Abstract
53BP1 (p53-binding protein 1) is a conserved nuclear protein that is phosphorylated in response to DNA damage and rapidly recruited to the site of DNA double strand breaks, demonstrating its role in the early events to DNA damage and repair of damaged DNA. In this study, we used the yeast two-hybrid system to identify proteins that interact with 53BP1. Identification and characterization of 53BP1 protein interactions may help to further elucidate the function and regulation of 53BP1. We identified protein phosphatase 5 (PP5), a serine/threonine phosphatase that has been implicated in multiple cellular function, as a 53BP1-binding protein. This interaction further confirmed that 53BP1 interacts with PP5 in PP5-overexpressing U2OS cells, after radiomimetic agent neocarzinostatin (NCS) treatment. 53BP1 dephosphorylation at Ser-25 and Ser-1778 was accelerated in PP5-overexpressing U2OS cells following NCS treatment, and its dephosphorylation was correlated with reduced phospho-53BP1 foci formation. In contrast, the overexpression of PP5 had no effect on NCS-activated BRCA1-Ser-1524 phosphorylation. Additionally, PP5 down-regulation inhibited the dephosphorylation of 53BP1 on Ser-1778 and the disappearance of phospho-53BP1 foci following NCS treatment. Moreover, non-homologous end-joining activity was reduced in PP5-overexpressing U2OS cells. These findings indicate that PP5 plays an important role in the regulation of 53BP1 phosphorylation and activity in vivo.
Protein phosphatase 5 (PP5)3 was identified later than other members of the PPP family of serine/threonine-specific phosphatases, including PP1, PP2A, and PP2B (1). Sequencing of PP5 and its yeast homolog PPT revealed the presence of tetratricopeptide repeat domains within their structures. Although many proteins use tetratricopeptide repeat domains as protein-protein interaction motifs, PP5 and PPT are the only phosphatases known to contain these structures. Through its tetratricopeptide repeat domain, PP5 interacts with a number of proteins and has been reported to be involved in regulating various biological processes such as glucocorticoid receptor activity (2), apoptosis (3), and cell growth (4). Additionally, PP5 appears to play a role in cell cycle progression in several ways. Cells treated with PP5 antisense RNA exhibit p53 hyperphosphorylation and a subsequent G1 growth arrest (5). PP5 also binds CDC16 and CDC27, which are members of the anaphase-promoting complex, a complex required for anaphase initiation and the exit from mitosis (5). Recently, it was shown that PP5 plays an important role in DNA damage repair and cell cycle arrest by attenuating the activities of two closely related checkpoint kinases, ataxia telangiectasia-mutated kinase (ATM) and ATM- and Rad3-related kinase (ATR). Around the same time, another report showed that PP5 interacts with and dephosphorylates DNA-dependent protein kinase catalytic subunit on at least two functional sites (6). More recent work utilizing cells from PP5-deficient mice has confirmed that PP5 participates in the ATM-mediated G2/M DNA damage checkpoint pathway (7).
The integrity of the information encoded in DNA is essential for cell survival. Endogenous and exogenous DNA-damaging agents are constantly challenging the stability of DNA. DNA double strand breaks (DSBs) are particularly dangerous for cells because they may lead to genomic instability, cancer development, or cell death (8–10). Generally, DSBs are repaired by two major pathways, homologous recombination (HR) and non-homologous end joining (NHEJ). HR utilizes DNA molecules with a significant length of sequence homology to prime DNA synthesis, allowing for accurate repair. In contrast, NHEJ rejoins DNA ends with little or no sequence homology, potentially leading to inaccurate joining. In higher animal cells, NHEJ is believed to play a predominant role in DSB repair. In addition to the repair of spontaneous DSBs, NHEJ is also used to repair programmed DSBs that arise during the rearrangement of immunoglobulin loci via V(D)J and class-switch recombination. The failure to repair these programmed DSBs results in a lack of immunoglobulin production, whereas the inappropriate repair of these DSBs has been linked to the establishment of lymphomas (11).
53BP1 was originally identified in a yeast two-hybrid screen for proteins interacting with the tumor suppressor p53 (12). 53BP1 contains two tightly packed tudor domains, which bind the methylated Lys-79 of histone H3 (13) or Lys-20 of histone H4 (14), and tandem repeat of BRCA C terminus (BRCT) domains. BRCT domains are thought to be protein-protein interaction domains and are found in many DNA damage response (DDR) proteins (15, 16). Upon exposure to ionizing radiation (IR), 53BP1 is rapidly redistributed to sites of DSBs where it is hyperphosphorylated in an ATM-dependent manner (17, 18). The N terminus of 53BP1 possesses several (S/T)Q motifs, which are preferentially phosphorylated by members of the PIKK family (19). Several studies have shown that 53BP1 is required for the accumulation of p53, the G2/M checkpoint arrest, the intra-S-phase checkpoint in response to IR, and the IR-stimulated phosphorylation of at least a subset of ATM substrates (20–22). 53BP1-null mice are viable but are highly tumor-prone, have defects in IgG class switching and V(D)J recombination, and are profoundly hypersensitive to IR, probably because of a defect in NHEJ (23–25). Despite these observations, the exact molecular functions of 53BP1 that mediate its biological roles are not understood. It is generally assumed that whatever the molecular role of 53BP1 is, it is specific to DSBs.
Although the phosphorylation of 53BP1 has been studied in detail, little is known about its corresponding dephosphorylation. One reason for this is that little information is available regarding the putative protein phosphatases responsible for the dephosphorylation of 53BP1. In this study, we found that PP5 participates in the dephosphorylation of 53BP1 in response to the radiomimetic drug neocarzinostatin (NCS). To investigate the role of PP5 in DDR, we established PP5 over- and underexpressing cell lines. Using these two cell types, we found that the phosphorylation of 53BP1 at Ser-25 and Ser-1778 were removed by PP5 after DNA damage, and that the 53BP1 foci also rapidly disappeared in PP5-overexpressing cells corresponding to their phosphorylation levels. Furthermore, our data show that NHEJ activity was significantly reduced in both the PP5 over- and underexpressing cells.
