Abstract
The crystal structure of the cyanobacterial cytochrome b6f complex has previously been solved to 3.0-Å resolution using the thermophilic Mastigocladus laminosus whose genome has not been sequenced. Several unicellular cyanobacteria, whose genomes have been sequenced and are tractable for mutagenesis, do not yield b6f complex in an intact dimeric state with significant electron transport activity. The genome of Nostoc sp. PCC 7120 has been sequenced and is closer phylogenetically to M. laminosus than are unicellular cyanobacteria. The amino acid sequences of the large core subunits and four small peripheral subunits of Nostoc are 88 and 80% identical to those in the M. laminosus b6f complex. Purified b6f complex from Nostoc has a stable dimeric structure, eight subunits with masses similar to those of M. laminosus, and comparable electron transport activity. The crystal structure of the native b6f complex, determined to a resolution of 3.0Å (PDB id: 2ZT9), is almost identical to that of M. laminosus. Two unique aspects of the Nostoc complex are: (i) a dominant conformation of heme bp that is rotated 180° about the α- and γ-meso carbon axis relative to the orientation in the M. laminosus complex and (ii) acetylation of the Rieske iron-sulfur protein (PetC) at the N terminus, a post-translational modification unprecedented in cyanobacterial membrane and electron transport proteins, and in polypeptides of cytochrome bc complexes from any source. The high spin electronic character of the unique heme cn is similar to that previously found in the b6f complex from other sources.
The hetero-oligomeric cytochrome b6f complex that functions between the reaction center complexes of the oxygenic photosynthetic electron transport chain contains eight tightly bound polypeptide subunits whose dimer molecular weight is ∼217,000. Plastoquinol-plastocyanin or -cytochrome c6 oxidoreductase activity is coupled to proton translocation, which generates the proton electrochemical potential gradient necessary for ATP synthesis and provides the electronic connection between the two photosynthetic reaction center complexes, photosystems I and II. A three-dimensional crystal structure of the b6f complex has been determined from the cyanobacterium Mastigocladus laminosus (1, 2) and also from the green alga Chlamydomonas reinhardtii (3). Cyanobacteria are potentially ideal sources for obtaining stable photosynthetic membrane proteins for structural studies (4, 5), because they are readily grown, and the protein yield from a reasonable volume of cell culture is sufficient to obtain crystals suitable for x-ray diffraction analysis. It is unexpected that stable intact dimeric b6f complex suitable for crystal growth has thus far not been obtained from unicellular cyanobacteria, but only from the filamentous moderately thermophilic M. laminosus. In our experience and that of other laboratories, the b6f complex extracted by detergent from unicellular cyanobacteria is always monomerized and inactivated. The reasons for the monomerization are not known. They may involve intrinsic instability of the protein complex in detergent micelles or the action of membrane-bound proteases. Because M. laminosus is not transformable at present, site-directed mutagenesis studies of the b6f complex in cyanobacteria have been carried out in the unicellular cyanobacteria, Synechococcus sp. 7002 (6, 7) or Synechocystis sp. PCC 6803 (8). This situation motivated a search for a cyanobacterium that could be used in structure and function studies of the b6f complex that would include the potential for mutagenesis.
To identify such cyanobacteria suitable for further analysis of the structure and function of the b6f complex, the amino acid sequences of the closely related homologues of the filamentous M. laminosus b6f complex were analyzed. In the present study, isolation of active dimeric cytochrome b6f complex from the transformable Nostoc sp. PCC 7120, which also contain the unique heme cn previously found in C. reinhardtii (3) and M. laminosus (1), is reported. Crystallographic analysis yielded crystals with 3.0-Å resolution and a structure similar to that previously obtained for the M. laminosus b6f complex (2). An additional unique aspect of the structure, resolved by mass spectrometry, was N-terminal acetylation of the Rieske iron-sulfur protein (ISP)3 subunit. N-terminal acetylation is common in proteins of eukaryotes and halophilic archaea but very rare in prokaryotes, including cyanobacteria. There has been only one previous report of N-terminal post-translational modification of a cyanobacterial protein, acetylation of a transcription factor in the cyanobacterium, Aphanizomenon ovalisporum (9).
EXPERIMENTAL PROCEDURES
Materials—Ammonium sulfate was purchased from MP Biomedicals (Solon, OH). Decyl-plastoquinol and propyl-agarose were from Sigma-Aldrich, SP-Sepharose from Amersham Biosciences, and 1,2-dioleoyl-sn-glycero-3-phosphocholine and n-undecyl-β-d-maltopyranoside from Anatrace (Maumee, OH). Unless otherwise specified, all other reagents were purchased from Mallinckrodt Baker, Inc. (Phillipsburg, NJ).
Cyanobacterial Strains and Growth Conditions—Nostoc sp. PCC 7120 strain has been kindly provided by R. Haselkorn (University of Chicago). The cyanobacteria were grown in BG-11 media (10) at 30 °C under a light intensity of 100–150 μEinsteins m–2 s–1 obtained from fluorescent lamps. Cells were harvested at early exponential growth phase (A730 ≈ 1–2) and were stored in the frozen state at –70 °C.
Purification of Nostoc Cytochrome b6f Complex—A conventional method for isolation of the complex was modified to optimize the procedure for detergent extraction of the b6f complex. Thylakoid membranes were isolated as described for the cyanobacterium, M. laminosus (11), resuspended in 10 mm Tricine-NaOH, pH 8.0, and washed with 2 m NaBr solution as described (12). The chlorophyll a concentration was determined as previously described (13). The final sediment was resuspended to a chlorophyll a concentration of 2 mg/ml in TNE (30 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm EDTA, with protease inhibitors, 2 mm benzamidine, and 2 mm ε-aminocaproic acid) supplemented with 300 mm sucrose. Cytochrome b6f complex was extracted by adding 0.1 volume of UDM solution to a final concentration of 10 mm with stirring of the suspension for 30 min on ice. The extract was centrifuged at 200,000 × g for 45 min in a 70-Ti rotor (Beckman-Coulter, Inc., Fullerton, CA). Further purification using ammonium sulfate precipitation, hydrophobic chromatography purification, and density centrifugation in sucrose gradient was performed as described (11), except that TNE buffer was used in all steps.
