Abstract
The fibrinogen-related protein family (FREP, also known as FBN) is an evolutionarily conserved immune gene family found in mammals and invertebrates. It is the largest pattern recognition receptor gene family in Anopheles gambiae, with as many as 59 putative members, while the Drosophila melanogaster genome has only 14 known FREP members. Our sequence and phylogenetic analysis suggest that this remarkable gene expansion in the mosquito is the result of tandem duplication of the fibrinogen domain. We found that the majority of the FREP genes displayed immune-responsive transcription after challenge with bacteria, fungi, or Plasmodium, and these expression patterns correlated strongly with gene phylogeny and chromosomal location. Using RNAi-mediated gene-silencing assays, we further demonstrated that some FREP members are essential factors of the mosquito innate immune system that are required for maintaining immune homeostasis, and members of this family have complementary and synergistic functions. One of the most potent anti-Plasmodium FREP proteins, FBN9, was found to interact with both Gram-negative and Gram-positive bacteria and strongly co-localized with both rodent and human malaria parasites in the mosquito midgut epithelium, suggesting that its defensive activity involves direct interaction with the pathogen. Interestingly, FBN9 formed dimers that bound to the bacterial surfaces with different affinities. Our findings indicate that the A. gambiae FREP gene family plays a central role in the mosquito innate immune system and provides an expanded pattern recognition and anti-microbial defense repertoire.
Mosquitoes transmit a broad range of human parasitic and viral diseases, of which malaria remains the most devastating disease with a worldwide prevalence of over 400 million cases and 2 million deaths per year. The mosquito innate immunity plays a pivotal role in the interaction between a pathogen and the insect vector, and it determines the mosquito vectorial capacity. This immune defense system is comprised of cellular and humoral mechanisms that are activated upon recognition of invading pathogens by the mosquito pattern recognition receptor (PRR) molecules. Recognition of pathogen-associated molecular patterns (PAMPs) can indirectly trigger a variety of defense mechanisms through the activation of serine protease cascades and intracellular immune signaling pathways that control the transcription of effectors, or it can directly invoke killing mechanisms such as encapsulation and phagocytosis (1–3).
Whereas microbial exposure of mosquitoes can be quite extensive, the molecular mechanisms responsible for recognizing such a diverse range of microbes is not yet well understood. Insect immune surveillance systems lack antibody-mediated pathogen recognition and must instead rely on a rather limited set of pattern recognition receptors that detect invariant and evolutionarily conserved molecules on the surface of pathogens (4). Such a system would conceptually only be capable of distinguishing between broad categories of pathogens, such as Gram-positive versus Gram-negative bacteria (5), but studies have shown a much greater degree of specificity whose mechanistic basis is unknown (6–9). One mechanism that has been proposed to increase the mosquito and fruit fly pattern recognition receptor repertoire relies on alternative splicing of the Down syndrome cell adhesion molecule (Dscam) gene that encodes a hypervariable immunoglobulin domain-containing receptor (10–12).
A common pattern recognition receptor gene family in invertebrates is the fibrinogen-related proteins (FREPs)2 gene family, also known as fibrinogen domain immunolectin (FBN) (13–16). There are ∼200 amino acid residues in the fibrinogen-like (FBG) domain with high sequence similarity to the C terminus of the fibrinogen β and γ chains. In mammals, fibrinogen participates in both the cellular and fluid phases of coagulation (17). Among the three distinct fibrinogen domain immunolectins that been identified, ficolins are the most important molecules in terms of their function as pattern recognition receptors in phagocytosis and complement activation (18–25). All of these FREPs contain a pathogen-binding FBG domain at their C terminus, and their N-terminal sequence is likely involved in interactions with the N termini of other FREPs, resulting in the formation of multimeric protein bundles with potentially increased affinity and specificity for the pathogens.
In invertebrates, several FREPs have been discovered in various species, and almost all of them are likely implicated in the innate immune systems (13–15, 26–30). The horseshoe crab (Tachypleus tridentatu) tachylectins (TL5A and TL5B) can specifically recognize acetyl group-containing substances (N-acetylglucosamine, GlcNAc) and agglutinate all types of human erythrocytes and Gram-positive and Gram-negative bacteria (13). Ficolins and TL5A share a conserved three-dimensional structure in their Ca2+-binding site, and the acetyl group ligand-binding pocket. TL5A and TL5B also form 3- or 4-bladed and 2-bladed propeller structure multimers, respectively, with each blade corresponding to a dimer formed through inter- and intra-chain disulfide linkages involving conserved cysteine residues (13). The ficolins form triple helices, and four of these triple helices associate to form a bouquet structure that is responsible for the GlcNAc and other sugar binding activity (20–22, 31). FREPs from the snail Biomphalaria glabrata are composed of two functional domains, with an N-terminal immunoglobulin (Ig) domain that may be repeated in tandem and a C-terminal FBG domain (15, 16, 32, 33). This family has at least 14 members, of which FREP2 is an immuneresponsive protein that plays a role in host-parasite interactions (34, 35). A very recent interesting observation is that the FREPs demonstrated the recognition specificities to different categories of pathogens with FREP4 bound to parasites specifically (36).
The FREP gene family is the largest immune- and pattern recognition gene family in Anopheles gambiae, with 59 putative members; in Anopheles aegypti, it has 37 members and in Drosophila melanogaster, only 14 members. Species-specific expansions have occurred, and only three mosquito orthologous pairs exist (37). Our previous studies have shown that several members of the FREP (or FBN) family are immune-responsive to challenge with bacteria or Plasmodium, and FBN8, FBN9, and FBN39 are involved in anti-Plasmodium defense (30). Here we present a comprehensive functional and molecular dissection of the A. gambiae FREP gene family with regard to immunity-related functions.
MATERIALS AND METHODS
Mosquito Rearing and RNA Sample Preparation—A. gambiae Keele strain mosquitoes were maintained on a 10% sugar solution in laboratory culture at 27 °C and 70% humidity with a 12-h light/dark cycle, according to standard rearing procedures (38). Tissues were dissected on ice, and RNA was extracted from the dissected tissues at the assayed time points using the RNeasy kit (Qiagen). Quantification of RNA concentrations was performed using an Eppendorf spectrophotometer.
Data Base Searches, Phylogenetic Analysis, and Secondary Structure Prediction—Using the BLAST program, a fibrinogen-like domain (FBG) seed sequence was aligned with the A. gambiae translation sequences. Proteins were considered if they were predicted to contain at least one FBG domain by Vector-Base and the Ensembl A. gambiae genome server (39) or a web-accessible resource, ImmunoDB (37). The resulting protein sequences from the first search were used to iterate the search and retrieve any FREPs that had been missed. The list was manually checked, and a non-redundant set of protein sequences was obtained. There are two available naming systems for the sequence identifiers (37, 40): we used the FBN naming system according to Ref. 40, and thereafter when we pointed out the specific genes we used FBN. Corresponding FREPs from Ref. 37 are included in the supplemental Table S1.
Full-length or partial predicted sequences of FREP homologues were aligned using the ClustalX program, and cladograms were constructed by neighbor-joining analysis and displayed through Treeview software. The chromosomal location of each FREP gene was manually searched through Vector-Base, and the relationship between the phylogenetic tree and chromosomal location were drawn manually using Adobe Photoshop 8.0. Secondary structure predictions were obtained using the PHD program, with multiple sequence alignment of FBG domains. The structural data for TL5A were obtained from the Protein Data Bank (PDB) (41).
