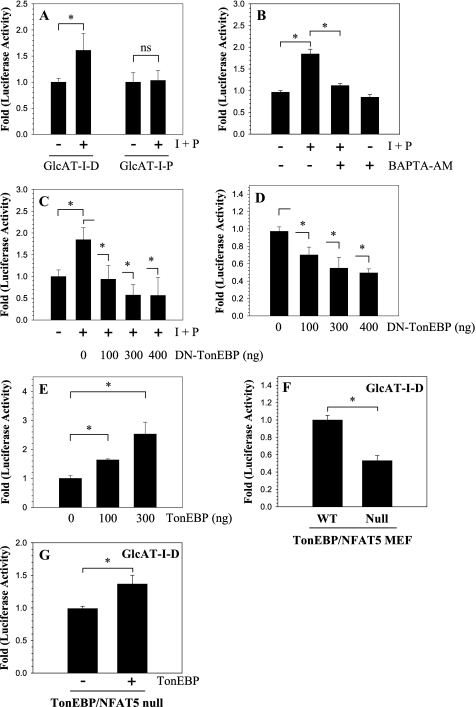
We next examined the effect of ionomycin treatment on GlcAT-I promoter activity in nucleus pulposus cells. When cells are treated with ionomycin and PMA, there is an increase in activity of –1170/+61 bp GlcAT-I promoter (pGlcAT-I-P); the shortest promoter fragment (pGlcAT-I-P), lacking TonE and NFAT sites, did not show any change in activity (Fig. 3A). We then determined if ionomycin-mediated activation of GlcAT-I required calcium ions. When cells were treated with the calcium chelator BAPTA-AM along with ionomycin, there was complete inhibition of GlcAT-I-D reporter activation (Fig. 3B). To investigate if TonEBP participated in ionomycin-mediated induction of GlcAT-I promoter activity, nucleus pulposus cells were transiently co-transfected with plasmids encoding DN-TonEBP or full-length TonEBP. Fig. 3C shows that forced expression of DN-TonEBP completely abolishes ionomycin induction of GlcAT-I promoter activity. Moreover, expression of DN-TonEBP also suppresses the basal activity of the GlcAT-I-D promoter fragment (Fig. 3D). A significant inhibitory effect of DN-TonEBP expression on GlcAT-I reporter activity is seen at a dose of 100 ng, which is further enhanced when the concentration of the DN-TonEBP is increased to 400 ng (Fig. 3D). On the other hand, overexpression of TonEBP using the pFLAG-TonEBP vector results in a dose-dependent increase in GlcAt-I-D promoter activity in the absence of ionomycin (Fig. 3E). We used MEFs derived from TonEBP/NFAT5 null and wild type mice to further validate the role of TonEBP in regulation of the GlcAT-I promoter. Fig. 3F shows that the basal GlcAT-I promoter activity in null cells is 50% lower than in the wild type cells. Moreover, co-transfection with TonEBP expression vector in null cells results in an increase in basal GlcAT-I promoter activity (Fig. 3G).
FIGURE 3.
Calcium regulates GlcAT-I promoter activity through TonEBP. A, GlcAT-I-D or GlcAT-I-P reporter activity measured following ionomycin and PMA (I + P) treatment. Treatment resulted in induction in GlcAT-I-D but not GlcAT-I-P reporter activity. B, effect of BAPTA-AM (10 μm) on GlcAT-I-D reporter activity. The calcium chelator completely blocks ionomycin-mediated activity but not basal activity of the GlcAT-I-D reporter in nucleus pulposus cells. C and D, effect of ionomycin treatment on cells transfected with GlcAT-I-D reporter construct along with DN-TonEBP or empty backbone FLAG-CMV2. Note that the expression of DN-TonEBP resulted in a complete suppression of ionomycin-mediated induction in GlcAT-I promoter activity. In addition, when TonEBP function was blocked, there was suppression of basal GlcAT-I-D activity. E, effect of TonEBP on GlcAT-I promoter activity. When pTonEBP was co-expressed, there was a dose-dependent increase in GlcAT-I reporter activity. F, GlcAT-I promoter activity of TonEBP/NFAT5 null and wild type cells. Null cells evidenced decreased basal activity of reporter compared with wild type cells. G, effect of co-expression of TonEBP and GlcAT-D in null cells. When co-expressed, TonEBP increased GlcAT-I-D reporter activity. Values shown are mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05.