Purification of Plastocyanin—The protocol was adapted from a previous study (14). Solid ammonium sulfate was added to the supernatant of broken Nostoc cells (in the first step of the thylakoid membrane isolation described above) to achieve 45% saturation and stirred for 2 h at 4 °C. The precipitate was removed by centrifugation at 10,000 × g for 20 min. The supernatant was collected, and ammonium sulfate was added to 100% saturation, and the mixture was stirred for 2 h and stored for 16 h at 4 °C. The precipitate containing plastocyanin was sedimented by centrifugation at 10,000 × g for 30 min. The pellet was resuspended in buffer A (1 mm MES, pH 6.5, 0.25 mm ferricyanide), and ammonium sulfate was removed using a Centriprep YM-10 concentrator (Millipore, Billerica, MA). This sample was loaded onto a SP-Sepharose column equilibrated with buffer A. After a wash with buffer A supplemented with 2 mm NaCl, pure plastocyanin was eluted with a linear gradient of 2–30 mm NaCl in buffer A.
Plastoquinol-plastocyanin Oxidoreductase Activity—The assay suspension for plastoquinol-plastocyanin oxidoreductase activity contained 2.5 μm plastocyanin, 5 nm cyt b6f complex, 0.25 mm potassium ferricyanide, 1 mm UDM, 50 mm Tris-HCl, pH 7.5. Absorbance changes were assayed on a Cary 4000 spectrophotometer (Varian Inc., Palo Alto, CA). The reaction was initiated by addition of 20 μm decyl-plastoquinol, and activity was monitored as the change of potassium ferricyanide absorbance at 420 nm based on εmm = 1.02 mm–1 cm–1 (15).
Absorbance Difference Spectra—Chemical difference spectra of cytochromes f and b6 were measured as described previously (16) using a Cary 300 spectrophotometer (Varian Inc.).
Mass Spectrometry—Liquid chromatography with electrospray-ionization mass spectrometry and concomitant fraction collection was performed as previously described for the M. laminosus b6f complex (17). Liquid chromatography with data-dependent tandem mass spectrometry was performed on a quadrupole time-of-flight electrospray mass spectrometer (QSTAR XL, Applied Biosystems, Foster City, CA) as described previously (18).
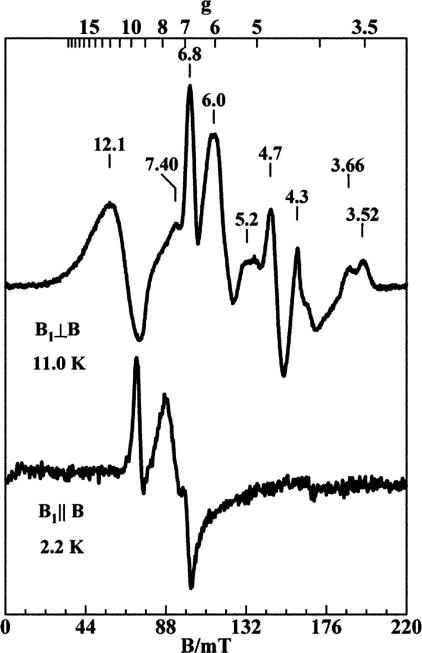
EPR—EPR spectra of cyt b6f complex samples were recorded on a Bruker 300 spectrometer (Bruker BioSpin Corp., Billerica, MA) equipped with an Oxford ESR-910 liquid helium cryostat and a Bruker bimodal cavity for generation of the microwave magnetic field parallel and perpendicular to the static orienting magnetic field (19). The quantitation of all signals was relative to a CuEDTA spin standard. The microwave frequency was calibrated with a frequency counter and the magnetic field with a NMR gaussmeter. The sample temperature of the cryostat was calibrated using a calibrated carbon-glass resistor (LakeShore CGR-1–1000) placed in an EPR tube to mimic a sample. A modulation frequency of 100 kHz was used for all EPR spectra. All experimental data were collected under non-saturating conditions. Specific experimental conditions are described in the legend to Fig. 4.
FIGURE 4.
X-band EPR spectra of cytochrome b6f complex from Nostoc for parallel and perpendicular orientations of the microwave magnetic field. Purified b6f complex was diluted to a concentration of 0.105 mm in TNE buffer supplemented with 1 mm UDM and 300 mm sucrose. Experimental conditions: microwave frequency, 9.65 GHz (B1 ⊥ B) and 9.37 GHz (B1 ∥ B); microwave power, 2 milliwatts (B1 ⊥ B) and 0.2 milliwatt (B1 ∥ B).
Protein Sequence Analysis—Multiple sequence alignments for the eight subunits of cyanobacterial cyt b6f complex were built using the ClustalW version 1.83 algorithm (20) with Gonnet matrix, gap opening penalty, and gap extension penalty equal to 10.0 and 0.2, respectively. A genomic DNA search to identify non-annotated small subunit homologues has been performed using the NCBI Genomic BLAST server (21). A Comparative Genomics server of the MEROPS peptidase database release 8.1 (22) was used to search unique peptidases characteristic for selected cyanobacteria strains.