Immune Challenges for Transcriptional Analysis—To prepare bacterial-challenged samples for transcriptional analyses, 4-day-old mosquitoes were first injected with ∼20,000 heat-inactivated Escherichia coli or Staphylococcus aureus (S. aureus). Approximately 10 mosquitoes were collected 4 h after challenge for each replicate, and at least three replicates were included. For the preparation of fungus-challenged samples, 4-day-old female mosquitoes were rolled over in a Petri dish with fully grown Beauveria bassiana (B. bassiana) with spores. Mosquitoes were then released into a paper cup and incubated for 24 h in the regular insectary. Plasmodium infections were carried out according to standard procedures and our established protocols (30, 42). Plasmodium berghei (P. berghei) infections were performed with a transgenic GFP P. berghei strain of wild-type ANKA at 21 °C, with mosquitoes fed on an uninfected mouse as a control. Plasmodium falciparum (P. falciparum) NF54 gametocyte cultures were prepared as previously described, and mosquitoes were fed on cultures through a membrane feeder at 37 °C and then maintained at 26 °C; mosquitoes fed on uninfected human blood were used as a control (30, 43). Mosquito midguts were dissected at 24 h after feeding, and unfed mosquitoes were removed; the remaining mosquitoes were kept at 21 °C for 13 days or 26 °C for 8 days for P. berghei and P. falciparum infections, respectively. The infection phenotypes were determined as previously described (30, 44).
Primers Design, Semi-quantitative RT-PCR, and qRT-PCR—The primers used for the RT-PCR transcriptional analysis were designed with the Primer 3 Program on a web-based server. Given that the FBG domain has high homology across all the FREP genes, the designed primer sequences were further BLAST-searched against the whole genome of A. gambiae. The primer sequences with no other hits with over 80% identity were selected. The primer specificity was tested with only one fragment having been amplified; the PCR products obtained were ∼300 bp (supplemental Table S2). Using the 39 FREP genes from the A. gambiae genome that have been well-annotated in Ensembl 26 (Nov. 2004), we designed a panel of 39 pairs of primers. The FBN15 primers amplified two fragments and were therefore excluded from further analysis. The primer sequences used are listed in supplemental Table S2.
Reverse transcription was carried out at 42 °C for 2 h using a SuperScript II kit (Invitrogen) and 20-μl reaction mixtures containing oligo (dT) primers and 2 μg of total RNA. The PCR cycles were controlled to produce a non-saturated band, indicating the linear amplification. Optimal cycle numbers were empirically determined from a test PCR run, and three replicates were obtained for each gene. The PCR products were separated by gel electrophoresis and the DNA bands documented using a Fuji camera after SYBR Green staining (Qiagen). Signal intensities from the gels were quantified using Science Lab 98 software (Fuji-film Imaging System). The fold induction or repression of immune-challenged samples was determined by comparing the results obtained to those for the control PBS treatment, normalizing the data to the PCR intensity obtained for the housekeeping ribosomal gene S7. The fold changes in expression were log2-transformed and hierarchically clustered into a color schematic profile using Cluster software, then organized according to transcription specificity by using TreeView software from Eisen Lab.
The efficiency of the gene silencing was assessed by real-time quantitative PCR (qRT-PCR), carried out essentially according to Ref. 30. Quantification was carried out using the QuantiTect SYBR Green PCR Kit (Qiagen) and ABI Detection System ABI Prism 7300.
RNAi Gene Silencing and Immune Challenge Assays—To make these assays as cost effective as possible, we employed a protocol based on using the same set of primers that were used in the semi-quantitative PCR for expression analysis to synthesize the required dsRNAs. The plasmid vector LITMUS 28i (New England Biolabs) was engineered as a TA cloning vector by digestion with NsiI and KpnI for 2 h, followed by Klenow treatment for 30 min. The vector was cleaned with the Mini-Elute Enzyme cleaning kit (Qiagen), then incubated with dTTP and Taq polymerase at 70 °C for 1 h. The product was cleaned again with the Qiagen kit before being used as a TA cloning vector, with the cloning site attached and T7 primers at both ends. The 38 FREP PCR fragments were amplified using the primers, then individually cloned into this TA cloning vector. FREP fragments with T7 sequences attached were obtained through a second round of PCR with the T7 primers. The sense and antisense RNAs were then synthesized from these fragments using the T7 MEGAscript kit (Ambion). About 69 nl of dsRNA (3 μg/μl) was introduced into the thorax of cold-anesthetized 4-day-old female mosquitoes using a nano-injector (Nanoject, Drummond) according to established methodology (45). For both the control (dsGFP-injected) and experimental (gene dsRNA-injected) groups, ∼80 4-day-old female mosquitoes were injected. Gene silencing was verified 3–4 days after injection by qRT-PCR using at least three biological replicates, with the A. gambiae ribosomal S7 gene as the internal control for the normalization. The primers used for silencing verification are presented in supplemental Table S2. 3–4 days after dsRNA injection, survival analysis of RNAi gene-silenced mosquitoes was carried out according to established protocol (30). Dead mosquitoes were counted and removed daily over a 7-day period after challenge with bacteria. Student's t test and two-way analysis of variance were used to assess the significance of the gene silencing effect on mosquito survival after challenge, with p values <0.05 being considered significant. P. berghei and P. falciparum infections of RNAi gene-silenced mosquitoes were done essentially according to our previous work (30).
Enumeration of Endogenous Bacteria from RNAi Gene-silenced Mosquitoes—Isolation of bacteria from gene-silenced mosquitoes and colony forming unit (CFU) enumeration and determination were done essentially according to Ref. 10. The hemolymph was obtained by perfusion of surface-sterilized mosquitoes 4 days after dsRNA injection, and CFU were determined by plating serial dilutions of the hemolymph on LB agar plates and incubating the plates at 27 °C for 2 days. Each assay was performed using pooled hemolymph from two mosquitoes, and at least 10 independent replicates were obtained for each gene. The species of the isolated bacteria were determined by amplifying a region of the 16S rDNA as described, using primers 27f and 1492r (46). PCR products were sequenced and BLAST-searched against the Nucleotide Collection (nr/nt) data base to verify the species identifications.
Anti-FBN9 Antibody—A PCR fragment of FBN9, spanning a region that coded for a peptide from amino acids 47–118 was cloned into the EK-LIC vector according to the manufacturer's instructions (Novagen). The kanamycin-resistant bacterial colonies were further screened for positive clones by PCR analysis with specific FBN9 primers, and the in-frame sequences with the histidine tag were also confirmed by DNA sequencing. The positive clones were then transformed in BL21 (DE3) pLysS-competent cells (Novagen) and induced with 0.4 m isopropyl-1-thio-β-d-galactopyranoside according to the manufacturer's instructions. The recombinant FBN9 protein was purified on an Ni-nitrilotriacetic acid column (Qiagen), and the eluted fractions were run on 4–12% SDS-PAGE gels (Invitrogen, Novex, Tris-glycine gel) to confirm that only a single band had been purified. A polyclonal antibody recognizing FBN9 was prepared by injection of the purified protein into rabbits. The titer and specificity of the antibody were assessed by Western blot analysis using the purified recombinant protein.