EXPERIMENTAL PROCEDURES
Cells Lines and Drug Treatment—The following cell lines were used in this study: U2OS, U2OS-PO (transfected with full-length human PP5), and U2OS-PS (transfected with an siPP5 construct). U2OS cells were cultured in McCoy's 5A medium supplemented with 10% fetal bovine serum (FBS), 10 μg/ml streptomycin, and 10 units/ml penicillin at 37 °C in the presence of 5% CO2. The U2OS-PO and -PS cells were grown in the same media as the U2OS cells except for the addition of 800 μg/ml neomycin. To induce DNA damage, exponentially growing cells were treated with 200 ng/ml NCS (Sigma) and harvested at different times after treatment.
Stable Cell Lines—Full-length human PP5 cDNA was directly cloned into pcDNA 3.1 TOPO using the PP5-specific primers 5′-ATGGCGATGGCGGAGGGCGA-3′ and 5′-GAATTCCATTCCTAGCTGCAGCAG-3′. A synthetic siRNA duplex for PP5 (5′-AACAUAUUCGAGCUCAACGGU-3′) was purchased from Bioneer and cloned into pSilencer™ neo (Ambion) (26). The resulting plasmids were transfected into U2OS cells using Lipofectamine 2000 (Invitrogen). The cells were then cultured in McCoy's 5A medium supplemented with 10% FBS. After 24 h, G418 (Sigma) was added to the culture medium at a concentration of 800 μg/ml. After 4 weeks of culture, during which the G418-containing medium was changed every 3 days, the G418-resistant colonies were isolated and confirmed by Western blotting.
Antibodies and Immunoblotting—Harvested NCS-treated cells were lysed with M-PER (Pierce) containing proteinase inhibitors (Complete Mini, Roche Applied Science) and separated by SDS-PAGE. The separated proteins were then transferred to a polyvinylidene difluoride membrane (Millipore), and the membrane was blocked for 1 h with 5% nonfat dry milk in TBS-T (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.1% Tween 20). The membrane was then incubated with primary antibodies against phospho-53BP1 (S1778) (Cell Signaling Technology), phospho-53BP1 (Ser-25) (Novus), 53BP1 (BD Biosciences and Santa Cruz Biotechnology), ATM (Oncogene), ATR (Cell Signaling Technology), phospho-BRCA1 (Thr-68) (Cell Signaling Technology), BRCA1 (Santa Cruz Biotechnology), PP5 (BD Biosciences), and α-tubulin (NeoMarkers). After primary antibody incubation, the membranes were incubated with the corresponding secondary antibodies and visualized by chemiluminescence (Intron, Seoul, Korea).
Immunoprecipitation—At 3 h post-NCS treatment, the harvested cells were lysed in RIPA buffer (25 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with proteinase inhibitors (Roche Applied Science), and the extracts were incubated with anti-53BP1 antibodies (Santa Cruz Biotechnology) overnight at 4 °C. Protein A-Sepharose beads (Santa Cruz Biotechnology) were then added, and the mixture was incubated for 2 h at room temperature. The beads were then gently washed three times with PBS, and the precipitated complexes were resuspended in 2× loading buffer. After boiling, the samples were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and immunoblotted.
Immunofluorescence Analysis—Standard immunofluorescence procedures were followed according to Nakamura et al. (27). Cells grown on cover slides were briefly rinsed with PBS and then fixed with freshly prepared 3.7% paraformaldehyde in PBS for 15 min. The slides were then either directly processed or exposed to 70% ethanol prior to storage at 4 °C. After being washed with PBS, the cover slides were blocked in 5% bovine serum albumin in PBS for 1 h at room temperature and then incubated overnight at 4 °C with the following primary antibodies: mouse monoclonal γ-H2AX (Ser-139) (1:200; Upstate Biotechnology) or rabbit polyclonal 53BP1 (Ser-1778) (1:100; Cell Signaling Technology). Secondary antibodies labeled with Alexa 488 or Alexa 594 (Molecular Probes) were added at 1:200 and incubated at room temperature for 1 h. The slides were mounted with a mounting solution containing 4′,6-diamidino-2-phenylindole (Vector Laboratories). Images were acquired using a Nikon ELIPSE 80i microscope.
NHEJ Assay—To document the role of 53BP1 phosphorylation in NHEJ, we used the plasmid pEGFP-Peml-Ad2 (28). Briefly, the NHEJ reporter plasmid was digested with HindIII overnight and purified using a Qiagen gel extraction kit. All plasmids were transfected into U2OS, U2OS-PO, and U2OS-PS cells using Lipofectamine 2000 according to the manufacturer's instructions. In a typical reaction, 5 × 105 cells were transfected with 0.5 μg of the predigested NHEJ reporter substrate along with 0.5 μg of pDsRed2-N1 (Clontech) as a transfection control. Green fluorescent protein and DsRed expression was monitored by fluorescence microscopy (Nikon Eclipse TE2000-U). After transfection, the cells were incubated for 48 h, harvested, resuspended in 0.5 ml of PBS, pH 7.4 (Invitrogen), and then analyzed by FACS.
RESULTS AND DISCUSSION
PP5 Binds with 53BP1 after DNA Damage—To search for factors that interact with 53BP1, yeast two-hybrid screening of a human prostate cDNA library was performed using the BRCT domain of human 53BP1 as bait. One of the positive clones isolated from 2 × 106 transformants turned out to be PP5. We confirmed the human PP5 could also interact with human 53BP1 by performing the yeast two-hybrid assay with a human PP5 cDNA.
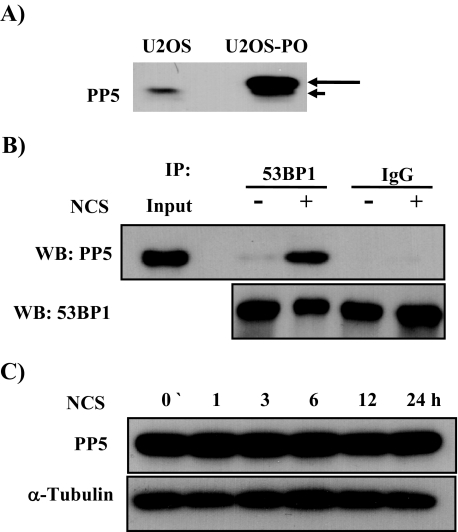
To investigate the functional relationship between 53BP1 and PP5, we established U2OS cell lines stably expressing the PP5 (U2OS-PO) (Fig. 1A). PP5 has been reported to dephosphorylate DNA-PK in response to ionizing radiation (6). Based on this, identification of PP5 as a 53BP1-associating protein made us postulate that PP5 might function as a phosphatase protein for 53BP1 dephosphorylation following ionizing radiation. Thus, we first examined whether the association between 53BP1 and PP5 was affected by radiomimetic agent NCS treatment. Coimmunoprecipitation experiments revealed that the specific interaction between overexpressed His-PP5 and endogenous 53BP1 increased significantly 3 h after treatment with 200 ng/ml NCS (Fig. 1B). Because 53BP1 was bound to PP5 after DNA damage, it could be critical for 53BP1 dephosphorylation as cells repair their damaged DNA. We could not detect any changes in PP5 expression after NCS treatment in U2OS-PO cells (Fig. 1C), suggesting that the increased association between 53BP1 and PP5 was not attributed to NCS-mediated induction of PP5 expression.