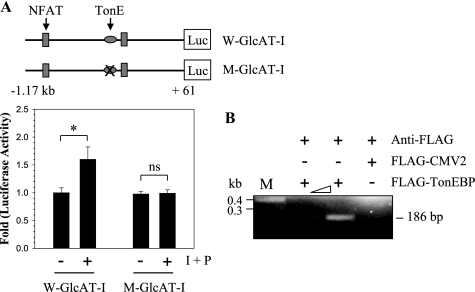
We then evaluated whether GlcAT-I promoter activity required that TonEBP be bound to TonE. For this purpose, we introduced a 4-base pair mutation in the TonE site of the GlcAT-I-D reporter plasmid (TTTCCA to TAAAAA). Following ionomycin treatment, nucleus pulposus cells transfected with wild type reporter plasmid evidence induction of activity (Fig. 4A). In contrast, when a mutant plasmid is used, induction of the reporter is completely blocked (Fig. 4A). Moreover, wild type promoter also exhibited a significantly higher basal activity than the TonE mutant reporter (not shown). We used the ChIP assay to evaluate the interaction of TonEBP protein with the TonE motif in the GlcAT-I promoter (Fig. 4B). TonEBP associated with the TonE motif under basal conditions. Binding of TonEBP to TonE was proportional to the amount of transfected FLAG-TonEBP vector. When an empty FLAG vector was co-transfected in place of FLAG-TonEBP, PCR analysis indicated that the amplicon is not formed. This was confirmed by cloning and sequencing of the PCR product.
FIGURE 4.
Ionomycin mediated induction of GlcAT-I promoter activity requires TonEBP binding to TonE. Effect of a 4-bp mutation introduced into the TonE motif of the GlcAT-I-D reporter plasmid. Nucleus pulposus cells were transfected with wild type GlcAT-I-D (W-GlcAT-I) or mutant GlcAT-I-D (M-GlcAT-I) reporter plasmids, and the induction of the luciferase activity was determined following ionomycin treatment. Treatment caused an induction of wild type reporter activity, whereas the mutant reporter failed to increase activity. Values shown are mean ± S.D. of three independent experiments. *, p < 0.05. B, interaction of TonEBP with the GlcAT-I promoter measured using a chromatin immunoprecipitation assay. COS7 cells were transfected with GlcAT-I-D along with either FLAG-TonEBP or FLAG-CMV2 empty vector. PCR amplification was performed using primer pairs that encompass TonE sequences of the GlcAT-I promoter. The use of anti-FLAG antibody resulted in generation of a PCR amplicon containing TonE only when FLAG-TonEBP was present. The addition of increasing amounts of FLAG-TonEBP vector evidenced enhanced binding to TonE. Pull-down using anti-FLAG antibody did not result in the formation of a PCR product when cells received empty FLAG-CMV2 vector.
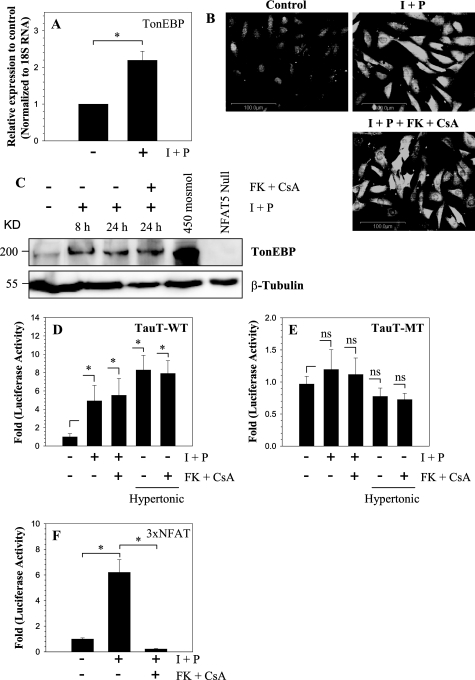
The mechanism of TonEBP activation by ionomycin in nucleus pulposus cells was investigated. Fig. 5A shows that ionomycin treatment results in increased TonEBP mRNA expression. Moreover, both immunofluorescence and Western blot analysis indicate that there is a concomitant increase in TonEBP protein expression (Fig. 5, B and C); the increase in protein level is evident as early as 8 h, and it remains high at 24 h (Fig. 5C). We explored the possibility that activation of Cn, a calcium-dependent phosphatase that mediates NFATc signaling, was responsible for TonEBP induction. Treatment of nucleus pulposus cells with Cn inhibitors, FK506 and cyclosporine A, in the presence of ionomycin did not block induction in TonEBP protein levels (Fig. 5, B and C). In addition, we measured the promoter activity of the TonEBP target gene, tauT. Significant activation in tauT reporter activity is evident following ionomycin treatment; simultaneous treatment with Cn inhibitors fails to suppress induction (Fig. 5D). In contrast to the wild-type tauT reporter, activity of mutant tauT construct is unaffected by ionomycin (Fig. 5E). To confirm that ionomycin promotes Cn-NFAT signaling in nucleus pulposus cells, we measured the activity of 3×NFAT reporter. Fig. 5F shows that ionomycin treatment significantly up-regulates 3×NFAT reporter activity, which is completely blocked by the addition of Cn inhibitors FK506 and cyclosporine A.