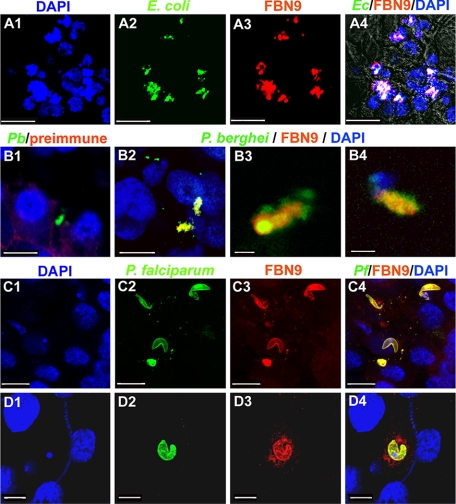
Immunohistochemical Staining and Confocal Microscopy—Immunofluorescence microscopy analysis of the interaction of the Sua5B cells with FITC conjugates of E. coli and S. aureus (Molecular Probes) was essentially based on the description given in (10). FBN9 antibody and preimmune serum as a control were used at 1:400 dilution. Immunostaining of mosquito midguts was performed essentially as described (47): In brief, 4-day-old Keele mosquitoes were fed on a transgenic GFP P. berghei strain (48) and kept at 21 °C or were membrane blood-fed with a P. falciparum gametocyte culture (1%) and kept at 26 °C. At 24–30 h after blood feeding, the midguts were dissected in 1% paraformaldehyde and washed with three times with PBS to remove the blood, then fixed in 4% paraformaldehyde (in PBS) for 1 h and washed twice with PBS. For confocal microscopy, the midguts were incubated with a blocking solution of 10% goat serum for 1 h, then incubated overnight at 4 °C with primary antibody at 1:400 dilution. Anti-Pbs28 (kindly provided by Dr. Sinden, Imperial College, London, UK) and mouse monoclonal anti-Pfs25 (MRA-28, ordered from MR4) were used for P. berghei and P. falciparum, respectively. After three PBS washes, the midguts were incubated with secondary antibody (Molecular Probes, 1:1000) for 2 h. AlexaFluor 488-conjugated (green) goat anti-mouse antibody (1:500 dilution) for the parasites and AlexaFluor 568-conjugated (red) goat anti-rabbit antibody (1:500 dilution) for FBN9. As a control, midguts were also incubated with preimmune FBN9 antibody at the same dilution. After another three PBS washes, the midguts were mounted using the ProLong Antifade Kit (Molecular Probes), after staining of the cell nuclei with DAPI. The samples were examined under a Zeiss LSM 510 confocal microscope, collecting 0.3- to 1-μm optical sections.
In Vitro Bacterial Binding Assays and Western Blotting—Bacterial binding assays were performed as described previously, with some modification (10, 49): In brief, 1 ml of the cell culture supernatant was also collected, and protein concentrations were determined using the BCA method (Pierce). A 10-ml sample of late-logarithmic phase cultures of E. coli, of Pseudomonas veronii (P. veronii) isolated from this study, or of Bacillus subtilis (B. subtilis) was centrifuged and resuspended in 1 ml of 0.2 m NaCl, followed by inactivation with 10% acetic acid and neutralization with five volumes of 1 m Tris-HCl (pH 8.0). The bacterial pellets were washed three times with PBS and then resuspended in 1:20 of the original volume in the same buffer. For the binding assay, equal amounts of bacterial suspensions (2.0 × 1010 CFU) were added to 1 ml of Sua5B cell culture supernatant. After incubation at 4 °C for 3–4 h with gentle agitation, samples were centrifuged at 8000 × g for 5 min at 4 °C, and the bacterial pellets were washed twice with 10 mm Tris-HCl (pH 8.0). Proteins bound to the bacteria were eluted in 50 μl of the same buffer containing increasing amounts of NaCl (0.1, 0.25, and 0.4 m), with a final elution using 50 μl of 0.5 m NaCl, 0.1 m NH4Ac (pH 5.0). Finally, 15 μl of the eluted samples from each treatment were subjected to 4–12% SDS-PAGE (Invitrogen, Novex, Tris-glycine gel) and incubated with anti-FBN9 (1:1000 dilution). The Sua5B cell culture supernatant was run as a positive control.
RESULTS AND DISCUSSION
Evolutionary Dynamics of the FREPs—A BLAST search using human ficolins against A. gambiae proteins has identified 59 FREP genes in A. gambiae, 37 members in A. aegypti, and 14 members in D. melanogaster (37). These three genomes share only two 1:1:1 trios of FREP (AgFREP1 and AgFREP16) and three orthologs between A. gambiae and A. aegypti (AgFREP9, AgFREP3, and AgFREP13). Recently available genome sequences of 12 Drosophila species have revealed that the gene number of FREP family in each Drosophila species varies from 14 to 43 genes (50), suggesting that in both mosquito and Drosophila species the FREP family diverged rapidly and has been under relaxed evolution constraints. Most of the A. gambiae FREP proteins include the FBG domain in their C terminus. All of the FREP proteins have only one FBG domain at their C terminus, while some 50% of these are truncated (supplemental Table S1). A majority of them do not have a typical domain at their N terminus, and only one FREP member, FREP1, also contains a thiamin diphosphate-binding (THDP binding) domain in addition to the FBG domain.
Secondary structure prediction and multiple sequence alignment of the FBG domains revealed a conserved structure profile throughout the FBG domain (supplemental Fig. S1). Most of these secondary structures are located in the conserved region, and their predicted structure is closely related to that of the horseshoe crab TL5A, indicating that the FBG domain is architecturally conserved between these species. The horseshoe crab TL5A and TL5B form multimers through inter- and intra-chain disulfide linkages that are based on the conserved cysteine residues. These four conserved cysteine residues are also present in the FBG domains of the FREP gene family, suggesting that the mosquito homologues also engage in multimerization (supplemental Fig. S1). Furthermore, TL5A contains four aromatic side chains (Tyr-210, Tyr-236, Tyr-248, and His-220) that form a funnel ligand-binding pocket for an acetyl group and are also conserved in the majority of the FBG domains of FREP proteins. However, these sites are absent from some of the FREP proteins; this diversity may reflect a potential for recognition of diverse carbohydrates on the surface of pathogens. Beyond the core structure, the FBG domains also show great diversity in terms of deletion and insertion within the conserved sequences, which may help these receptors provide different binding specificities.
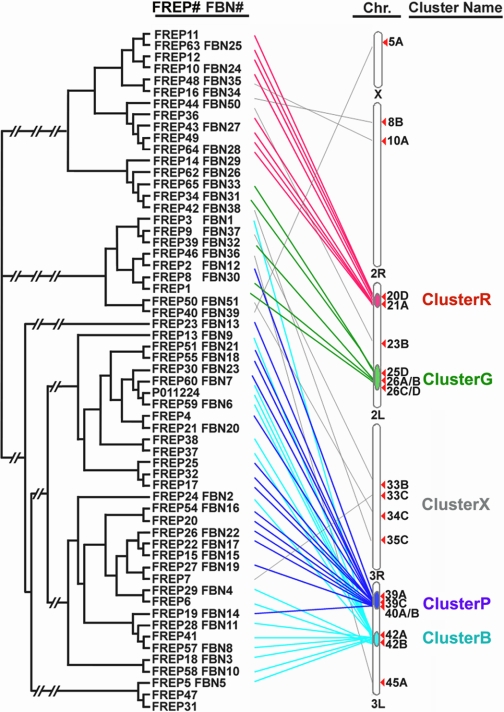
Correlation of the FREP phylogenetic map with the chromosomal locations of the genes showed a strong correlation between phylogeny clusters and cytogenetic locations (Fig. 1). The majority of the FREP genes were found in clusters on chromosomes 2L and 3L. As many as 33 FREP genes are located on chromosome 3L in two large clusters, ClusterB (chr. 3L: 42A,B) and ClusterP (chr. 3L: 39A,C/40A,B), and another 8 genes are located on chromosome 2L, where they form two small clusters, ClusterR (chr. 2L: 20D/21A) and ClusterG (chr. 2L: 25D/26A-D). The rest of the genes are scattered throughout the entire genome (grouped together into ClusterX and indicated by gray lines in Fig. 1).
FIGURE 1.