Crystallization of the b6f Complex—Crystals of the native cytochrome b6f complex were obtained using the modified hanging drop, vapor-diffusion method described before (2). Buffer was exchanged to 100 mm Tris-HCl, pH 7.5, containing 1 mm DOPC lipid, and 3 mm UDM detergent. The protein was concentrated to 135 μm, and 4-μl droplets for crystallization were set up by diluting 1:1 with reservoir solution containing 100 mm Tris-HCl, pH 7.5, 200 mm MgCl2, 5 mm CdCl2, 1 mm DOPC, 3 mm UDM, and 12% polyethylene glycol monomethyl ether 550. Crystals were grown at 6 °C for 2 days. Prior to flash freezing, crystals were transferred to cryo-protectant solutions containing the same components as the reservoir solution except that the polyethylene glycol monomethyl ether 550 concentration was 14% and the glycerol concentration was increased in steps of 5%, to 25%.
Crystallography and Analysis of Diffraction Data—Crystals were screened for quality of diffraction at 100 K. Diffraction quality, resolution, and mosaicity were first assessed using in-house x-ray equipment and subsequently at beam line 19-ID at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). The structure of the Nostoc b6f complex was determined by rigid body refinement using REFMAC5 (23) in CCP4 (24), with the native structure from M. laminosus (2E74, PDB accession code) as the initial model. The structure refinement used programs REFMAC5 and O (25).
RESULTS
Comparative Sequence Analysis of the Subunits of Cyanobacterial b6f Complexes—The amino acid sequences of the subunits of the b6f complex from several cyanobacteria were compared with that of M. laminosus. Representative cyanobacterial strains whose complete genome sequence has been deposited in the NCBI Genome database were selected for the multiple sequence alignment analysis. The results displayed in Table 1 demonstrate that the highest degree of identity among the subunits of the M. laminosus b6f complex is displayed by the phylogenetically and morphologically related filamentous heterocyst-forming cyanobacteria that reside in the phylogenetically defined subsection IV of the cyanobacteria (10), such as Anabaena variabilis ATCC 29413 and Nostoc sp. PCC 7120. These results are in agreement with phylogenetic data derived from 16S rRNA sequence analysis (26). Compared with M. laminosus, the sequences of the subunits that have been shown to be essential for the function of the complex (cyt b6, cyt f, SuIV and Rieske ISP, petG, and petN) have 81–95% identity. Subunits petL and petM, which appear not to be essential (27, 28), were 48 and 79% identical.
TABLE 1.
Amino acid sequence identity of subunits of cyanobacterial cytochrome b6f complex compared to b6f complex from M. laminosus
The subclass of cyanobacteria strain is specified in parentheses, according to Ref. 10. M. laminosus belongs to subclass V. Group IV strains, including Nostoc, which is the subject of the present study, are written in boldface.
| Strain | cyt f | cyt b6 | ISP | SuIV | petG | petL | petM | petN |
|---|---|---|---|---|---|---|---|---|
| Acaryochloris marina MBIC 11017 (I) | 60 | 88 | 70 | 75 | 66 | 50 | 13 | 65 |
| Gloeobacter violaceus PCC 7421 (I) | 45 | 83 | 48 | 70 | 54 | n.i.a | 42b | 57 |
| Prochlorococcus marinus MIT 9301 (I) | 59 | 83 | 64 | 79 | 56 | n.i. | 40 | 62 |
| Synechococcus elongatus PCC 6301 (I) | 67 | 88 | 77 | 79 | 60 | 30 | 42 | 68 |
| Synechococcus sp. PCC 7002 (I) | 68 | 86 | 70 | 73 | 76 | n.i. | 25 | n.i. |
| Synechocystis sp. PCC 6803 (I) | 68 | 84 | 75 | 76 | 62 | 34 | 25 | 72 |
| Thermosynechococcus elengatus BP-1 (I) | 69 | 88 | 70 | 81 | 64 | 30 | 39 | 65 |
| Trichodesmium erythraeum IMS 101 (III) | 71 | 85 | 73 | 83 | 72 | 21b | 51b | 68b |
| Anabaena variabilisATCC 29413 (IV) | 83 | 95 | 85 | 91 | 81 | 74 | 79 | 86 |
| Nostocsp. PCC 7120 (IV) | 83 | 95 | 84 | 91 | 81 | 74b | 79 | 86 |
n.i., the gene has not been identified in a BLAST search of genomic DNA sequences of homologous cyanobacterial proteins.
The gene has been identified by BLAST search of genomic DNA using sequences of homologous cyanobacterial proteins.
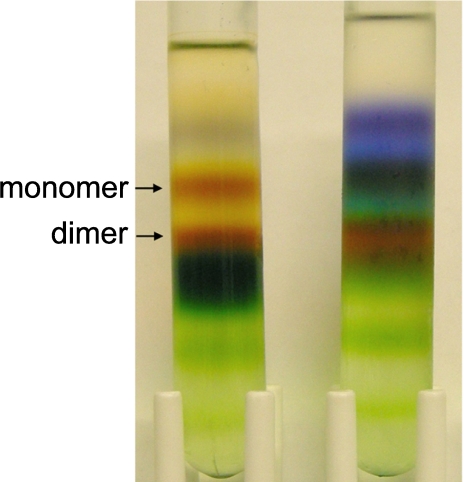
Purification of b6f Complex from Nostoc sp. PCC 7120; Electron Transport Activity—During extraction of the complex from the membranes of Nostoc, a rapid loss of oxidoreductase activity was observed when the same procedure was employed that was used for isolation from M. laminosus (11). It is likely that extraction of thylakoid membranes with a mixture of ionic and glycoside detergents (sodium cholate and octylglucoside) results in lipid depletion of, and/or dissociation of, integral components from the complex. To avoid such denaturation of the protein, a milder maltoside detergent, undecyl-maltoside, was used for extraction. The oxidoreductase activity of the b6f complex was retained when thylakoid membranes containing a cyt b6f equivalent of 2 mg/ml chlorophyll a were extracted on ice with 10 mm UDM. The extraction efficiency was ∼65%. Better extraction efficiency was obtained by increasing concentrations of the detergent to 15–20 mm, although this resulted in a significant reduction of the protein purity ultimately obtained. The UDM concentration was further reduced to 1 mm during purification on a propyl-agarose column. A pure form of dimeric b6f complex was obtained after density centrifugation through a sucrose gradient (Fig. 1). The dimeric protein fraction from Nostoc represented 70–80% of the total purified b6f complex. The density centrifugation was repeated twice to enrich the complex. The final yield of purified b6f complex was ∼7 mg of pure dimer from 25 g of wet cell pellet.