FIGURE 5.
Effect of calcium ions on TonEBP expression. A, nucleus pulposus cells were treated with the calcium ionophore ionomycin (I; 1 μm), along with PMA (P; 100 ng), and TonEBP expression was measured. Ionomycin treatment resulted in significant increase in TonEBP mRNA expression. B and C, immunofluorescence and Western blot analysis of cells as treated in A. note the increase in TonEBP protein after ionomycin treatment. The addition of Cn inhibitors FK506 (10 ng/ml) and cyclosporine A (CsA; 1 μg/ml) did not suppress synthesis of TonEBP protein. When nucleus pulposus cells were cultured under hyperosmotic conditions (450 mosmol/kg), there was high induction in TonEBP, whereas TonEBP protein was undetectable in TonEBP null cells. D and E, reporter activity of nucleus pulposus cells transfected with wild type (D) or mutant (E) tauT reporter and treated with ionomycin and PMA with or without FK506 and cyclosporine. Note that treatment with ionomycin increased the activity of the WT but not the TonE-MT tauT reporter. FK506 and CsA did not inhibit ionomycin-mediated increase in tauT-WT promoter activity. As expected under hyperosmotic conditions, there is a robust induction in activity of tauT-WT but not tauT-MT reporter. F, induction of the activity of 3×NFAT reporter in NP cells following ionomycin and PMA treatment. The reporter is highly induced, whereas the addition of FK506 and cyclosporine completely blocks activation, indicating a requirement for Cn in this process. Values shown are mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05.
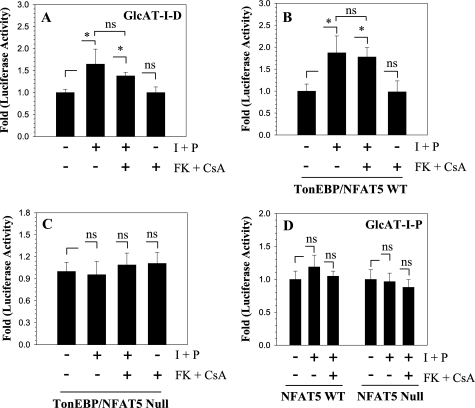
To ascertain if Cn signaling together with TonEBP plays a role in GlcAT-I expression, we treated nucleus pulposus cells with ionomycin and PMA together with FK506 and cyclosporine A. Fig. 6A shows that ionomycin treatment results in an increase in GlcAT-I promoter activity. The presence of FK506 and cyclosporine did not block ionomycin-dependent induction in promoter activity. To further validate these findings, we used NFAT5 null and wild type MEFs and measured activities of GlcAT-I reporters following ionomycin treatment. Fig. 6B shows that ionomycin causes an increase in GlcAT-I reporter activity. Again, inclusion of FK506 and cyclosporine did not affect promoter induction in wild type MEFs. In contrast, in NFAT5 null MEFs, ionomycin did not alter GlcAT-I reporter activity (Fig. 6C). Moreover, neither wild type nor null cells exhibit an increase in GlcAT-I-P reporter (Fig. 6D).
FIGURE 6.
Ionomycin mediates GlcAT-I promoter activation, although TonEBP is independent of calcineurin. Nucleus pulposus cells (A), TonEBP/NFAT5 wild type (B), and TonEBP/NFAT5 null (C) MEFs were transfected with GlcAT-I-D reporter, and activity was measured following treatment with ionomycin with or without FK506 and cyclosporine A. Unlike TonEBP/NFAT5, null MEFs GlcAT-I-D reporter activity was induced in both nucleus pulposus and NFAT5 wild type cells. Calcineurin inhibitors did not suppress ionomycin-induced or basal activity of GlcAT-I reporter in any of the cell types. D, effect of ionomycin treatment on GlcAT-I-P reporter activity. The reporter was nonresponsive to ionomycin treatment in both TonEBP/NFAT5 wild type and null MEFs. Values shown are mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05.