Phylogenetic relationships between the FBG domains and the chromosomal location of FREPs. FREP genes are grouped into 4 large clusters, and the chromosomal locations (Chr.) of each FREP have been manually drawn, with red, green, purple, and blue lines indicating ClusterR, ClusterG, ClusterP, and ClusterR, respectively. FREP genes scattered throughout all the chromosomes are indicated by gray lines and grouped into ClusterX. The FREP and FBN are indicated, and the accession numbers are listed in supplemental Table S1. The tree was constructed with ClustalX and TreeView using NJ-joining methods.
Through a comparison to the A. aegypti FREP phylogenetic tree, we found that some clusters were composed of FBG domains from A. gambiae alone and not from A. aegypti, suggesting that the FREP gene family evolved by expansions that occurred in A. gambiae after its divergence from other mosquito species (37). This conclusion is consistent with the hypothesis that recent gene duplications have occurred considerably more often in Anopheles than in fruit flies (40). The tandem orientation of the FBG domains is likely to provide a target for mispairing and unequal crossover, which can result in gene duplication and divergence over time. However, some of these tandemly duplicated FBG domains could have become physically separated through chromosomal rearrangement and/or translocation. Conversely, some FBG domains that were located in the same genomic regions were scattered in different phylogenetic tree clusters (Fig. 1), suggesting that the dynamic history of the FBG domains is likely to involve shuffling among chromosomes. At least a subset of FBG domains has putative function in sensing the carbohydrate on pathogen surfaces. The expansion of the FREP gene family in A. gambiae may represent the result from exposure to the diversity of pathogens during evolution processes, resulting in the utilization of varieties of FBG domains to recognize a broad range of different carbohydrates on the surfaces of different pathogenic organisms.
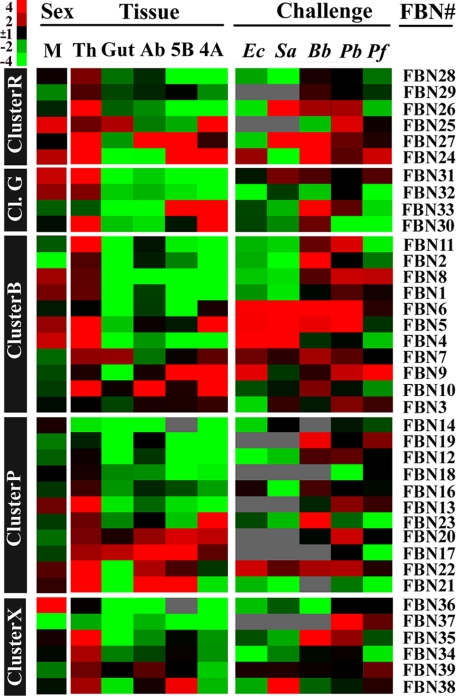
Diverse Sex- and Tissue-specific Expression Patterns Dictate Diverse Roles for FREPs in Mosquito Biology—To gain insight into the functional attributes of FREP gene members in A. gambiae, we investigated their sex- and tissue-specific expression patterns. Specifically, we assessed the mRNA abundance of 38 FREP gene members in whole adult males and females; in 4-day-old adult female head, thorax, abdomen, and midgut tissues; and in the hemocyte-like immune-competent A. gambiae cell lines Sua5B and Sua4A (Fig. 2 and supplemental Fig. S2). Eight FBN genes (4, 5, 8, 24, 25, 31, 32, and 36) were specifically expressed in males, and the expression of FBN36 was the most abundant in male mosquitoes. These members are less likely to play major roles in the defense against Plasmodium, since only female mosquitoes transmit malaria parasites. Ten genes were specifically expressed in the females, of which FBN2 was the most abundant followed by FBNs 37, 29, 19, 39, 20, 7, 12, 33, and 11.
FIGURE 2.
The sex, tissue-specific, and immune-responsive expression of FREP genes. At the left side are the FREP clusters, and the right side gives the corresponding FBN. M indicates the gene expression of that FREP in male mosquitoes compared with female mosquitoes and is followed by FREP expression within different tissues (Tissue exp.), compared with whole female mosquitoes. Th, Gut, Ab, 5B, 4A indicates thorax, gut, abdomen, cell line Sua5B, cell line Sua4A, respectively. The expression profiles of individual FREPs upon immune challenge of E. coli (Ec), S. aureus (Sa), B. bassiana (Bb), and Plasmodium-(P. berghei (Pb) and P. falciparum (Pf)) are given by comparing to naïve samples. Red and green color indicates higher and lower expression, respectively; black color indicates no difference within expression levels and gray indicates the value was not available. The expression profiles within each cluster are clustered based on the tissue expression analysis.
Our tissue-specific expression analysis showed that a majority of 14 FREP genes were highly expressed in the thoracic part, in which the fat body and hemocytes are located; a fairly large proportion were also expressed at high levels in the cell lines and abdomen. Almost all the genes that were significantly expressed in the cell lines were also expressed in the thorax, except FBN33. Hemocytes are abundant in the thorax, and the Sua5B and Sua4A cell lines are hemocyte-like immune-competent cell lines, suggesting that FREP proteins play important roles in hemocyte functions. Only three FREP genes were specifically expressed in the gut tissue, and this relatively small number suggests that this gene family is not playing a major role as an anticoagulant for the blood meal.
The Immune-responsive Expression Patterns of FREPs Suggest Diverse Functions within the Innate Immune System—Previous global transcriptomic analyses have shown that several FREP genes are regulated in response to bacterial infections; FBN9 and FBN25 are up-regulated after E. coli challenge, and FBN8, -9, and -39 are strongly induced locally in the midgut during ookinete invasion (30, 51). Rather than using less-sensitive microarray analyses, we chose to make use of semi-quantitative RT-PCR with specific primer pairs to investigate the expression of the 38 family members after immune challenge. There were seven genes induced in response to Gram-negative bacterial (E. coli) challenge and eight genes induced after Gram-positive bacterial (S. aureus) infection. FBN4, -5, -6, and 22 were induced upon both Gram-negative and Gram-positive bacterial immune challenges. In general, FREP members responded differently to these two elicitors (the correlation coefficient for overall gene expression profile for these two elicitors was Pearson's r = 0.31, n = 38). Conversely, the response of the FREP genes was quite similar upon challenge with two species of Plasmodium parasites, P. berghei and P. falciparum (Pearson's r = 0.61, n = 38). In total, there were seven FBNs (3, 6, 8, 9, 13, 22, and 37) that were induced by infections with both parasites. Fifteen genes were up-regulated in response to P. berghei challenge, while only eight genes were induced by P. falciparum infection. This difference could reflect the fact that P. berghei infection levels are in general higher than those of P. falciparum (30).
It is interesting to note that FBN5, 6, 9, 22, and 24 were up-regulated in response to four of the five pathogens, suggesting that these genes might be playing an important and more general role in the innate immune system. FBN7, 8, 26, and 27 were up-regulated by three of the five elicitors. Surprisingly, the expression of FBN39, a previously identified anti-Plasmodium molecule, was only induced by P. falciparum challenge and not by other pathogens. This gene was female-specific and is located on the X chromosome. Functional assays in the present study and in a previous study showed that FBN39, indeed, defended the mosquitoes only against P. falciparum, and not P. berghei (30).
Functional Amplification of FREPs through Gene Duplication— Gene expression is tightly linked to function, and this relationship led us to predict similarity in the transcription patterns between members that recently originated from a common ancestor. To look for such similarities, we examined the degree to which the phylogeny, chromosomal location, and expression pattern of the genes were correlated (Figs. 1 and 2). FREP genes were grouped into five major clusters (Clusters R, G, P, B, and X), and the expression profiles within each cluster were grouped on the basis of sex- and tissue-specific gene expression patterns. Genes with similar expression patterns within each group were therefore clustered in the same branch (approximate location).