FIGURE 1.
Density centrifugation of Nostoc cytochrome b6f fractions collected from the hydrophobic chromatography column. The two cytochrome b6f containing fractions, which eluted in the first fractions from the column (left), and in the late fractions (right), were concentrated, loaded onto 10-ml 8–35% sucrose gradient in the TNE buffer supplemented with 1 mm UDM, and centrifuged at 200,000 × g (16 h) in an SW-41 rotor (Beckman-Coulter). Cytochrome b6f monomer and dimer bands are indicated by arrows.
The heme ratio of the isolated b6f complex, that of cyt b6 to cyt f, was determined to be 2.1 ± 0.07 by redox difference spectrophotometry using extinction coefficients described previously (16). Decyl-plastoquinol-plastocyanin oxidoreductase activity of the b6f complex was 277 ± 14 s–1·cyt f –1.
Comparative Genomics of Cyanobacterial Peptidases—Considering the hypothesis that the absence of endogenous peptidase activity is a major determinant of the stability of the dimeric complex, a proteolytic enzyme composition analysis of unicellular cyanobacteria and the Nostoc sp. PCC 7120 genome was performed using the comparative genomics search engine (MEROPS peptidase data base). The peptidase composition was analyzed for selected cyanobacterial strains (Thermosynechococcus elongatus, Anabaena variabilis, Synechococcus elongatus, and Synechocystis sp. PCC 6803) that we or others had previously used for purification of the b6f complex. Compared with Nostoc, 4–20 unique peptidases were found in the other cyanobacterial strains (Table 2).
TABLE 2.
Comparative genomics analysis of cyanobacterial proteases
Cyanobacterial subclass is specified in parentheses, according to Ref. 10.
| Strain | Genome size | Total, known, or putative peptidases | Unique peptidases compared to Nostoc sp. PCC 7120 |
|---|---|---|---|
| Mbp | |||
| Nostoc sp. PCC 7120 (IV)a | 7.21 | 125 | |
| A. variabilis (IV)b | 7.07 | 157 | 20 |
| S. elongatus (I)b | 2.7 | 100 | 20 |
| Synechocystis sp. PCC 6803 (I)b | 3.95 | 78 | 11 |
| T. elongatus (I)b | 2.59 | 52 | 4 |
Active dimeric cyt b6f complex has been purified.
Cytochrome b6f complex purification attempts have been made but resulted in an unstable complex (J. Yan and W. A. Cramer, unpublished).
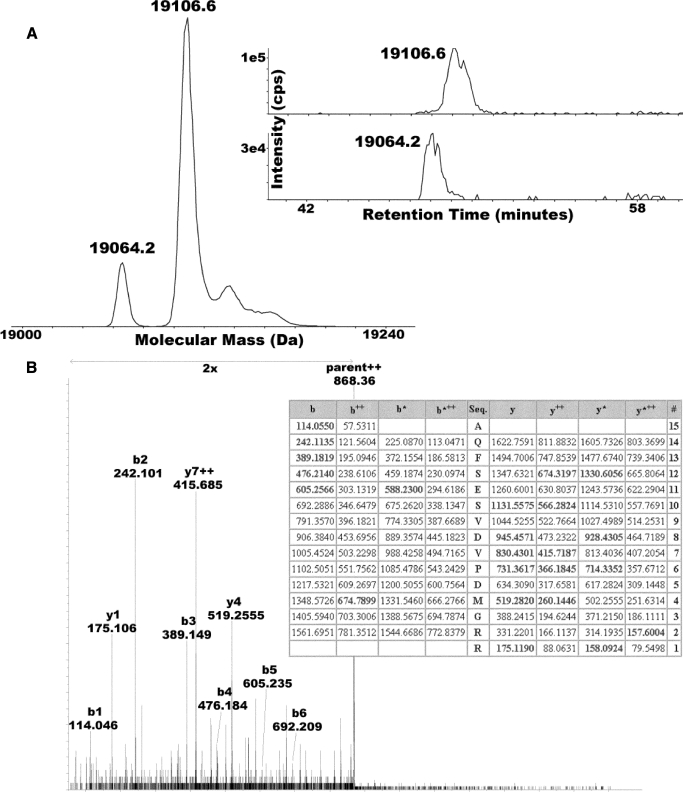
Chemical Modification of the Rieske Iron-Sulfur Protein; N-terminal Acetylation—The polypeptide composition of the purified complex was analyzed by electrospray-ionization mass spectrometry, revealing the eight subunit composition of the complex (Table 3). The analysis also determined that the N terminus of the Rieske ISP is partly acetylated (Fig. 2, A and B, and Table 3). This is seen as a spectral peak in the mass spectrum that is 42 mass units greater than predicted from the amino acid sequence. Two forms of the Rieske iron-sulfur protein (petC; ISP) were clearly resolved by chromatography and their mass in Nostoc (Fig. 2A). These species have been assigned as a smaller species with its initiating Met-1 removed (molecular mass = 19,064.2 Da) and a larger species that is acetylated at the N terminus (molecular mass = 19,106.6 Da). A column fraction containing the larger species was treated with trypsin, and tandem mass spectrometry was performed on the N-terminal peptide (nano-liquid chromatograph coupled to MSMS using Qstar XL). An N-terminal ion series (b1–b6) and all other MS and MS/MS mass measurements were all consistent with acetylation at position 1 (Fig. 2B).