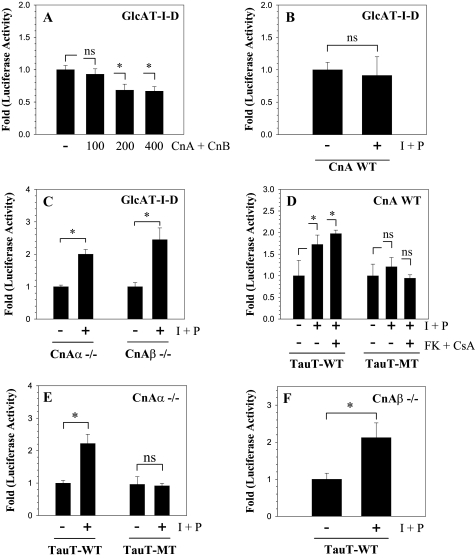
The contribution of Cn signaling in GlcAT-I expression was studied by co-transfecting nucleus pulposus cells with plasmids expressing CnA and CnB along with GlcAT-I-D reporter. Fig. 7A shows that nucleus pulposus cells receiving 200 ng or more of Cn plasmids display suppression in GlcAT-I reporter activity. To further explore if Cn signaling played a regulatory role, we used primary medullary fibroblasts derived from CnAα null, CnAβ null, and wild type mice. Treatment of wild type fibroblasts with ionomycin results in little change in GlcAT-I reporter activity (Fig. 7B). Interestingly, in both CnAα null and CnAβ null cells, ionomycin treatment enhances activation (2-3-fold) of the GlcAT-I-D reporter (Fig. 7C). We then measured the activity of both wild type (containing the WT TonE motif) and mutant tauT reporter (mutant TonE motif) in these cells. Wild type cells show a small increase in the activity of the WT-tauT reporter; activity is unaffected by the calcineurin inhibitors (Fig. 7D). A significant induction in tauT reporter is also seen in both CnAα (Fig. 7E) and CnAβ (Fig. 7F) null cells. Mutant tauT reporter is not induced in either the wild type, CnAα, or CnAβ null fibroblasts.
FIGURE 7.
Calcineurin suppresses GlcAT-I promoter function. A, the GlcAT-I reporter activity following cotransfection with calcineurin A/B constructs or empty vector pBJ5 in nucleus pulposus cells. Co-expression of calcineurin subunits suppressed GlcAT-I reporter activity in transfected cells. B–F, reporter activity of medullary fibroblasts derived from CnA WT or CnAα or CnAβ null (–/–) mice transfected with GlcAT-I or tauT (WT or MT) reporter and treated with ionomycin and PMA with or without FK506 and CsA. Ionomycin did not increase GlcAT-I reporter activity in wild type cells (B), but a robust induction was observed in both the CnAα or CnAβ null cells (C). A similar high induction in the activity of tauT-WT reporter was observed in CnAα or CnAβ null cells; a relatively small inductive effect was seen in wild type cells, which remained constant after the addition of calcineurin inhibitors FK506 and CsA. Neither wild type (D) nor the null cells (E and F) displayed induction in tauT-MT reporter activity. Values shown are mean ± S.D. of three independent experiments performed in triplicate. *, p < 0.05.
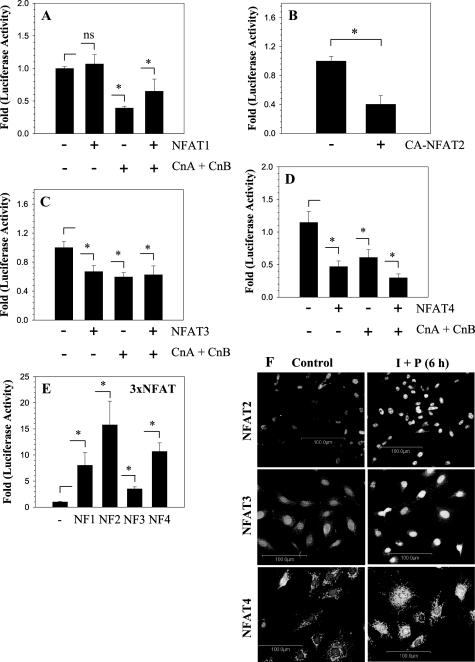
To investigate the relationship between GlcAT-I and NFAT signaling, we transfected nucleus pulposus cells with plasmids encoding NFAT1–4 and measured the activity of the GlcAT-I-D reporter. Fig. 8A shows that co-expression of NFAT1 alone has no effect on GlcAT-I reporter activity. However, when Cn (CnA and CnB) was co-expressed with or without NFAT1, there is a significant suppression in reporter activity. Unlike NFAT1, co-expression of CA-NFAT2 (Fig. 8B), NFAT3 (Fig. 8C), or NFAT4 (Fig. 8D), with or without Cn plasmids, suppresses GlcAT-I promoter activity in nucleus pulposus cells. To confirm that transfected NFAT plasmids expressed functional proteins in nucleus pulposus cells, we measured activation of an NFAT-responsive 3×NFAT reporter. Fig. 8E shows that there is significant activation of 3×NFAT luciferase reporter when co-transfected with NFAT1 or CA-NFAT2, NFAT3, or NFAT4 expression plasmids. We used immunofluorescence microscopy to confirm expression and activation of endogenous NFAT2, -3, and -4 in rat nucleus pulposus cells. Fig. 8F shows that under basal conditions, NFAT2, -3, and -4 are expressed by the nucleus pulposus cells and mostly localize to the cytoplasm. Increased localization of these proteins to nuclei is seen following a 6-h treatment of cells with ionomycin (Fig. 8F).