For the tandemly duplicated genes, there was a strong correlation between phylogeny, chromosomal location, and expression profile (Fig. 2). For example, FBN4, -5, and -6 were clustered in the same phylogenetic grouping and the same chromosomal location, and the expression of these genes in response to immune challenges also showed a high level of similarity (Pearson's r = 0.74, n = 5). However, genes located in the chromosone 2L region (ClusterG) showed significant diversity in their gene expression patterns. The evolution of various groups of organisms is mainly a reflection of chromosomal rearrangements. In anopheline mosquitoes, synteny has been highly conserved, while gene order has been extensively shuffled, primarily through paracentric chromosomal inversions. In A. gambiae mosquitoes, the major gene duplication of chromosome 2L derived from the 2La inversion and A. gambiae s.s. is the only member of the complex in which the 2La inversion is polymorphic (52). FREP gene ClusterG is located in the chromosome 2La inversion region, a highly polymorphic region that is likely contributing to the diversity of FREP gene expression in chromosome 2L (ClusterG).
The FREPs Are Essential for Antibacterial Defense and Immune Homeostasis (or Basal Immune Surveillance)—Given the immune-responsiveness of some FREPs to bacterial challenge, we hypothesized that this gene family may play a major role in anti-bacterial defense. RNAi-mediated gene silencing of candidate FREP genes that were found to be regulated in response to bacterial challenge did indeed impair the mosquito capacity to defend itself against experimental and opportunistic bacterial infections (Figs. 2 and 3). The lack of an apparent infection phenotype after the silencing of some FREP members does, however, not necessarily mean that they are not involved in anti-microbial defense, but simply that the RNAi did not efficiently silence these genes. For example, some of the FREP genes may be expressed in cell types with less accessibility to the injected dsRNAs.
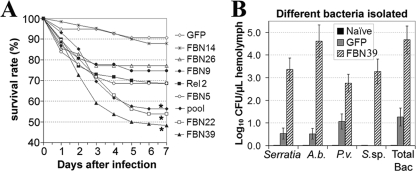
FIGURE 3.
The involvement of FBN22 and FBN39 in the defense against opportunistic bacterial infections. A, survival rates of the mosquitoes treated with dsRNA of FBN22-, FBN39-, and the pool of dsRNA-treated (pool) mosquitoes, dsGFP-injected (GFP) mosquitoes were served as control. The experiments shown represent at least three replicates; standard errors are not shown here to allow for clearer visualization (2-way analysis of variance; *, p < 0.01). B, CFUs of opportunistic bacteria isolated from FBN39 gene-silenced mosquito hemolymph at 4 days post dsRNA injection, non-treated mosquitoes (naive), and dsGFP-injected mosquitoes (GFP) were served as control. The isolated bacterial species are Serratia sp. (Serratia), Asaia bogorensis (A.b.), Pseudomonas veronii (P.v.), and Sphingomonas sp. (S.sp). Total Bac indicates the total number of bacteria been isolated.
Given the high degree of identity among some FREP genes, we investigated the possibility that dsRNAs that were designed to target specific members could also silence closely related members. Our results showed that dsRNA-mediated RNAi gene silencing was quite specific; only genes with 90% identity to the injected dsRNA could, in some cases, become partially silenced, to about 20% (supplemental Fig. S3). Gene silencing of FBN4, -5, -6, and –26 caused a significant reduction (∼56%) in survival rates during S. aureus infection. This level of reduction was comparable to that obtained for silencing of the antimicrobial peptide Gambicin, which served as a positive control (10, 53). Depletion of FBN4, -5, -6, -9, or -22 was associated with a significant decrease in survival (∼65%) at several time points during E. coli infection. An established positive control for E. coli challenge was not available.
The innate immune system is involved defending the mosquito against continuous exposure to opportunistic microbes, and it is therefore required to maintain a basal level activity to ensure immune homeostasis throughout the lifetime of an insect. In the absence of immune challenge, gene silencing of a pool of 13 FREP members (FBN 4, 5, 6, 8, 9, 11, 20, 22, 24, 25, 26, 37, and 39) that had been up-regulated after immune challenge resulted in a significantly increased level of mortality, with half of the mosquitoes dying by day 3 or 4; in contrast, 87% of the GFP dsRNA-treated control mosquitoes were still alive on day 7. A survival analysis was done for each individual gene to identify those gene(s) responsible for defense against opportunistic bacterial infection (Fig. 3A). Individual gene silencing of FBN22 and FBN39 resulted in similar levels of mortality, which were lower than that obtained for dsRNA pool injection, whereas silencing of FBN14 resulted in comparable survival rate as that obtained from GFP negative control. Gene silencing of the rest of the individual genes had similar survival rates as compared with Rel2 gene-silencing mosquitoes.
We next wanted to determine the species specificity of the bacteria against which the FREPs were active. Sequence analyses of the 16S ribosomal genes from the proliferating bacteria suggested a close phylogenetic relationship to the four Gram-negative bacterial species Serratia sp. (99%), Asia bogorensis (99%), Pseudomonas veronii (99%), and Sphingomonas sp. (99%). These bacteria together increased by 4 × log10 CFUs per microliter in the FBN39-silenced mosquito hemolymph 4 days after dsRNA injection (Fig. 3B), a similar bacterial distribution was observed for FBN22 gene-silenced mosquitoes (data not shown), supporting a role for FBN39 and FBN22 in immune homeostasis and general bacterial clearance. Therefore, we concluded that FBN22 and FBN39 are essential participants in the mosquito immune defense against bacteria, and necessary to maintain a basal immune surveillance level. Most likely, other species of opportunistic bacteria also proliferated in these FREP gene-silenced mosquitoes but were not detected by the culturing approach we utilized.
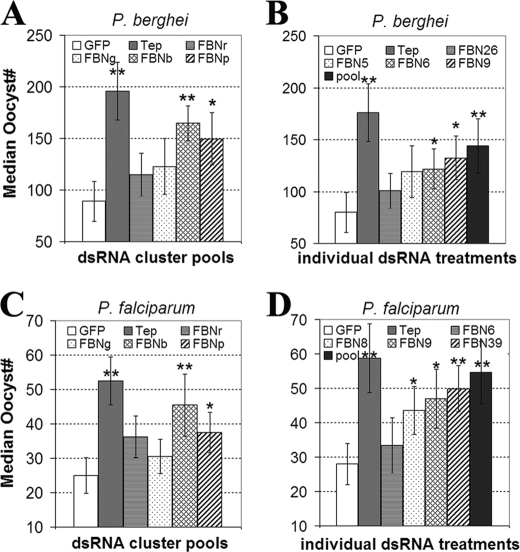
Synergistic and Complementary Functions of FREPs in the Defense against Plasmodium—To address the contribution of individual FREP gene clusters to the defense against malaria parasites, we used a pool of dsRNAs to target all the genes in each cluster. Silencing of a pool of FREP genes from ClusterB (FBNb) and ClusterP (FBNp) resulted in a statistically significant overall increase of 85 and 68%, respectively, in the number of P. berghei oocysts in the midgut and a larger proportion of mosquitoes with exceptionally high oocyst counts (>150) when compared with negative control GFP-dsRNA injected mosquitoes (GFP). In contrast, depletion of genes within ClusterR (FBNr) and ClusterG (FBNg) produced no significant change in oocyst number (Fig. 4A). The effect of gene silencing of ClusterB was ∼85% of that recorded for gene silencing of the highly potent anti-Plasmodium factor Tep1 (54). We also addressed the function of FREP (or FBN) clusters in the defense against human malaria parasite, P. falciparum. Depletion of the genes in ClusterB and ClusterP led to a significant overall increase of 82 and 50%, respectively in oocyst number (Fig. 4C). Gene silencing of ClusterB resulted in the increasing of oocysts numbers which was ∼87% of that for positive control anti-Plasmodium factor Tep1.