TABLE 3.
Masses of subunits of Nostoc cyt b6f complex measured by electrospray-ionization MS
| Subunit | Measured massa | Calculated massb | Modificationsc |
|---|---|---|---|
| Cytochrome f | 31,769 | 31,768 | + heme |
| Cytochrome b6 | 24,757 | 24,758 | - Met-1, + heme |
| Rieske ISP | 19,107 | 19,106 | - Met-1, +/- Ac |
| 19,064 | 19,064 | + Disulfided | |
| SuIV | 17,404 | 17,405 | - Met-1 |
| petG | 4,023 | 4,023 | N-Formyl |
| petM | 3,574 | 3,574 | N-Formyl |
| petN | 3,262 | 3,262 | N-Formyl |
| petL | 3,253 | 3,253 | N-Formyl |
Average mass, mean of two measurements.
Calculated average mass based upon natural isotopic abundances and available sequence modification data.
Modifications include removal of initiating methionine, N-formylation of N terminus (retention of initiating formylmethionine), acetylation (Ac), presence of heme, and disulfide bonds.
Calculated 19,066.43 - 2(1.0078) = 19064.41, and for the acetylated form: 19106.42.
FIGURE 2.
Partial N-terminal acetylation of the Rieske iron-sulfur protein. LC-MS of the cytochrome b6f complex from Nostoc reveals two populations of Rieske ISP. A, electrospray-ionization mass spectrum of Rieske ISP after zero-charge deconvolution reveals sub-populations at 19,064.2 and 19,106.6 Da, consistent with partial acetylation (+42 Da). Specific ion chromatograms for each species are shown in the inset, revealing that the putative acetylated form is more highly retained, consistent with it being more hydrophobic due to having one less charge. B, analysis of an N-terminal tryptic peptide of Rieske ISP by tandem mass spectrometry using collisionally activated dissociation. The collisionally activated dissociation spectrum shown was annotated with respect to the important b-ion series (b1–b5) that are all consistent with N-terminal acetylation. The table was generated by the Mascot algorithm (Matrix Sciences), which picked the N-terminally acetylated N-terminal tryptic peptide of Nostoc Rieske ISP out of the complete protein data base (MSDB at Matrix Sciences on 11/11/08; search run in “no enzyme” mode) as the best match to the experimental dataset with a score of 40 (see Ref. 18). The matched ions are shown in boldface and localize the delta 42-Da modification to the N-terminal amino acid residue.
Crystal Structure of the Nostoc b6f Complex—Crystallization trials that utilized conditions previously optimized for crystal growth of M. laminosus b6f complex (2) resulted in visible bipyramidal crystals after growth at 6 °C for 2 days (Fig. 3, inset). The diffraction data collected under cryo-conditions defined a resolution limit of 3.0 Å (Table 4). The processed data showed that the crystals belong to the hexagonal space group P6122, as in M. laminosus, with very similar unit-cell parameters a = b = 159.2 Å, c = 365.9 Å. The r.m.s.d. between the Cα atom positions of the intact eight subunit structure of M. laminosus (PDB id: 2E74) and Nostoc sp. PCC 7120 (PDB id: 2ZT9) is 0.83 Å. Other r.m.s.d. values are: (i) p-side prosthetic groups, cyt f heme and Rieske [2Fe-2S] cluster, 0.66 and 0.51 Å, respectively; (ii) heme bn, 0.28 Å; (iii) for the unique prosthetic groups in the b6f complex, the n-side heme cn, chlorophyll a, and β-carotene, the r.m.s.d. is 0.33, 0.61, and 0.90 Å. For the p-side heme bp, however, the r.m.s.d. between the 3.0-Å crystal structures from M. laminosus and Nostoc is large, 5.74 Å. The explanation for the latter discrepancy between the two cyanobacterial b6f structures is that the major or exclusive conformation of heme bp in the Nostoc b6f complex is rotated 180° around the axis of the α- and γ-meso carbon atoms relative to the heme orientation in the M. laminosus b6f complex.
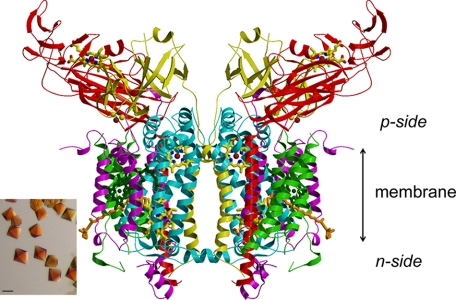
FIGURE 3.
Ribbon diagram of the symmetric dimeric structure of the cytochrome b6f complex from Nostoc sp. PCC 7120 determined from the 3.0-Å structure. The color code for the p-side of complex are: red, cyt f (petA); yellow, Rieske ISP (petA); blue, cytb (petB); purple, subunit IV; and green, petG, -L, -M, and -N. p-side heme of cyt f and [2Fe-2S] cluster are shown, along with two trans-membrane b-hemes, n-side heme cn represented as stick diagrams. The bound chlorophyll a and β-carotene are shown in stick format, in dark green and in orange, respectively. The inset shows bipyramidal crystals of the b6f complex from Nostoc obtained using the hanging drop, vapor-diffusion method described under “Experimental Procedures” (bar size, 100 μm).
TABLE 4.