FIGURE 8.
GlcAT-I promoter activity suppressed by NFAT signaling in nucleus pulposus cells. Effect of NFAT vectors (NFAT1, -2, -3, and -4) and/or CnA and CnB on GlcAT-I-D reporter activity of nucleus pulposus cells. A, NFAT1 alone did not affect GlcAT-I reporter activity. When calcineurin was added alone or together with NFAT1, there was a suppression in reporter activity. CA-NFAT2 (B), NFAT3 (C), or NFAT4 (D) alone or when added with CnA/B significantly suppressed the GlcAT-I-D reporter activity in nucleus pulposus cells. E, nucleus pulposus cells were transfected with 3×NFAT luciferase construct with or without NFAT1, CA-NFAT2, NFAT3, or NFAT4, and reporter activity was measured. Co-expression of NFAT1 to -4 resulted in a significant increase in activity of 3×NFAT reporter plasmid, indicating functionality of expressed proteins. Values shown are mean ± S.D., of three independent experiments. *, p < 0.05; ns, nonsignificant. F, activation and nuclear localization of NFAT2/3/4 following ionomycin treatment of nucleus pulposus cells.
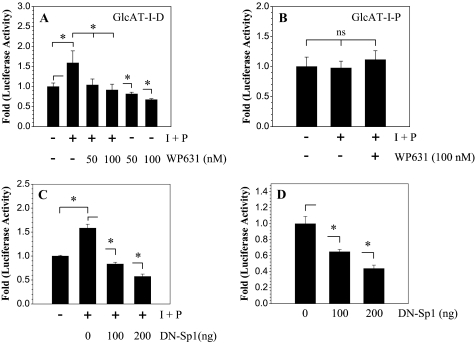
Finally, we investigated if Sp1 activity is required for TonEBP regulation of GlcAT-I promoter function in nucleus pulposus cells. We treated nucleus pulposus cells with WP631, an inhibitor of Sp1 activity, with or without ionomycin and measured the activity of GlcAT-I reporters. Treatment with WP631 at 50–100 nm completely abolishes ionomycin-mediated induction in GlcAT-I-D reporter activity as well as suppressing basal promoter activity (Fig. 9A). On the other hand, ionomycin did not induce GlcAT-I-P reporter (–123/+61 bp), and the activity remained unaffected by WP631 (Fig. 9B). To further validate these findings, nucleus pulposus cells were co-transfected with plasmid encoding DN-Spl. Fig. 9C shows that expression of DN-Spl completely abolishes ionomycin induction of GlcAT-I promoter activity. Moreover, expression of DN-TonEBP also suppresses the basal activity of the GlcAT-I-D promoter fragment (Fig. 9D). A significant inhibitory effect of DN-TonEBP expression on GlcAT-I reporter activity is seen at a dose of 100 ng, which is further enhanced when the concentration of the DN-Spl is increased to 200 ng (Fig. 9D).
FIGURE 9.
TonEBP and Spl are both required for calcium-mediated regulation of GlcAT-I promoter activation in nucleus pulposus cells. Cells were transfected with GlcAT-I-D (A) or GlcAT-I-P (B) promoter constructs and treated with ionomycin with or without WP631 (50–100 nm), an Spl inhibitor. Note that suppression of Spl function resulted in complete loss of ionomycin-mediated induction of GlcAT-I reporter. B, ionomycin failed to induce activity of GlcAT-I-P construct lacking the TonE motif in nucleus pulposus cells; WP631 has no effect on the activity of this reporter. Values shown are mean ± S.D. of three independent experiments. *, p < 0.05; ns, nonsignificant.