FIGURE 1.
PP5 interacts with 53BP1 after NCS-induced DNA damage. A, to make PP5-overexpressing cells, U2OS cells were transfected with pcDNA 3.1-PP5. The transfected cells were then selected with G418 (800 g/ml) for 4 weeks and screened by Western blotting using a PP5-specific antibody. The long arrow indicates V5/His-tagged PP5, and the short arrow indicates endogenous PP5. B, U2OS and U2OS-PO cells were treated with NCS (200 ng/ml) for 3 h and lysed. Proteins were immunoprecipitated (IP) from the lysates with anti-53BP1 antibody, and the immunoprecipitates were subjected to Western blot (WB) analysis with an antibody specific for PP5 or 53BP1. 1st and 2nd lanes contain 10% input. Normal mouse IgG was used as an immunoprecipitation control. C, U2OS cells were treated with 200 ng/ml NCS. At 0, 1, 3, 6, 12, and 24 h after NCS treatment, cell were harvested for Western blotting and probed with antibodies against PP5 and α-tubulin.
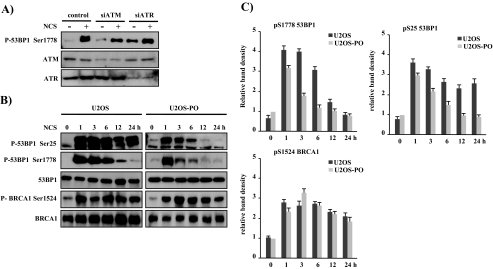
Overexpression of PP5 Specifically Induced the Earlier Dephosphorylation of 53BP1—The appearance of the 53BP1-PP5 complex after NCS treatment raised the possibility that phosphatase activity of PP5 may regulate 53BP1 phosphorylation after NCS-induced DNA damage. To explore this possibility (hypothesis), we analyzed 53BP1 phosphorylation in U2OS and U2OS-PO cells. There are only two commercially available antibodies to detect the phosphorylated form of 53BP1 at Ser-25/Ser-29 and Ser-1778. The phosphorylation of 53BP1 at Ser-25/Ser-29 has been well characterized by numerous groups (29, 30); however, the role of phosphorylation at Ser-1778 on the BRCT domain of 53BP1 is unknown. Thus, we first assessed the effect of NCS on 53BP1 phosphorylation at Ser-1778. We observed that exposing U2OS cells to 200 ng/ml NCS for 3 h resulted in a significant increase in the amount of phosphorylated 53BP1 at Ser-1778 (Fig. 2A). Because ATM and ATR proteins participate in the phosphorylation of 53BP1, we examined 53BP1 phosphorylation at Ser-1778 in U2OS cells deficient in ATM or ATR by transient transfection of their specific siRNAs. Transfection of either ATM siRNA and ATR siRNA reduced the expression level of the targeted ATM and ATR by ∼57 and ∼63%, respectively, compared with control siRNA-transfected cells. Immunoblot analysis revealed that the phosphorylation of 53BP1 at Ser-1778 after NCS treatment was mediated at least in part by ATM (Fig. 2A). Further kinetic analysis revealed that the increase of Ser-25 and Ser-1778 phosphorylation was clearly detected as early as 1 h after NCS treatment (Fig. 2B, left panel). However, the phosphorylation patterns between Ser-25 and Ser-1778 of 53BP1 are significantly different. NCS-induced Ser-25 phosphorylation remained constant for 24 h, whereas NCS-induced Ser-1778 phosphorylation reached peak levels at 1 h and decreased progressively in a time course manner, becoming significantly diminished at 24 h. This result suggests that there may be different roles for the different phosphorylation sites of 53BP1. In response to ionizing radiation, ATM phosphorylates 53BP1 on Ser-25 and Ser-29 (29, 30). However, mutation of these sites does not affect the function of 53BP1 in the DNA damage response (29), and the mutant 53BP1 protein, in which Ser-25 and Ser-29 are mutated to alanine residues, is still hyperphosphorylated after DNA damage (19). Additionally, 53BP1 is phosphorylated on multiple residues in response to different typed of DNA damage (31, 32). Thus, although the phosphorylation of 53BP1 at Ser-25 and Ser-29 in response to DNA damage has been linked to its activation, several novel phosphorylation sites of 53BP1 such as Ser-1788 may be important for 53BP1 function.
FIGURE 2.
PP5 participates in the dephosphorylation of 53BP1 at Ser-25 and Ser-1778 after the DNA repair process. A, to identify the kinase responsible for the phosphorylation of 53BP1 at Ser-1778, U2OS cells were transfected with siRNAs against control, ATM, and ATR using Lipofectamine reagent. After 48 h, the cells were treated with 200 ng/ml NCS for 1 h, and the phosphorylation status of 53BP1 at Ser-1778 in cell extracts was determined with anti-phospho-53BP1-Ser-1778. In the bottom two panels, depletion of ATM or ATR by siRNA was demonstrated by immunoblotting of cell lysates with antibodies against ATM or ATR. B, dephosphorylation of 53BP1 was mediated specifically by PP5. U2OS and U2OS-PO cells were treated with NCS (200 ng/ml) and then harvested 1, 3, 6, 12, or 24 h later. The phosphorylation patterns of 53BP1 and BRCA1 were analyzed by Western blotting using the indicated antibodies. C, graphs show the quantification of the levels of 53BP1 phosphorylated on Ser-25 and Ser-1778 and BRCA1 phosphorylated on Ser-1524 shown in B at the indicated time point. The data were normalized to the untreated U2OS-PO cells (as the value of 1) and are the mean ± S.D. of three independent experiments.