Intensity data and statistics for a native crystal of Nostoc cytochrome b6f complex
Values in parentheses apply to the highest resolution shell.
| Crystal | Native |
|---|---|
| Space group | P6122 |
| Cell constants | |
| a, b (Å) | 159.2 |
| c (Å) | 365.9 |
| Data collection | |
| Resolution (Å) | 3.00 (3.11-3.00) |
| Measured reflections | 295,712 (29,625) |
| Unique reflections | 54,614 (5,362) |
| Redundancy | 5.4 (5.5) |
| I/σ(I) | 21.9 (4.1) |
| Completeness (%) | 98.3 (99.3) |
| Rmergea | 0.080 (0.337) |
| Refinement | |
| Rb | 0.230 |
| Rfreec | 0.259 |
| rms deviation from ideal | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.77 |
Rmerge = ΣhklΣi|Ii(hkl) – 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii is the intensity of the measured reflection.
R = Σ ∥Fo| – |Fc∥/Σ|Fo|, where Fc and Fo are the calculated and observed structure factors, respectively.
Rfree is calculated for a randomly chosen 5.0% of reflections omitted from refinement.
This rotation results in an exchange of the positions of the methyl and vinyl side chains of the heme pyrrole rings I and II. As discussed below, this rotameric reversal has been described in crystal structures of some soluble b-type heme proteins (discussed below). However, it has not previously been noted in structures of the integral membrane cytochrome bc complexes, although, upon inspection of these structures, it was also found to exist in their cytochrome b subunits.
EPR Spectra; Electronically Coupled Hemes bn and cn; g = 12 Signal—X-band EPR spectra of b6f complex purified from Nostoc, displayed in Fig. 4 for perpendicular and parallel orientations of the microwave magnetic field, show the very large g value, g = 12, described previously in spectra derived from the M. laminosus b6f complex, which clearly show that hemes bn and cn are spin-coupled. A weak signal from the reduced state of the FeS cluster (data not shown) is observed at g = 2.03, 1.90, and 1.73, which corresponds to <5% of the protein concentration. The signals at g = 3.66 and 3.52 arise from the low spin hemes bp and f, respectively. The signal at g = 4.3 and part of the signal at g = 6.0 arise from impurity iron species in concentrations <1% of the protein. The signals at g = 12, 7.4, and 4.7 (perpendicular, ⊥) and g = 9.3, 7.7, and 6.7 (parallel, ∥) arise from the electronically interacting hemes bn and cn.
DISCUSSION
Purification of the b6f Complex from Nostoc sp. PCC 7120—Development of a cyanobacterial expression system for the cytochrome b6f complex that would produce a protein complex suitable for studies of structure and function relationships using both mutagenesis and crystallographic analysis at higher resolution (< 3.0 Å) has been difficult because of instability of the isolated b6f complex. The filamentous moderately thermophilic M. laminosus had been the only cyanobacterium from which stable b6f complex could be obtained. The method originally used to isolate cyt b6f complex from spinach chloroplasts (12) was modified to isolate an active complex from M. laminosus (11). However, the method has thus far not been applicable for isolation of stable b6f complex from unicellular cyanobacterial strains such as Synechocystis sp. PCC 6803 or Synechococcus sp. PCC7102, for which assays of function were hampered by loss of integrity of the b6f complex dimer and low levels of activity. The reasons for the instability of the complex are not understood. Proteolytic degradation of the purified protein from M. laminosus has been observed (29), suggesting that preparations from other cyanobacteria might contain more damaging proteolytic activity. It is also likely that detergent compatibility was an important factor. However, further development of a purification procedure for a histidine-tagged b6f complex from Synechococcus sp. PCC 7002, which tested the quality of the b6f complex extracted from membranes with a range of detergents, including the relatively “mild” undecyl- or dodecyl-maltopyranoside, did not improve the stability of the extracted complex. In these tests, detergent extraction resulted in a severe loss of activity.4
The difficulties in the purification of the b6f complex may be a result of combination of intrinsic structural features and extrinsic factors such as the interaction of the detergent with the complex in the particular organism. Therefore, it could be assumed that cyanobacteria containing closely related sequences of the subunit polypeptides compared with M. laminosus are likely to retain the characteristics important for protein dimer stability. Based on the protein sequence conservation of cyanobacterial b6f complexes (Table 1), type IV cyanobacteria, such as Anabaena variabilis ATCC 29413 and Nostoc sp. PCC 7120, appeared to be the best candidates for purification of native active cytochrome b6f complex.
An extract of active cytochrome b6f complex from the cyanobacterium A. variabilis has been described previously (30). However, >70% of oxidoreductase activity was lost during the purification, which was possibly a consequence of the lability of the protein dimer in the cholate/octylglucoside and Triton X-100 detergent mixture that was used (30). We chose Nostoc sp. PCC 7120 for further studies. Nostoc has been used for genetic and physiological studies of oxygenic photosynthesis, nitrogen fixation, cellular differentiation (31, 32), and techniques for genetic manipulation, including a highly efficient conjugation system, have been developed for the bacterium (33). A complete sequence of the Nostoc chromosome and six plasmids has been published (34). It is noted that the set of cyanobacterial genome sequences has led to inferences about the origin of photosynthesis (35).
A purification method previously used for the isolation of b6f complex from M. laminosus (11) was modified to employ an extraction with maltoside detergent. The change of detergent resulted in an efficient purification of the stable dimeric b6f complex from Nostoc with a yield similar to previously obtained from M. laminosus.
Crystal Structure—The crystal structure of the Nostoc cyt b6f complex (PDB id: 2ZT9 (Fig. 3 and Table 4)) obtained at 3.0-Å resolution was very similar to that previously determined for the complex from M. laminosus (2). Several exceptions include (i) the rotameric change in orientation of heme bp, (ii) acetylation of Rieske ISP, and (iii) differences in amino acid side-chain composition.