DISCUSSION
The experiments described in this investigation demonstrated for the first time that GlcAT-I, a key enzyme required for chondroitin sulfate biosynthesis, was regulated by TonEBP (NFAT5). This transcription factor would serve to promote the expression of a critical enzyme required for GAG chain synthesis as well as enhancing the expression of the aggrecan core protein (4). Our studies revealed that the regulated expression of GlcAT-I in nucleus pulposus cells was calcium-dependent. This finding is of critical functional importance, since shifts in calcium ions serve as key transducers of applied biomechanical loads. Accordingly, loading would promote calcium flux and enhance the synthesis of hydrated proteoglycans. A second major observation was that members of the NFAT family were expressed by nucleus pulposus cells; in this case, calcium-dependent Cn-NFAT signaling functioned as a negative regulator of GlcAT-1 expression. Thus, calcium ions regulate GlcAT-I expression through a signaling network comprising both suppressor (Cn-NFAT) and activator (TonEBP) molecules. In this way, by controlling both GAG and aggrecan synthesis, cells of the nucleus pulposus can autoregulate their own osmotic environment and accommodate mechanical loading.
Since cell function in the osmotically and mechanically stressed environment of the nucleus pulposus is dependent on the biosynthesis of aggrecan (2), we first evaluated the expression of GlcAT-I. This protein completes the synthesis of the common linker region of GAGs by transferring glucuronic acid to the core protein Gal-Gal-Xyl-O-Ser trisaccharide. Predictably, at both the mRNA and protein level, there was robust expression of GlcAT-I in nucleus pulposus cells of both the neonatal and mature rat discs; the protein was also expressed by cells of the annulus fibrosus. This latter observation was not unexpected, since the inner annulus, like the nucleus, is rich in proteoglycans (1, 2). Gain and loss of function experiments were performed to learn if calcium flux induced transcriptional activation of GlcAT-I reporter activity and if this activation was regulated by TonEBP. In the presence of ionomycin, suppression of TonEBP activity blocked induction of GlcAT-I; pTonEBP enhanced GlcAT-I reporter expression under basal conditions. Moreover, suppression of TonEBP in the absence of ionomycin also blocked GlcAT-I promoter activity, indicating that this transcription factor controlled basal gene expression. When GlcAT-I reporters that either lacked TonE site or contained a mutant TonE were treated with ionomycin, there was failure to increase activity. This observation lent strength to the findings reported above and indicated that TonEBP regulated GlcAT-I expression. We were able to confirm these findings using ChIP analysis and cells from TonEBP/NFAT5 null mice. ChIP analysis confirmed binding of TonEBP to TonE motif. Null MEFs displayed decreased basal GlcAT-I promoter activity and failure to induce reporter activity even when treated with ionomycin. Together, the results of these functional studies indicated that, in addition to regulating tauT, other osmolytes, and HSP-70 (4, 14, 21), by controlling the expression of GlcAT-I, TonEBP adapts nucleus pulposus cells to the mechanically and osmotically restrained conditions of the disc (4, 22).
The mechanism of activation of TonEBP is not completely understood, especially whether it is mediated by protein phosphorylation (23). In T cells and kidney cells, there is some evidence to indicate that regulation may be mediated by a phosphatase, Cn, which is activated by calcium ions (24, 25). The results of the studies described herein suggest that this mechanism may be cell type-specific. Treatment of nucleus pulposus cells with cyclosporine and FK506, agents that inhibit Cn signaling, failed to block induction of TonEBP or change the promoter activities of the TonEBP target gene tauT. Further support of the notion that Cn signaling was not required for TonEBP-dependent GlcAT-I promoter activation came from studies performed on TonEBP/NFAT5 null MEFs. We noted that, following ionomycin treatment, the null MEFs failed to induce GlcAT-I-D reporter activity, whereas both wild type and null cells did not induce activation of GlcAT-I-P construct. Although these results suggested that Cn signaling did not play a role in regulation of basal GlcAT-I expression, they did not elucidate role of Cn in the absence of TonEBP activity in nucleus pulposus cells.