To investigate whether 53BP1 is a target for the PP5 phosphatase in vivo, the U2OS and U2OS-PO cells were treated with 200 ng/ml NCS for different times and then analyzed for 53BP1 phosphorylation at Ser-25 and Ser-1788. As shown in Fig. 2B, overexpression of PP5 was reduced significantly in the phosphorylation of 53BP1 at Ser-25 and Ser-1788 after NCS treatment, as compared with those of U2OS cells. The phosphorylation of Ser-25 and Ser-1778 of 53BP1 peaked at 1 h and then decreased over time, becoming significantly diminished by 12 h in U2OS-PO cells. To evaluate that the PP5-induced dephosphorylation of 53BP1 at Ser-25 and Ser-1778 was not because of a nonspecific phosphorylation, we analyzed the Ser-1524-phosphorylated BRCA1, which contains the same BRCT domain as 53BP1 and is also a known DDR protein. We did not observe a substantial difference in the phosphorylation of BRCA1 at Ser-1524 in U2OS versus U2OS-PO cells, suggesting that the dephosphorylation of 53BP1 at Ser-25 and Ser-1778 appears to occur via a specific interaction mediated by PP5. Therefore, we conclude that PP5 is the phosphatase responsible for removing the phosphate group from Ser-25 and Ser-1778 of 53BP1 after DNA double strand breaks.
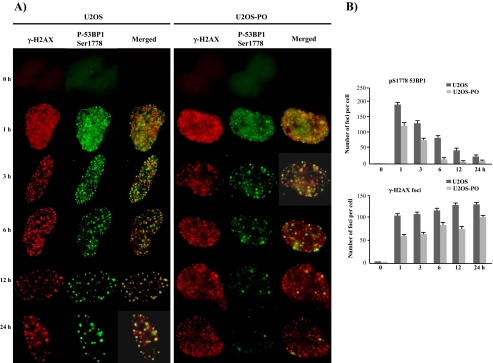
Rapid Dephosphorylation at Ser-1778 Also Influences Phospho-53BP1 Focus Formation—In response to IR, 53BP1 rapidly colocalizes with γ-H2AX. H2AX, a variant of histone H2A, becomes phosphorylated and forms foci at sites of double strand breakage after DNA damage. The number, appearance, and disappearance of the foci were nearly identical between 53BP1 and γ-H2AX (33, 34). Although the phosphorylation activity has been studied in detail, little is known about the corresponding dephosphorylation and detachment from DNA DSBs. Generally, it is thought that either the phosphorylated protein must be degraded or the phosphate group must be removed. Therefore, we analyzed the relationship between focus formation and the dephosphorylation of 53BP1 by immunofluorescence using U2OS and U2OS-PO cells (Fig. 3). The patterns of pS1778–53BP1 and γ-H2AX focus formation were evaluated from 0 to 24 h after NCS-induced DNA damage. As shown in Fig. 3, the DNA focus pattern of pS1778–53BP1 differed between the U2OS and U2OS-PO cells. In the U2OS cells at 1 h, the DNA foci of pS1778–53BP1 were numerous, small, and fused; by 24 h, the foci had grown and decreased in number. This pattern of focus formation corresponded to our Western blotting results for pS1778–53BP1 (Fig. 2). In the U2OS-PO cells, the pattern of the pS1778–53BP1 foci was similar to that in the U2OS cells during the first 3 h, and the pS1778–53BP1 foci were well matched with those of γ-H2AX, even though there were fewer pS1778–53BP1 foci in the U2OS-PO cells than in the U2OS cells (Fig. 3). After 3 h, however, the number of pS1778–53BP1 foci in the U2OS-PO cells was greatly decreased, and the pS1778–53BP1 foci did not match with those of γ-H2AX. This indicates that the detachment of DDR proteins from DNA breakage sites is the result of dephosphorylation. Generally, after DNA injury, members of the phosphatidylinositol 3-kinase kinase family, including ATM, ATR, and DNA-PKcs, phosphorylate many DDR proteins, which are subsequently recruited to participate in the repair process at sites of DNA breakage (35, 36). The phosphorylation of 53BP1, which is mediated by ATM and ATR after DNA damage, has been studied using mass spectrometry (19). The tandem tudor domains of 53BP1 are required for the recruitment of 53BP1 to sites of DNA DSBs. The BRCT domain is not required for 53BP1 phosphorylation (14) or DNA DSB repair (37). Although 53BP1 is regarded as an important protein in the DDR pathway (20, 38), its exact function is not clearly understood.
FIGURE 3.
The maintenance of 53BP1 foci is affected by the phosphorylation status caused by PP5 overexpression. A, U2OS and U2OS-PO cells were grown on cover slides and then treated with NCS (200 ng/ml). At 0, 1, 3, 6, 12, and 24 h after 200 ng/ml NCS treatment, the cells were fixed and immunostained with antibodies to γ-H2AX and 53BP1-Ser-1778. B, statistical evaluation of the experiments shown in A. The graph shows the average number of γ-H2AX and 53BP1-Ser-1778 foci based on ∼150 nuclei per sample. The values represent the mean ± S.D. of three separate experiments.
In U2OS-PO cells after NCS-induced DNA damage, the number and size of the γ-H2AX foci were almost identical to those seen in the U2OS cells (Fig. 3). To confirm the specificity of PP5 for 53BP1, we analyzed the formation of pS1524-BRCA1 and pS1981-ATM foci after DNA damage; as for γ-H2AX, there were no differences in the patterns of the foci between the cells (data not shown). These data confirm that the interaction between PP5 and phosphorylated 53BP1 is specific.
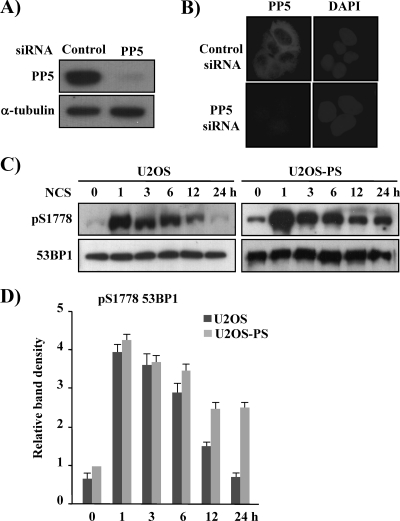
Decreased PP5 Expression Delays the Dephosphorylation of 53BP1—To confirm the involvement of PP5 in 53BP1 dephosphorylation in response to DNA damage, PP5 expression was knocked down by stable transfection with siRNA for 53BP1 as reported previously (26). We cloned an siRNA duplex for PP5 into the pSilencer vector and transfected the siPP5 plasmid into U2OS cells. Over a period of 4 weeks, G418 selection was used to obtain several PP5-suppressed U2OS clones. These clones were then screened by Western blotting, and one was chosen for further analysis. As shown in Fig. 4A, PP5 protein expression was inhibited more than 90% by PP5 siRNA transfection. Immunofluorescence analysis of this clone also demonstrated that PP5 expression was suppressed (Fig. 4B).