The Heme bp Rotamer—Remarkably, the heme bp in the structure of b6f complex from Nostoc is rotated 180° around the axis of the α- and γ-meso carbon atoms relative to the heme conformation in the b6f complex from M. laminosus. The overall position of the heme and its environment is not changed.
Inspection of the 3.1-Å structure of the b6f complex in C. reinhardtii (PDB id: 1Q90 (3)) shows that heme bp has the same orientation as that in the Nostoc b6f complex. The existence of the two rotamers of a non-covalently bound b-type heme in a soluble, or soluble fragment, of a heme protein has been documented previously: (i) in the comparison of bovine cytochrome b5, soluble in erythrocytes and solubilized from microsomes (36); (ii) cytochrome P450 from M. tuberculosis (37). The exchanged position of the heme side chains, methyl-vinylmethyl-vinyl to vinyl-methyl-vinyl-methyl, has been depicted by Walker et al. (36), who also determined that the midpoint redox potential of the two cytochrome b5 rotamers differed by ∼30 mV.
The present case is the first noted observation of the two heme b rotamers in an integral membrane cytochrome complex. Furthermore, it can be seen from inspection of the structures that the two rotamers exist in the cytochrome bc1 complex. The orientation of heme bp in the crystal structure of the bovine mitochondrial bc1 complex crystallized in the presence of the quinone analogue inhibitors methoxyacrylate stilbene and 5-n-undecyl-6-hydroxy-4,7-dioxobenzothiazole (38) is the same as that in Nostoc and C. reinhardtii, and rotated by 180° relative to that of heme bp in (i) the b6f complex of M. laminosus, (ii) the bovine bc1 complex formed in the presence of the quinone analogue inhibitors, azoxystrobin, myxathiazol, or stigmatellin (38), and (iii) the yeast (39) or (iv) photosynthetic bacterial (40) bc1 complex with stigmatellin. The two rotameric states of heme bp presumably have a role in the energy transduction function and/or assembly of the cytochrome bc complex, which is/are presently not understood.
EPR Spectroscopy—EPR spectra of the isolated b6f complex from Nostoc were similar overall to those previously observed for b6f complex from M. laminosus and spinach (19, 41). In addition, a similar conclusion on coupling of hemes cn and bn was subsequently obtained from EPR analysis of the C. reinhardtii b6f complex, using just the perpendicular orientation of the magnetic field (42). These spectra demonstrate that spin coupling between hemes bn and cn in the b6f complex is a quantitatively conserved property. Heme cn is important in the n-side pathway of quinone reduction (2). Additional information on residues necessary for the reduction pathway is available through mutagenesis studies (43).
Acetylation of Rieske ISP; Limited Prokaryotic Precedent—MS analysis revealed the same subunit composition, four “large” (cyt f, cyt b6, Rieske ISP, and subuit IV) and four “small” (PetG, -M, -L, and -N) (Table 2), as previously found for M. laminosus (17). The analysis also determined that the N terminus of the Rieske ISP is partly acetylated (Fig. 2, A and B, and Table 2). The mass of Rieske ISP in the M. laminosus b6f complex had been determined previously to be 19,294.8 Da (17), although at that time the M. laminosus sequence was unavailable and the Nostoc sequence (19,202.8 Da, including Met-1 and no modifications) was used. Based upon the M. laminosus sequence (Swiss-Prot id: P83794),5 an average mass of 19,270.0 Da was calculated, suggesting either a sequence error to account for the mass difference or possibly acetylation of the N terminus. The latter modification would, however, bring the calculated mass of Rieske ISP to 19,312 Da, which, with an expected error of ± 2 Da, is not consistent with the measured mass. In this study, high resolution Fourier transform mass spectrometry data (data not shown), including intact mass measurements and a top-down collisionally activated dissociation experiment on the 19,107 Da species, which measured the mass of smaller b-ions, were both more consistent with N-terminal acetylation (42.010565 Da; COCH2) rather than a mutation that changed Ala-2 to Leu/Ile (delta mass 42.04695 Da; C3H6). It is believed that this is the most complete evidence yet presented for N-terminal protein acetylation in a prokaryote. It was not possible to resolve the N-terminal acetylation of the Rieske ISP in the crystal structure, because the N-terminal eight residues on the electrochemically negative (n) side of the complex in the membrane are disordered in the crystal structure.
N-terminal post-translational protein modification by acetylation is common in eukaryotes and halophilic archaea (44), but very rare in prokaryotes. In eukaryotes, acetylation has been correlated with increased binding of histones to DNA, altered binding of transcription factors to DNA (45), and increased protein stability of transcription factor E2F1 (46) and α-tubulin (47). Acetylation influences protein-protein interactions (45), which inhibit association of T-cell factor with Armadillo in Drosophila (48) and the binding of activator of retinoid and thyroid receptors cofactor to nuclear receptors (49), and affect the interaction of histones with acetylases (50). Acetylation is required for photosynthesis in Arabidopsis thaliana, and it was proposed that the N-acetylation of certain chloroplast precursor protein(s) is necessary for the efficient accumulation of the mature proteins in chloroplasts (51). Although there is established precedent for N-terminal acetylation of electron-transport proteins in the eukaryotic photosynthetic apparatus of plants, particularly in photosystem II reaction center and light-harvesting polypeptides (51–54), acetylation of a cyanobacterial electron transport protein, in particular the PetC subunit of the b6f complex documented in the present study, is novel.
There are three reports of protein acetylation in prokaryotes: (i) acetylation is often mediated by the enzyme acetyl-CoA synthetase that is itself acetylated in Salmonella enterica (55) as well as in Escherichia coli, where it has been recognized as an autoacetylation (56). (ii) Acetylation of CheY, a protein associated with flagella rotation, signals cellular adaptation of motility to metabolic state (56). (iii) Acetylation of the protein AbrB, which is implicated in the regulation of growth stages, in the cyanobacterium Aphanizomenon ovalisporum alters its ability to bind DNA (9). The latter is the only known precedent for acetylation of a cyanobacterial protein. It has been demonstrated that bacterial acetyltransferases acetylate antibiotics (57).