To delineate the import of Cn in cells of the disc, we overexpressed catalytic (CnA) and regulatory (CnB) subunits and determined GlcAT-I reporter activity. Surprisingly, GlcAT-I promoter activity was suppressed; moreover, unlike wild type cells, ionomycin treatment of fibroblasts derived from Cn (CnAα and CnAβ) null mice strongly induced GlcAT-I reporter activity. Of interest was the observation that, unlike in nucleus pulposus cells and wild type NFAT5 MEFs, ionomycin treatment did not increase GlcAT-I-D reporter activity in Cn wild type fibroblasts, indicating cell type-specific differences. It is not unreasonable to assume that in these fibroblasts simultaneously with TonEBP, there is a strong induction of factors that negatively regulate the promoter activity, thereby neutralizing TonEBP-mediated induction.
Based on these studies, it is concluded that calcineurin signaling may be a negative regulator of GlcAT-I expression. The mechanism of suppression was not immediately obvious, since ionomycin simultaneously increased TonEBP-dependent GlcAT-I promoter activity. One possibility was that other members of the NFAT family may be involved in regulating GlcAT-I promoter activity. Relevant to this issue, it is well documented that Cn-mediated dephosphorylation of SP motifs results in the nuclear import of NFAT and stimulation of transcription (26). To address this issue, we overexpressed individual NFATs and determined GlcAT-I reporter activity. Since NFAT2, -3, and -4 but not NFAT1 decreased GlcAT-I promoter activity, it was concluded that GlcAT-I promoter activity was negatively regulated by these three NFATs but was independent of NFAT1. Activation of 3×NFAT luciferase reporter after co-transfection with NFAT expression plasmids confirmed that the expressed NFATs were functional in nucleus pulposus cells. Based on these findings and immunofluorescence experiments that document expression and activation of endogenous NFATs, we conclude that Cn-mediated activation of NFAT2, NFAT3, and NFAT4 serve an inhibitory role in governing GlcAT-I promoter activity in cells of the nucleus pulposus.
A recent study indicates that there are a number of Spl binding sites close to the transcription start site in the human GlcAT-I promoter (13). This study further showed that calcium promotes Spl activation through MEK/ERK signaling and subsequent binding to the motif between –65 and –56 bp influencing GlcAT-I expression (see Fig. 2) (13). Our studies confirmed that inhibition of Spl activity suppressed ionomycin-mediated induction of GlcAT-I promoter activity. Interestingly, ionomycin did not influence the activity of the GlcAT-I-P construct that lacked TonE but contained Spl motifs. Based on these findings, it is clear that Spl together with TonEBP regulates GlcAT-I promoter function. Moreover, specific to nucleus pulposus cells, our study shows that Spl activation alone is not sufficient for induction of GlcAT-I promoter activity, suggesting a cell type-specific regulation (13).
In summary, we demonstrate that calcium regulates expression of GlcAT-I, a critical enzyme required for GAG synthesis through TonEBP/NFAT5 and Cn-NFAT signaling pathways. Ongoing studies are being performed to test the hypothesis that changes in the activity of GlcAT-I are linked to the development of degenerative disc disease.
This work was supported, in whole or in part, by National Institutes of Health Grants AR050087 and AR055655. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GAG, glycosoaminoglycan; GlcAT-I, β1,3-glucuronosyltransferase-I; NFAT, nuclear factor of activated T cells; Cn, calcineurin; BAPTA, 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid; BAPTA-AM, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid acetoxymethyl ester; PMA, phorbol 12-myristate 13-acetate; DN, dominant negative; CA, constitutively active; MEF, mouse embryo fibroblast; PBS, phosphate-buffered saline; ChIP, chromatin immunoprecipitation; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; ERK, extracellular signal-regulated kinase.