FIGURE 4.
The phosphorylation of 53BP1 at Ser-1778 is enhanced in U2OS-PS cells. A, U2OS cells were transfected with a control pSilencer vector (control) or pSilencer vector, including PP5 siRNA. The transfected cells were then selected with G418 (800 g/ml) for 4 weeks and screened by Western blotting using a PP5-specific antibody. B, control and PP5 siRNA-transfected U2OS cells were fixed and immunostained with a polyclonal antibody to PP5. 4′,6-Diamidino-2-phenylindole (DAPI) staining was performed to indicate the position of nuclei. C, U2OS and U2OS-PS cells were treated with NCS (200 ng/ml) and harvested 0, 1, 3, 6, 12, or 24 h later. The phosphorylation status of 53BP1 at Ser-1778 in cell extracts was determined with anti-phospho-53BP1-Ser-1778 and 53BP1. D, graph shows the quantification of the levels of 53BP1 phosphorylated on Ser-1778 shown in C at the indicated time point averaged with an additional, independent experiment. The value given for the amount of protein present in the to the untreated U2OS-PS sample was set as 1 (n = 3).
Using the PP5-suppressed cell line U2OS-PS, we examined the activity of PP5 as a phosphatase at pS1778 of 53BP1. As shown in Fig. 4C, in the U2OS cells, the level of pS1778–53BP1 peaked at 1 h and then decreased slowly until 24 h after NCS treatment, at which point pS1778–53BP1 had completely disappeared. In the U2OS-PS cells, however, the level of pS1778–53BP1 was elevated 1 h after NCS treatment and was slightly lower after 3 h; this level of pS1778–53BP1 was maintained for up to 12 h post-treatment and then decreased again at 24 h. These results strongly suggest that PP5 is involved in the dephosphorylation of pS1778 in 53BP1; however, we cannot assume that PP5 is the only enzyme involved. Recently, Travesa et al. (39) reported on the dephosphorylation of Rad53, which plays a central role in preventing genomic instability and maintaining viability in Saccharomyces cerevisiae. According to their data, Rad53 kinase is dephosphorylated by different enzymes, Ptc2/3 and Pph3, depending on the nature of the DNA damage; under DNA replication stress (i.e. under hydroxyurea), Pph3 dephosphorylated Rad53, whereas Ptc2/3 participated after DNA methylation damage (i.e. under methylmethane sulfonate) or replication stress. Similar to Rad53, 53BP1 may be dephosphorylated by different enzymes depending on the type of DNA damage.
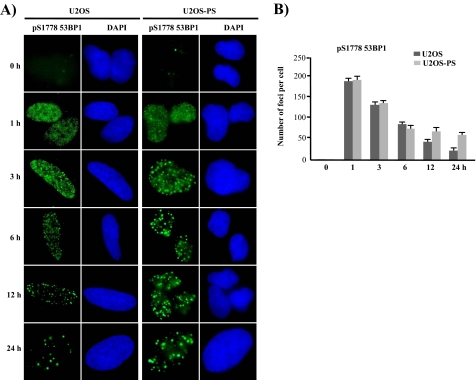
Next, we investigated the influence of PP5 suppression on focus formation by pS1778–53BP1 after DNA damage in U2OS-PS cells. As shown in Fig. 5, there were differences in pS1778–53BP1 focus formation between the U2OS and U2OS-PS cells. At 1 h after DNA damage, the pS1778–53BP1 foci were similar between the U2OS and U2OS-PS cells; however, after 3 h, the pattern between the two was different. In the U2OS cells, the number of pS1778–53BP1 foci decreased whereas the size increased in a time-dependent manner; in contrast, the number and size of the pS1778–53BP1 foci remained constant from 3 to 24 h in the U2OS-PS cells. This means that pS1778–53BP1 foci are influenced by the level of PP5 activity, whereas the focus patterns are almost identical to the patterns produced by Western blotting after DNA damage. Thus, the maintenance of 53BP1 foci is strongly related to the phosphorylation status of the protein, which may also influence the repair process after DNA damage.
FIGURE 5.
53BP1 foci are also influenced by the PP5 expression level after NCS-induced DNA damage. A, U2OS and U2OS-PS cells cultured on cover slides were treated with NCS (200 ng/ml). Each slide was then fixed with 3.7% paraformaldehyde at various time points and stained with an antibody specific for Ser(P)-1778 (pS1778) in 53BP1. The nuclei were visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining. B, statistical evaluation of the experiments shown in A. The graph shows the average number of 53BP1-Ser-1778 foci based on ∼150 nuclei per sample. The values represent the mean ± S.D. of three separate experiments.