FIGURE 4.
P. berhgei and P. falciparum oocysts infection levels of the pools or individual FREPs gene-silenced mosquitoes. A, median P. berghei oocysts infection level of mosquitoes been gene-silenced of a pool of FREPs from ClusterB (FBNb) and ClusterP (FBNp), dsGFP-injected (GFP) mosquitoes were served as control. Tep1 dsRNA-treated mosquitoes (Tep) served as the positive control. The standard errors for three biological replicates are shown; * indicates p < 0.05 and ** indicates p < 0.01. B, median P. berghei oocysts infection level of mosquitoes been gene-silenced of individual FREPs (FBN5, FBN6, FBN9) and a pool of these three FBNs and FBN26 (pool). C, P. falciparum oocysts counts from the groups of mosquitoes with same treatments as from A. D, P. falciparum oocysts counts from the groups of mosquitoes with same treatments as from B.
Although the midgut is the primary site of the response to invading ookinetes and parasitic infections, most of the FREPs are not found in the midgut tissue but rather in the abdominal parts, which contain hemolymph and hemocytes. Hence, the observed anti-Plasmodium activities are most likely originating from the fatbody, midgut, hemocytes, and other tissues.
The genes that were significantly up-regulated in response to either P. berhgei or P. falciparum infection (Fig. 2) were tested individually to assess their involvement in the defense against both rodent and human malaria parasites. As indicated in Fig. 4B, depletion of FBN6 and FBN9 led to an overall increase of 55 and 68%, respectively, in oocyst number upon P. berghei infection, while gene silencing of FBN5 or FBN26 had no effect on infection. However, gene silencing that targeted a pool of these four FBNs (pool) resulted in increased permissiveness to P. berghei infection, as indicated by a significant 82% increase in oocyst number, comparable to the effect obtained for the positive control (dsTep1). With regard to the involvement of FREPs in the defense against human malaria parasites (P. falciparum), we found that gene silencing of FBN6 had no significant effect on the oocyst number (Fig. 4D). Gene silencing of individual FREPs (FBN8, FBN9, and FBN39) resulted in an increased permissiveness to P. falciparumi infection, as indicated by a significant 58–81% increase in oocyst number, while targeting of these genes together, including FBN6, resulted in a 95% increase in oocyst number. Thus, the effect of individually silencing genes was not comparable to that obtained when we simultaneously silenced multiple genes, suggesting that FREP proteins are functioning synergistically in the defense against Plasmodium. Conversely, the effect of the simultaneous silencing of multiple FREP genes was less than the sum of the individual gene-silencing effects, further pointing to a certain degree of complementarity in anti-Plasmodium function.
The FREPs Associate with Plasmodium in the Midgut Epithelium, where the Anti-Plasmodium Defense Occurs—The importance of FREPs for the antibacterial defense and the pathogen binding nature of the FBG domain together suggested that FREPs may be interacting directly with bacteria. Interactions may also occur with parasite in the midgut epithelium, where most of the killing takes place (55–57). FBN9 has been shown to be involved in the defense against both Gram-negative and Gram-positive bacteria, and also in both malaria parasite species, making it a good candidate for the interaction assays. FBN9 displayed an even distribution on the surface of non-challenged immune competent Sua5B cells (data not shown) and became strongly co-localized to the sites of interaction of challenged cells with FITC-labeled E. coli (Fig. 5, A1–A4). A less intense co-localization was observed with S. aureus cells (data not shown). The lack of co-localization between FBN9 and yeast cell (Saccharomyces cerevisiae) served as a good negative control, indicating specificity of interaction (data not shown). Immunohistochemical analysis of FBN9 in both P. berghei- and P. falciparum-infected midguts revealed a strong co-localization with the ookinete stage of both parasite species at 24–30 h after ingestion of infected blood, the time at which the ookinetes invade the midgut epithelium (Fig. 5, B–D). This pattern of reactivity is likely to reflect a direct interaction or an association through other factors. The anti-Plasmodium activity of FBN9 and other FREP members is most likely mediated through this association.
FIGURE 5.
Co-localization of FBN9 with bacteria in the Sua5B cell line, and with Plasmodium in the mosquito midgut. A, FITC-labeled E. coli (green) that had been co-incubated with Sua5B cells co-localized with FBN9 (red). Co-localization is indicated in white; cell nuclei were stained with DAPI (blue). Scale bars:10 μm. B, co-localization of FBN9 with P. berghei ookinetes in the mosquito midgut epithelium 24–30-h post-infection (B2-B4). The ookinetes in the midgut stained with preimmune of anti-FBN9 antibody showed no co-localization, which served as negative control (B1). Blue indicates DAPI-stained cell nuclei, green indicates P. berghei ookinetes, and red indicates anti-FBN9 staining. Scale bars, B1-B2: 10 μm; B3: 1 μm; B4: 2 μm. C and D, FBN9 co-localizes with P. falciparum ookinetes in the mosquito midgut epithelium. Double staining of midgut tissues of mosquito 24–30-h post-infection with P. falciparum using the anti-FBN9 (red) and anti-Pfs25 (green) antibodies. Scale bars, C1-C4: 10 μm; D1-D4: 5 μm.
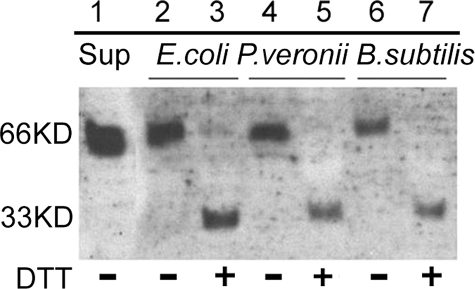
FBN9 Can Interact with Bacterial Surfaces as Homodimers—Sequence analyses and the predicted secondary structure of the A. gambiae FREPs suggested that they could form multimers through disulfide linkage, in a manner similar to that of horseshoe crab TL-5 (13). To test this prediction, we used an in vitro bacterial binding assay, involving incubation of bacteria with Sua5B cell line supernatant containing secreted FREPs. FBN9 could be eluted from the surface of E. coli, P. veronii, and B. subtilis at high salt concentrations after incubation and several subsequent washes (Fig. 6). A single 66-kDa protein band was observed in the binding assays involving Gram-negative E. coli or P. veronii and Gram-positive B. subtilis (Fig. 6). This band is likely to represent a dimer form of the FBN9 protein. In the presence of a strong reducing agent, DTT, the 66-kDa protein bands from the binding assays were converted into 33-kDa bands, confirming this interpretation. Our assays also suggested that FBN9 has a different level of affinity for the Gram-negative and Gram-positive bacteria. It showed a higher affinity for E. coli than for P. veronii and a significantly lower affinity for the Gram-positive bacterium B. subtilis. Although the binding assay here could be considered as preliminary, our data agree with a recent study of the FREPs from snail suggesting that FREPs are able to bind to encountered different pathogens with different specificities (36).
FIGURE 6.
FBN9 forms dimers when binding to either Gram-negative or Gram-positive bacteria. Western analysis by using anti-FBN9 antibody shows the results from in vitro bacterial binding assays. In the absence of additional DTT (100 mm final concentration) in the loading buffer, a 66-kDa protein complex recovered from the surface of E. coli, P. veronii, and B. subtilis (lanes 2, 4, and 6, respectively). The Sua5B cell supernatant used in the binding assay showed the same size of protein (66 kDa), as shown in lane 1. Addition of 100 mm of DTT to the loading buffer resulted in a 33-kDa band instead of the 66-kDa band (lanes 3, 5, 7, for E. coli, P. veronii, and B. subtilis, respectively). + or – indicates the supplement or the absence of DTT, respectively.