References
- 1.Feng, H., Danfelter, M., Strömqvist, B., and Heinegård, D. (2006) J. Bone Joint Surg. Am. 88 25–29 [DOI] [PubMed] [Google Scholar]
- 2.Setton, L. A., and Chen, J. (2006) J. Bone Joint Surg. Am. 88 52–57 [DOI] [PubMed] [Google Scholar]
- 3.Ng, L., Grodzinsky, A. J., Patwari, P., Sandy, J., Plaas, A., and Ortiz, C. (2003) J. Struct. Biol. 143 242–257 [DOI] [PubMed] [Google Scholar]
- 4.Tsai, T. T., Danielson, K. G., Guttapalli, A., Oguz, E., Albert, T. J., Shapiro, I. M., and Risbud, M. V. (2006) J. Biol. Chem. 281 25416–25424 [DOI] [PubMed] [Google Scholar]
- 5.Kitagawa, H., Ujikawa, M., and Sugahara, K. (1996) J. Biol. Chem. 271 6583–6585 [DOI] [PubMed] [Google Scholar]
- 6.Venkatesan, N., Barré, L., Benani, A., Netter, P., Magdalou, J., Fournel-Gigleux, S., and Ouzzine, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 18087–18092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai, X., Wei, G., Sinha, A., and Esko, J. D. (1999) J. Biol. Chem. 274 13017–13024 [DOI] [PubMed] [Google Scholar]
- 8.Gouze, J. N., Bordji, K., Gulberti, S., Terlain, B., Netter, P., Magdalou, J., Fournel-Gigleux, S., and Ouzzine, M. (2001) Arthritis Rheum. 44 351–360 [DOI] [PubMed] [Google Scholar]
- 9.Alford, A. I., Yellowley, C. E., Jacobs, C. R., and Donahue, H. J. (2003) J. Cell. Biochem. 90 938–944 [DOI] [PubMed] [Google Scholar]
- 10.Parvizi, J., Parpura, V., Greenleaf, J. F., and Bolander, M. E. (2002) J. Orthop. Res. 20 51–57 [DOI] [PubMed] [Google Scholar]
- 11.Vijayagopal, P., and Subramaniam, P. (2001) Atherosclerosis 157 353–360 [DOI] [PubMed] [Google Scholar]
- 12.Fagnen, G., Phamantu, N. T., Bocquet, J., and Bonnamy, P. J. (1999) J. Cell. Biochem. 76 322–331 [DOI] [PubMed] [Google Scholar]
- 13.Barré, L., Venkatesan, N., Magdalou, J., Netter, P., Fournel-Gigleux, S., Ouzzine, M. (2006) FASEB J. 20 1692–1694 [DOI] [PubMed] [Google Scholar]
- 14.Ito, T., Fujio, Y., Hirata, M., Takatani, T., Matsuda, T., Muraoka, S., Takahashi, K., and Azuma, J. (2004) Biochem. J. 382 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam, A. K., Ko, B. C., Tam, S., Morris, R., Yang, J. Y., Chung, S. K., and Chung, S. S. (2004) J. Biol. Chem. 279 48048–48054 [DOI] [PubMed] [Google Scholar]
- 16.Monticelli, S., and Rao, A. (2002) Eur. J. Immunol. 32 2971–2978 [DOI] [PubMed] [Google Scholar]
- 17.Ichida, M., and Finkel, T. (2001) J. Biol. Chem. 276 3524–3530 [DOI] [PubMed] [Google Scholar]
- 18.Clipstone, N. A., and Crabtree, G. R. (1992) Nature 357 695–697 [DOI] [PubMed] [Google Scholar]
- 19.Risbud, M. V., Guttapalli, A., Stokes, D. G., Hawkins, D., Danielson, K. G., Schaer, T. P., Albert, T. J., and Shapiro, I. M. (2006) J. Cell. Biochem. 98 152–159 [DOI] [PubMed] [Google Scholar]
- 20.Zeng, Y., Danielson, K. G., Albert, T. J., Shapiro, I. M., and Risbud, M. V. (2007) J. Bone Miner. Res. 22 1851–1861 [DOI] [PubMed] [Google Scholar]
- 21.Woo, S. K., Lee, S. D., Na, K. Y., Park, W. K., and Kwon, H. M. (2002) Mol. Cell. Biol. 22 5753–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai, T. T., Guttapalli, A., Agrawal, A., Albert, T. J., Shapiro, I. M., and Risbud, M. V. (2007) J. Bone Miner. Res. 22, 965–974 [DOI] [PubMed] [Google Scholar]
- 23.Jeon, U. S., Kim, J. A., Sheen, M. R., and Kwon, H. M. (2006) Acta Physiol. (Oxf.) 187 241–247 [DOI] [PubMed] [Google Scholar]
- 24.Trama, J., Lu, Q., Hawley, R. G., and Ho, S. N. (2000) J. Immunol. 165 4884–4894 [DOI] [PubMed] [Google Scholar]
- 25.Li, S. Z., McDill, B. W., Kovach, P. A., Ding, L., Go, W. Y., Ho, S. N., and Chen, F. (2007) Am. J. Physiol. 292 C1606–C1616 [DOI] [PubMed] [Google Scholar]
- 26.Hogan, P. G., Chen, L., Nardone, J., and Rao, A. (2003) Genes Dev. 17 2205–2232 [DOI] [PubMed] [Google Scholar]