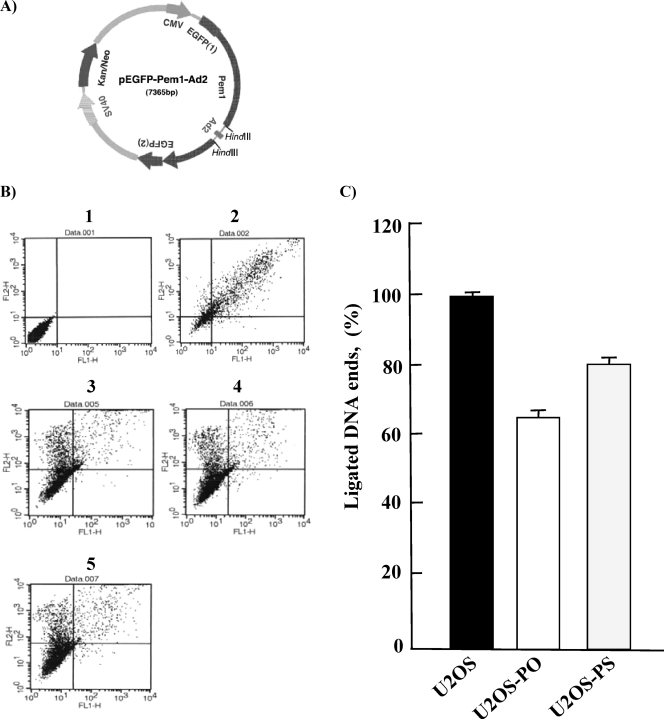
Both the Hyperphosphorylation and Hypophosphorylation of 53BP1 Influence NHEJ—Although its function is not clearly understood, 53BP1 has been reported to participate in NHEJ, but not HR, after DNA damage (23, 40–44). We therefore hypothesized that DNA repair activity may be differentially influenced by the inadequate dephosphorylation of 53BP1 in U2OS-PO and U2OS-PS cells. To test this, we introduced three different plasmids, pEGFP-Pem1-Ad2, pEGFP-Pem1, and pDsRed2-N1, into U2OS, U2OS-PO, and U2OS-PS cells to allow direct in vivo reporting using EGFP. The pEGFP-Pem1-Ad2 plasmid (Fig. 6A) was linearized by HindIII digestion, which removed the Ad2 exon; upon successful circularization of this plasmid in the cells, EGFP expression was detected and quantified by FACS. Supercoiled pEGFP-Pem1 was used to evaluate the EGFP signal without the need for end-joining, whereas pDsRed2-N1 was used as a transfection control. The results of two control transfections are shown in Fig. 6B, and panel 1 shows the autofluorescence of sham-transfected cells, and panel 2 shows the signal generated in cells simultaneously transfected with 0.5 μg of pEGFP-Pem1 and 0.5 μg of pDsRed2-N1. To evaluate the efficiency of NHEJ, U2OS, U2OS-PO, and U2OS-PS, cells were transfected with linearized pEGFP-Pem1-Ad2, and the fluorescent cells were quantified by FACS 24 h later (Fig. 6B, panels 3–5). NHEJ activity was decreased in both the U2OS-PO and U2OS-PS cells compared with that in the U2OS cells (Fig. 6C). In five independent experiments, the NHEJ efficiency was 64.9 ± 1.7% in the U2OS-PO cells and 80.3 ± 0.6% in the U2OS-PS cells. These results suggest that both overexpression of PP5 and expression of PP5 siRNA lead to decreased NHEJ activity. Generally, phosphorylation of 53BP1 has been linked to its function. Thus, impaired phosphorylation or premature dephosphorylation caused by PP5 overexpression might suppress 53BP1 function, leading to decreased NHEJ activity. In addition, dephosphorylation of 53BP1 may allow re-use of 53BP1 in subsequent repair events. Thus, in the absence of efficient dephosphorylation, caused by less PP5 enzyme, NHEJ activity may also be decreased.
FIGURE 6.
NHEJ activity is lower in U2OS-PO and -PS cells than in U2OS cells. A, map of the pEGFP-Pem1-Ad2 vector. Note the Pem1 intron, Ad2 exon, and location of the HindIII excision sites. B, dot plots of untransfected U2OS cells (panel 1), U2OS cells co-transfected with pEGFP-Pem1 and DsRed (panel 2), U2OS cells co-transfected with HindIII-linearized pEGFP-Pem1-Ad2 and DsRed (panel 3), U2OS-PO cells co-transfected with HindIII-linearized pEGFP-Pem1-Ad2 and DsRed (panel 4), and U2OS-PS cells co-transfected with HindIII-linearized pEGFP-Pem1-Ad2 and DsRed (panel 5). The numbers of green (green fluorescent protein) and red (DsRed) cells were determined by FACS analysis, and typical FACS traces are shown. The ratio of green fluorescent protein to DsRed was used as a measure of NHEJ efficiency. All measurements of the rejoining efficiency were carried out 24 h after transfection with the corresponding plasmids. C, percentage of ligated DNA ends was determined by using the data in B. All experiments were repeated at least four times, and the S.D. are shown.
It is generally accepted that the repair of IR-induced DSBs is performed mainly by a core NHEJ complex, which is composed of the DNA ligase IV/Xrcc4, Ku70/Ku80, DNA-PKcs, and Artemis (46). Iwabuchi et al. (44) proposed that 53BP1 may also play a role via its tudor domain, which can bind chromatin and stimulate end-joining by DNA ligase IV/Xrcc4 via a Ku70-dependent pathway. However, several reports have shown that 53BP1 uses a different pathway for NHEJ (23, 36, 47); a recent study revealed that 53BP1 participates in a pathway distinct from the Ku- and Artemis-dependent NHEJ pathways but still requires DNA ligase IV (48). In this study, the efficiency of NHEJ was lower in the PP5-overexpressing cells than in the PP5-suppressed cells. This can be explained by the fact that PP5 is involved in ATM/ATR-dependent repair after DNA damage (7, 26, 49) and that it functions as a phosphatase with DNA-PKcs (6). Thus, many more proteins involved in checkpoint signaling pathways should be affected by PP5 overexpression than by PP5 suppression.
Although phosphorylation in response to DNA damage has been well documented, little is known about the corresponding dephosphorylation. Among the members of the PP family, PP2A and PP4 are known to be involved in the dephosphorylation of γ-H2AX (31, 50). To date, there are a few reports that PP5 participates in DNA damage response; three of these mentioned that PP5 is involved as a regulatory proteins of ATM (7, 26) and ATR (49) but is not a phosphatase. Only one study reported that PP5 dephosphorylated the Thr(P)-2609 and Ser(P)-2056 of DNA-PKcs after DNA damage in HeLa cells (6).