The formation of FREP dimers that interact with bacteria could explain the synergistic action of the different FREP members, as discussed above. Synergism in the interactions and activities of various pattern recognition receptors involved in invertebrate immunity has been observed in previous studies (58–63). For example, in D. melanogaster, the recognition of Gram-negative bacteria depends on a synergistic interaction between PGRP-LC and PGRP-LE, through the homo- and heterodimerization of these molecules (64). In the horseshoe crab, the five FREPs in the lectin-agglutinin system cooperate extensively with three C-type lectins to synergistically detect and inactivate pathogens (61, 65). The homodimerization of FBN9 suggests that multimerization, or at least dimerization, of FREPs may provide a basis for creating broader recognition specificities. However, this hypothesis still needs to be experimentally addressed.
CONCLUSIONS
Our phylogenetic and cytogenetic analyses of the FREP family members suggest that the expansion of this gene family is mainly accounted for by a major expansion of FBG domains, through both tandem duplications and shuffling mechanisms. We observed a strong correlation between the phylogeny, chromosomal locations, expression, and immune function of the FREP genes. Some FREP clusters appeared to play a prominent part in the antimicrobial defense, while others had no detectable role in immunity. The antibacterial and anti-Plasmodium FREPs appeared to have both synergistic and complementary functions in the defense against pathogens. FBN9 is a key player in the mosquito innate immune system, with defensive activity against both P. berghei and P. falciparum. The association between FBN9 and the parasite in the midgut epithelium suggest that its anti-Plasmodium activity is mediated through contact, either directly or in association with other immune factors. FBN9 forms homodimers when binding to the bacterial surface suggests that the dimerization could be used as a mechanism to increase the pattern recognition repertoire.
Supplementary Material
Acknowledgments
We thank the parasitology core facility and the insectary personnel at the Johns Hopkins Malaria Research Institute for assistance with P. falciparum culture and mosquito rearing. We also thank Dr. Deborah McClellan at the Editing Referral Service, William H. Welch Medical Library, Johns Hopkins University School of Medicine.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01AI061576-01A1 from NIAID. This work was also supported by the Ellison Medical Foundation, the World Health Organization/TDR, a Johns Hopkins School of Public Health Faculty Innovation Grant, and the Johns Hopkins Malaria Research Institute.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
Footnotes
The abbreviations used are: FREP, fibrinogen-related protein; FBN, fibrinogen immunolectin, FBG, fibrinogen-like domain; GlcNAc, N-acetylglucosamine; qRT-PCR, real-time quantitative PCR; GFP, green fluorescent protein; dsRNA; double-stranded RNA; CFU, colony-forming unit, TL5A, tachylectins 5A; Tep1, thioester-containing protein 1; DTT, dithiothreitol; PBS, phosphate-buffered saline.
References
- 1.Dimopoulos, G. (2003) Cell Microbiol. 5 3–14 [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann, J. A., and Reichhart, J. M. (2002) Nat. Immunol. 3 121–126 [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann, J. A., Reichhart, J. M., and Hetru, C. (1996) Curr. Opin. Immunol. 8 8–13 [DOI] [PubMed] [Google Scholar]
- 4.Janeway, C. A., Jr., and Medzhitov, R. (2002) Annu. Rev. Immunol. 20 197–216 [DOI] [PubMed] [Google Scholar]
- 5.Lemaitre, B., Reichhart, J. M., and Hoffmann, J. A. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 14614–14619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurtz, J., and Armitage, S. A. (2006) Trends Immunol. 27 493–496 [DOI] [PubMed] [Google Scholar]
- 7.Schmid-Hempel, P. (2005) Annu. Rev. Entomol. 50 529–551 [DOI] [PubMed] [Google Scholar]
- 8.Little, T. J., and Cobbe, N. (2005) Insect. Mol. Biol. 14 599–605 [DOI] [PubMed] [Google Scholar]
- 9.Pham, L. N., Dionne, M. S., Shirasu-Hiza, M., and Schneider, D. S. (2007) PLoS Pathog. 3 e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, Y., Taylor, H. E., and Dimopoulos, G. (2006) PLoS Biol. 4 e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson, F. L., Puttmann-Holgado, R., Thomas, F., Lamar, D. L., Hughes, M., Kondo, M., Rebel, V. I., and Schmucker, D. (2005) Science 309 1874–1878 [DOI] [PubMed] [Google Scholar]
- 12.Meijers, R., Puettmann-Holgado, R., Skiniotis, G., Liu, J. H., Walz, T., Wang, J. H., and Schmucker, D. (2007) Nature 449 487–491 [DOI] [PubMed] [Google Scholar]
- 13.Gokudan, S., Muta, T., Tsuda, R., Koori, K., Kawahara, T., Seki, N., Mizunoe, Y., Wai, S. N., Iwanaga, S., and Kawabata, S. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 10086–10091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, X., Zhao, Q., and Christensen, B. M. (2005) BMC Genomics 6 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adema, C. M., Hertel, L. A., Miller, R. D., and Loker, E. S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 8691–8696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang, S. M., and Loker, E. S. (2003) Dev. Comp. Immunol. 27 175–187 [DOI] [PubMed] [Google Scholar]
- 17.Gorkun, O. V., Veklich, Y. I., Weisel, J. W., and Lord, S. T. (1997) Blood 89 4407–4414 [PubMed] [Google Scholar]
- 18.Matsushita, M., and Fujita, T. (2002) Immunobiology 205 490–497 [DOI] [PubMed] [Google Scholar]
- 19.Erickson, H. P. (1993) Curr. Opin. Cell Biol. 5 869–876 [DOI] [PubMed] [Google Scholar]
- 20.Endo, Y., Matsushita, M., and Fujita, T. (2007) Immunobiology 212 371–379 [DOI] [PubMed] [Google Scholar]
- 21.Lu, J., and Le, Y. (1998) Immunobiology 199 190–199 [DOI] [PubMed] [Google Scholar]
- 22.Lu, J., Teh, C., Kishore, U., and Reid, K. B. (2002) Biochim. Biophys. Acta 1572 387–400 [DOI] [PubMed] [Google Scholar]
- 23.Teh, C., Le, Y., Lee, S. H., and Lu, J. (2000) Immunology 101 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi, R., Mizutani, A., and Hidaka, H. (1994) Biochem. Biophys. Res. Commun. 198 1262–1266 [DOI] [PubMed] [Google Scholar]
- 25.Fujita, T. (2002) Nat. Rev. Immunol. 2 346–353 [DOI] [PubMed] [Google Scholar]
- 26.Kenjo, A., Takahashi, M., Matsushita, M., Endo, Y., Nakata, M., Mizuochi, T., and Fujita, T. (2001) J. Biol. Chem. 276 19959–19965 [DOI] [PubMed] [Google Scholar]
- 27.Dimopoulos, G., Casavant, T. L., Chang, S., Scheetz, T., Roberts, C., Donohue, M., Schultz, J., Benes, V., Bork, P., Ansorge, W., Soares, M. B., and Kafatos, F. C. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6619–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perovic-Ottstadt, S., Adell, T., Proksch, P., Wiens, M., Korzhev, M., Gamulin, V., Muller, I. M., and Muller, W. E. (2004) Eur. J. Biochem. 271 1924–1937 [DOI] [PubMed] [Google Scholar]