In this study, we show that the PP5 binds 53BP1 after DNA damage and dephosphorylates Ser-25 and Ser-1778 in vivo. A deficiency of PP5 phosphatase results in increased 53BP1 phosphorylation, whereas its overexpression is sufficient to reduce phosphorylation of 53BP1 in response to radiomimetic drug NCS. Thus, it seems likely that 53BP1 is a direct target for PP5 phosphatase in vivo. Although dephosphorylation of 53BP1 is profoundly delayed in PP5 knockdown cells (U2OS-PS), 53BP1 phosphorylation at Ser-1778 was decreased by ∼50% at 24 h after NCS treatment, suggesting that another phosphatase might exist to regulate 53BP1 dephosphorylation. One of the earliest signals in the DNA damage response is phosphorylation of the histone variant H2AX at Ser-139. Two recent studies, one in mammals and the other in S. cerevisiae, identified roles for PP2A family phosphatases in γ-H2AX dephosphorylation (45, 50), indicating that PP2A directly dephosphorylates γ-H2AX formed by exogenous DNA-damaging agents. However, the same groups also showed that another phosphatase, PP4C, efficiently dephosphorylates γ-H2AX (31). A similar situation might have been observed with 53BP1 and PP5. Because different stresses and/or different degrees of DNA damage induce phosphorylation of checkpoint components, distinct phosphatases might differentially regulate phosphorylated protein
The PP5 phosphatase, which associates with 53BP1 following DNA damage, could be one of the factors regulating the magnitude of 53BP1 phosphorylation and its activation. On the other hand, under the condition where PP5 is depleted or overexpressed, the level of 53BP1 phosphorylation and its duration after DNA damage are changed. Thus, future studies will be needed to unravel the mechanisms by which dephosphorylation of 53BP1 contributes to DNA damage response, and to determine whether PP5 may contribute to DNA damage response under certain pathologic conditions through the dephosphorylation of 53BP1. Uncovering how PP5 regulates 53BP1 dephosphorylation after DNA damage will contribute to our understanding of the mechanisms that control the 53BP1 signaling pathway after DNA damage.
This work was supported in part by Grants M1063901 and M20706000032 from the Ministry of Science and Technology, the Korean Government.
Footnotes
The abbreviations used are: PP5, protein phosphatase 5; 53BP1, p53-binding protein; NCS, neocarzinostatin; ATM, ataxia telangiectasia-mutated kinase; DSBs, double strand breaks; DDR, DNA damage response; ATR, ATM- and RAD-3-related; NHEJ, non-homologous end joining; HR, homologous recombination; EGFP, enhanced green fluorescent protein; FACS, fluorescence-activated cell sorter; PBS, phosphate-buffered saline; siRNA, small interfering RNA; IR, ionizing radiation; BRCT, BRCA C terminus; DDR, DNA damage response.
References
- 1.Russell, L. C., Whitt, S. R., Chen, M. S., and Chinkers, M. (1999) J. Biol. Chem. 274 20060–20063 [DOI] [PubMed] [Google Scholar]
- 2.Chen, M. S., Silverstein, A. M., Pratt, W. B., and Chinkers, M. (1996) J. Biol. Chem. 271 32315–32320 [DOI] [PubMed] [Google Scholar]
- 3.Morita, K., Saitoh, M., Tobiume, K., Matsuura, H., Enomoto, S., Nishitoh, H., and Ichijo, H. (2001) EMBO J. 20 6028–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Kriegsheim, A., Pitt, A., Grindlay, G. J., Kolch, W., and Dhillon, A. S. (2006) Nat. Cell Biol. 8 1011–1016 [DOI] [PubMed] [Google Scholar]
- 5.Chinkers, M. (2001) Trends Endocrinol. Metab. 12 28–32 [DOI] [PubMed] [Google Scholar]
- 6.Wechsler, T., Chen, B. P., Harper, R., Morotomi-Yano, K., Huang, B. C., Meek, K., Cleaver, J. E., Chen, D. J., and Wabl, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1247–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yong, W., Bao, S., Chen, H., Li, D., Sanchez, E. R., and Shou, W. (2007) J. Biol. Chem. 282 14690–14694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeiffer, P., Goedecke, W., and Obe, G. (2000) Mutagenesis 15 289–302 [DOI] [PubMed] [Google Scholar]
- 9.Khanna, K. K., and Jackson, S. P. (2001) Nat. Genet. 27 247–254 [DOI] [PubMed] [Google Scholar]
- 10.Bartkova, J., Horejsi, Z., Koed, K., Kramer, A., Tort, F., Zieger, K., Guldberg, P., Sehested, M., Nesland, J. M., Lukas, C., Orntoft, T., Lukas, J., and Bartek, J. (2005) Nature 434 864–870 [DOI] [PubMed] [Google Scholar]
- 11.Lieber, M. R., Yu, K., and Raghavan, S. C. (2006) DNA Repair 5 1234–1245 [DOI] [PubMed] [Google Scholar]
- 12.Iwabuchi, K., Bartel, P. L., Li, B., Marraccino, R., and Fields, S. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6098–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huyen, Y., Zgheib, O., Ditullio, R. A., Jr., Gorgoulis, V. G., Zacharatos, P., Petty, T. J., Sheston, E. A., Mellert, H. S., Stavridi, E. S., and Halazonetis, T. D. (2004) Nature 432 406–411 [DOI] [PubMed] [Google Scholar]
- 14.Botuyan, M. V., Lee, J., Ward, I. M., Kim, J. E., Thompson, J. R., Chen, J., and Mer, G. (2006) Cell 127 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callebaut, I., and Mornon, J. P. (1997) FEBS Lett. 400 25–30 [DOI] [PubMed] [Google Scholar]
- 16.Koonin, E. V., Altschul, S. F., and Bork, P. (1996) Nat. Genet. 13 266–268 [DOI] [PubMed] [Google Scholar]
- 17.Schultz, L. B., Chehab, N. H., Malikzay, A., and Halazonetis, T. D. (2000) J. Cell Biol. 151 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rappold, I., Iwabuchi, K., Date, T., and Chen, J. (2001) J. Cell Biol. 153 613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jowsey, P., Morrice, N. A., Hastie, C. J., McLauchlan, H., Toth, R., and Rouse, J. (2007) DNA Repair 6 1536–1544 [DOI] [PubMed] [Google Scholar]
- 20.Wang, B., Matsuoka, S., Carpenter, P. B., and Elledge, S. J. (2002) Science 298 1435–1438 [DOI] [PubMed] [Google Scholar]
- 21.DiTullio, R. A., Jr., Mochan, T. A., Venere, M., Bartkova, J., Sehested, M., Bartek, J., and Halazonetis, T. D. (2002) Nat. Cell Biol. 4 998–1002 [DOI] [PubMed] [Google Scholar]
- 22.Mochan, T. A., Venere, M., DiTullio, R. A., Jr., and Halazonetis, T. D. (2003) Cancer Res. 63 8586–8591 [PubMed] [Google Scholar]
- 23.Ward, I. M., Reina-San-Martin, B., Olaru, A., Minn, K., Tamada, K., Lau, J. S., Cascalho, M., Chen, L., Nussenzweig, A., Livak, F., Nussenzweig, M. C., and Chen, J. (2004) J. Cell Biol. 165 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward, I. M., Difilippantonio, S., Minn, K., Mueller, M. D., Molina, J. R., Yu, X., Frisk, C. S., Ried, T., Nussenzweig, A., and Chen, J. (2005) Mol. Cell. Biol. 25 10079–10086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales, J. C., Xia, Z., Lu, T., Aldrich, M. B., Wang, B., Rosales, C., Kellems, R. E., Hittelman, W. N., Elledge, S. J., and Carpenter, P. B. (2003) J. Biol. Chem. 278 14971–14977 [DOI] [PubMed] [Google Scholar]
- 26.Ali, A., Zhang, J., Bao, S., Liu, I., Otterness, D., Dean, N. M., Abraham, R. T., and Wang, X. F. (2004) Genes Dev. 18 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura, A., Sedelnikova, O. A., Redon, C., Pilch, D. R., Sinogeeva, N. I., Shroff, R., Lichten, M., and Bonner, W. M. (2006) Methods Enzymol. 409 236–250 [DOI] [PubMed] [Google Scholar]
- 28.Seluanov, A., Mittelman, D., Pereira-Smith, O. M., Wilson, J. H., and Gorbunova, V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7624–7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward, I. M., Minn, K., Jorda, K. G., and Chen, J. (2003) J. Biol. Chem. 278 19579–19582 [DOI] [PubMed] [Google Scholar]
- 30.Fernandez-Capetillo, O., Chen, H. T., Celeste, A., Ward, I., Romanienko, P. J., Morales, J. C., Naka, K., Xia, Z., Camerini-Otero, R. D., Motoyama, N., Carpenter, P. B., Bonner, W. M., Chen, J., and Nussenzweig, A. (2002) Nat. Cell Biol. 4 993–997 [DOI] [PubMed] [Google Scholar]
- 31.Chowdhury, D., Xu, X., Zhong, X., Ahmed, F., Zhong, J., Liao, J., Dykxhoorn, D. M., Weinstock, D. M., Pfeifer, G. P., and Lieberman, J. (2008) Mol. Cell 31 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, H., Kwak, H., Cho, I. T., Park, S. H., and Lee, C. H. (2009) Biochem. Biophys. Res. Commun. 378 32–36 [DOI] [PubMed] [Google Scholar]
- 33.Rogakou, E. P., Boon, C., Redon, C., and Bonner, W. M. (1999) J. Cell Biol. 146 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paull, T. T., Rogakou, E. P., Yamazaki, V., Kirchgessner, C. U., Gellert, M., and Bonner, W. M. (2000) Curr. Biol. 10 886–895 [DOI] [PubMed] [Google Scholar]
- 35.Ward, I., and Chen, J. (2004) Curr. Top. Dev. Biol. 63 1–35 [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi, J., Iwabuchi, K., Miyagawa, K., Sonoda, E., Suzuki, K., Takata, M., and Tauchi, H. (2008) J. Radiat. Res. 49 93–103 [DOI] [PubMed] [Google Scholar]
- 37.Ward, I., Kim, J. E., Minn, K., Chini, C. C., Mer, G., and Chen, J. (2006) J. Biol. Chem. 281 38472–38477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mochan, T. A., Venere, M., DiTullio, R. A., Jr., and Halazonetis, T. D. (2004) DNA Repair 3 945–952 [DOI] [PubMed] [Google Scholar]
- 39.Travesa, A., Duch, A., and Quintana, D. G. (2008) J. Biol. Chem. 283 17123–17130 [DOI] [PubMed] [Google Scholar]
- 40.Difilippantonio, S., Gapud, E., Wong, N., Huang, C. Y., Mahowald, G., Chen, H. T., Kruhlak, M. J., Callen, E., Livak, F., Nussenzweig, M. C., Sleckman, B. P., and Nussenzweig, A. (2008) Nature 456 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimitrova, N., Chen, Y. C., Spector, D. L., and de Lange, T. (2008) Nature 456 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie, A., Hartlerode, A., Stucki, M., Odate, S., Puget, N., Kwok, A., Nagaraju, G., Yan, C., Alt, F. W., Chen, J., Jackson, S. P., and Scully, R. (2007) Mol. Cell 28 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura, K., Sakai, W., Kawamoto, T., Bree, R. T., Lowndes, N. F., Takeda, S., and Taniguchi, Y. (2006) DNA Repair 5 741–749 [DOI] [PubMed] [Google Scholar]
- 44.Iwabuchi, K., Basu, B. P., Kysela, B., Kurihara, T., Shibata, M., Guan, D., Cao, Y., Hamada, T., Imamura, K., Jeggo, P. A., Date, T., and Doherty, A. J. (2003) J. Biol. Chem. 278 36487–36495 [DOI] [PubMed] [Google Scholar]
- 45.Keogh, M. C., Kim, J. A., Downey, M., Fillingham, J., Chowdhury, D., Harrison, J. C., Onishi, M., Datta, N., Galicia, S., Emili, A., Lieberman, J., Shen, X., Buratowski, S., Haber, J. E., Durocher, D., Greenblatt, J. F., and Krogan, N. J. (2006) Nature 439 497–501 [DOI] [PubMed] [Google Scholar]
- 46.Riballo, E., Kuhne, M., Rief, N., Doherty, A., Smith, G. C., Recio, M. J., Reis, C., Dahm, K., Fricke, A., Krempler, A., Parker, A. R., Jackson, S. P., Gennery, A., Jeggo, P. A., and Lobrich, M. (2004) Mol. Cell 16 715–724 [DOI] [PubMed] [Google Scholar]
- 47.Manis, J. P., Morales, J. C., Xia, Z., Kutok, J. L., Alt, F. W., and Carpenter, P. B. (2004) Nat. Immunol. 5 481–487 [DOI] [PubMed] [Google Scholar]
- 48.Iwabuchi, K., Hashimoto, M., Matsui, T., Kurihara, T., Shimizu, H., Adachi, N., Ishiai, M., Yamamoto, K., Tauchi, H., Takata, M., Koyama, H., and Date, T. (2006) Genes Cells 11 935–948 [DOI] [PubMed] [Google Scholar]
- 49.Zhang, J., Bao, S., Furumai, R., Kucera, K. S., Ali, A., Dean, N. M., and Wang, X. F. (2005) Mol. Cell. Biol. 25 9910–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chowdhury, D., Keogh, M. C., Ishii, H., Peterson, C. L., Buratowski, S., and Lieberman, J. (2005) Mol. Cell 20 801–809 [DOI] [PubMed] [Google Scholar]