- 29.Wang, X., Rocheleau, T. A., Fuchs, J. F., Hillyer, J. F., Chen, C. C., and Christensen, B. M. (2004) Insect. Mol. Biol. 13 273–282 [DOI] [PubMed] [Google Scholar]
- 30.Dong, Y., Aguilar, R., Xi, Z., Warr, E., Mongin, E., and Dimopoulos, G. (2006) PLoS Pathog. 2 e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, L. H., Roberts, T., Shahabuddin, M., and McCutchan, T. F. (1993) Mol. Biochem. Parasitol. 59 1–14 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, S. M., Adema, C. M., Kepler, T. B., and Loker, E. S. (2004) Science 305 251–254 [DOI] [PubMed] [Google Scholar]
- 33.Zhang, S. M., and Loker, E. S. (2004) Gene 341 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang, Y., Loker, E. S., and Zhang, S. M. (2006) Dev. Comp. Immunol. 30 855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, S. M., Nian, H., Zeng, Y., and Dejong, R. J. (2008) Dev. Comp. Immunol. 32 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, S. M., Zeng, Y., and Loker, E. S. (2008) Innate. Immun. 14 175–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterhouse, R. M., Kriventseva, E. V., Meister, S., Xi, Z., Alvarez, K. S., Bartholomay, L. C., Barillas-Mury, C., Bian, G., Blandin, S., Christensen, B. M., Dong, Y., Jiang, H., Kanost, M. R., Koutsos, A. C., Levashina, E. A., Li, J., Ligoxygakis, P., Maccallum, R. M., Mayhew, G. F., Mendes, A., Michel, K., Osta, M. A., Paskewitz, S., Shin, S. W., Vlachou, D., Wang, L., Wei, W., Zheng, L., Zou, Z., Severson, D. W., Raikhel, A. S., Kafatos, F. C., Dimopoulos, G., Zdobnov, E. M., and Christophides, G. K. (2007) Science 316 1738–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benedict, M. Q. (1997) in The Molecular Biology of Disease Vectors: A Methods Manual (Crampton, J. M., Beard, C. B., and Louis, C., eds) pp. 3–12, Champman and Hall, London
- 39.Stalker, J., Gibbins, B., Meidl, P., Smith, J., Spooner, W., Hotz, H. R., and Cox, A. V. (2004) Genome Res. 14 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zdobnov, E. M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R. R., Christophides, G. K., Thomasova, D., Holt, R. A., Subramanian, G. M., Mueller, H. M., Dimopoulos, G., Law, J. H., Wells, M. A., Birney, E., Charlab, R., Halpern, A. L., Kokoza, E., Kraft, C. L., Lai, Z., Lewis, S., Louis, C., Barillas-Mury, C., Nusskern, D., Rubin, G. M., Salzberg, S. L., Sutton, G. G., Topalis, P., Wides, R., Wincker, P., Yandell, M., Collins, F. H., Ribeiro, J., Gelbart, W. M., Kafatos, F. C., and Bork, P. (2002) Science 298 149–159 [DOI] [PubMed] [Google Scholar]
- 41.Guex, N., and Peitsch, M. C. (1997) Electrophoresis 18 2714–2723 [DOI] [PubMed] [Google Scholar]
- 42.Vlachou, D., Schlegelmilch, T., Christophides, G. K., and Kafatos, F. C. (2005) Curr. Biol. 15 1185–1195 [DOI] [PubMed] [Google Scholar]
- 43.Carter, R., Ranford-Cartwright, L., and Alano, P. (1993) Methods Mol. Biol. 21 67–88 [DOI] [PubMed] [Google Scholar]
- 44.Vlachou, D., Zimmermann, T., Cantera, R., Janse, C. J., Waters, A. P., and Kafatos, F. C. (2004) Cell Microbiol. 6 671–685 [DOI] [PubMed] [Google Scholar]
- 45.Blandin, S., Moita, L. F., Kocher, T., Wilm, M., Kafatos, F. C., and Levashina, E. A. (2002) EMBO Rep. 3 852–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lane, D. J. (1991) in Nucleic Acid Techniques in Bacterial Systematics (Stackebrandt, E., and Goodfellow, M., eds) pp. 115–175, John Wiley and Sons, New York
- 47.Blandin, S., and Levashina, E. A. (2004) Curr. Opin. Immunol. 16 16–20 [DOI] [PubMed] [Google Scholar]
- 48.Franke-Fayard, B., Trueman, H., Ramesar, J., Mendoza, J., van der Keur, M., van der Linden, R., Sinden, R. E., Waters, A. P., and Janse, C. J. (2004) Mol. Biochem. Parasitol. 137 23–33 [DOI] [PubMed] [Google Scholar]
- 49.Lee, W. J., Lee, J. D., Kravchenko, V. V., Ulevitch, R. J., and Brey, P. T. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 7888–7893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Middha, S., and Wang, X. (2008) BMC Genomics 9 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christophides, G. K., Zdobnov, E., Barillas-Mury, C., Birney, E., Blandin, S., Blass, C., Brey, P. T., Collins, F. H., Danielli, A., Dimopoulos, G., Hetru, C., Hoa, N. T., Hoffmann, J. A., Kanzok, S. M., Letunic, I., Levashina, E. A., Loukeris, T. G., Lycett, G., Meister, S., Michel, K., Moita, L. F., Muller, H. M., Osta, M. A., Paskewitz, S. M., Reichhart, J. M., Rzhetsky, A., Troxler, L., Vernick, K. D., Vlachou, D., Volz, J., von Mering, C., Xu, J., Zheng, L., Bork, P., and Kafatos, F. C. (2002) Science 298 159–165 [DOI] [PubMed] [Google Scholar]
- 52.Sharakhov, I. V., White, B. J., Sharakhova, M. V., Kayondo, J., Lobo, N. F., Santolamazza, F., Della Torre, A., Simard, F., Collins, F. H., and Besansky, N. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 6258–6262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vizioli, J., Bulet, P., Hoffmann, J. A., Kafatos, F. C., Muller, H. M., and Dimopoulos, G. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12630–12635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blandin, S., Shiao, S. H., Moita, L. F., Janse, C. J., Waters, A. P., Kafatos, F. C., and Levashina, E. A. (2004) Cell 116 661–670 [DOI] [PubMed] [Google Scholar]
- 55.Dimopoulos, G., Richman, A., Muller, H. M., and Kafatos, F. C. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 11508–11513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dimopoulos, G., Seeley, D., Wolf, A., and Kafatos, F. C. (1998) EMBO J. 17 6115–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tahar, R., Boudin, C., Thiery, I., and Bourgouin, C. (2002) EMBO J. 21 6673–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang, C. I., Chelliah, Y., Borek, D., Mengin-Lecreulx, D., and Deisenhofer, J. (2006) Science 311 1761–1764 [DOI] [PubMed] [Google Scholar]
- 59.Chang, C. I., Ihara, K., Chelliah, Y., Mengin-Lecreulx, D., Wakatsuki, S., and Deisenhofer, J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10279–10284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaneko, T., Yano, T., Aggarwal, K., Lim, J. H., Ueda, K., Oshima, Y., Peach, C., Erturk-Hasdemir, D., Goldman, W. E., Oh, B. H., Kurata, S., and Silverman, N. (2006) Nat. Immunol. 7 715–723 [DOI] [PubMed] [Google Scholar]
- 61.Kuo, T. H., Chuang, S. C., Chang, S. Y., and Liang, P. H. (2006) Biochem. J. 393 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiravanichpaisal, P., Lee, B. L., and Soderhall, K. (2006) Immunobiology 211 213–236 [DOI] [PubMed] [Google Scholar]
- 63.Endo, Y., Takahashi, M., and Fujita, T. (2006) Immunobiology 211 283–293 [DOI] [PubMed] [Google Scholar]
- 64.Wang, L., and Ligoxygakis, P. (2006) Immunobiology 211 251–261 [DOI] [PubMed] [Google Scholar]
- 65.Iwanaga, S., and Lee, B. L. (2005) J. Biochem. Mol. Biol. 38 128–150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.