Changes in Amino Acid Side-chain Composition—Compared with M. laminosus, the sequences of the subunits of the b6f complex from Nostoc have 74–95% identical amino acid residues (Table 1). Of the total of 126 residue replacements, 63 occur in the soluble domain of cyt f and the Rieske ISP, and most of them are oriented on the surfaces not involved in intramolecular or subunit interactions. In addition, 34 of the remaining residue replacements could be characterized as nonessential for the structural interactions (no significant change of charge and size of the side chain). Several examples that may appear of importance for side chain interactions are: the replacement Asp-67 → Asn of suIV and Lys-59 → Glu of Rieske ISP (at a distance of ∼3.7 Å); Lys-40 → Asn and Ser-46 → Ala of Rieske ISP and Ser-77 → Asn of cyt b6 (5.3 and 7.4 Å, respectively); Met-258 → Leu of cyt f and Ala-10 → Ser of petN (4.4 Å); Tyr-7 → Asn of petM and Ile-2 → Met at the one residue-truncated N terminus of petL; and Asp-4 → Leu of petN at a distance of ∼4.3 Å from Leu-41 of cyt f. None of these residue replacements are located close to functionally important cofactors. However, it could be proposed that they may play a significant role in protein complex stability, which could be assessed in future mutagenesis studies.
The Problem of Proteolysis—The 3.0-Å data set obtained from the first set of crystal structure data for the Nostoc b6f complex, and the concomitant ability to manipulate the Nostoc complex, bodes well for the potential of this system to obtain higher resolution structures of the b6f complex. Biochemical analyses of the purified complex demonstrated properties similar those of the intact dimeric b6f complex from M. laminosus. The dominant monomeric form seen in the unicellular cyanobacteria is indicative of the degradative proteolytic activity that can also be seen over prolonged incubation times in M. laminosus (29). If the minimal assumption is made that the same peptidase or the same set of peptidases is responsible for cyt b6f cleavage in all the cyanobacteria, then it must be among the four peptidases that are unique to T. elongatus (Table 4). These four include (note that the number in parentheses is a MEROPS identifier) glucosamine-fructose-6-phosphate aminotransferase (C44.971), vanY d-Ala-d-Ala carboxypeptidase (M15.010), family S49 unassigned peptidases (S49), and the microcin-processing peptidase 2 (U62.002). Among these four peptidases, only glucosamine-fructose-6-phosphate aminotransferase (C44.971) and microcin-processing peptidase 2 (U62.002) are found in all four cyanobacteria strains used in the purification of the b6f complex that results in monomerization of the complex. Therefore, it is suggested that activation of either or both of the proteases may be responsible for the cleavage of subunits of the b6f complex during detergent extraction and purification. However, it could not be excluded that there may be other relevant peptidases that have thus far not been identified.
Acknowledgments
We thank R. Haselkorn for the suggestion to use Nostoc sp. PCC 7120 for further studies on the cytochrome b6f complex, and J. Golden, O. Sharma, and T. Zakharova for helpful discussions. Much of the x-ray structure analysis was carried out at beam line SBC-19ID at the Advanced Photon Source, Argonne National Laboratory, where S. Ginell, J. Lazarz, and F. Rotella provided important advice and technical support.
This work was supported, in whole or in part, by National Institutes of Health Grants GM-38323 (to W. A. C.) and GM-077387 (to M. P. H.) and by NIH/NCRR Grant R21RR021913-01A2 (to J. P. W.). The x-ray structure analysis was carried out at the Advanced Photon Source, Argonne National Laboratory was supported by U. S. DOE Grant W31-109-ENG-389.
The atomic coordinates and structure factors (code 2ZT9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Footnotes
The abbreviations used are: ISP, iron-sulfur protein; cyt, cytochrome; heme bp/bn, cyt b6 hemes located on electrochemically positive/negative sides of membrane; heme cn, heme covalently bound by one thioether bond adjacent to heme bn; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; εmM, millimolar extinction coefficient; MEROPS, peptidase database; MES, 4-morpholinoethanesulfonic acid; Nostoc, Nostoc (Anabaena) sp. PCC 7120; r.m.s.d., root mean square deviation; UDM, n-undecyl-β-d-maltopyranoside; MS, mass spectrometry; MS/MS, tandem MS.
J. Yan and W. A. Cramer, unpublished data.
From J. Yan, H. Zhang, and W. A. Cramer.
References
- 1.Kurisu, G., Zhang, H., Smith, J. L., and Cramer, W. A. (2003) Science 302 1009–1014 [DOI] [PubMed] [Google Scholar]
- 2.Yamashita, E., Zhang, H., and Cramer, W. A. (2007) J. Mol. Biol. 370 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stroebel, D., Choquet, Y., Popot, J. L., and Picot, D. (2003) Nature 426 413–418 [DOI] [PubMed] [Google Scholar]
- 4.Jordan, P., Fromme, P., Witt, H. T., Klukas, O., Saenger, W., and Krauss, N. (2001) Nature 411 909–917 [DOI] [PubMed] [Google Scholar]
- 5.Zouni, A., Witt, H. T., Kern, J., Fromme, P., Krauss, N., Saenger, W., and Orth, P. (2001) Nature 409 739–743 [DOI] [PubMed] [Google Scholar]
- 6.Yan, J., and Cramer, W. A. (2003) J. Biol. Chem. 278 20925–20933 [DOI] [PubMed] [Google Scholar]
- 7.Yan, J. S., and Cramer, W. A. (2004) J. Mol. Biol. 344 481–493 [DOI] [PubMed] [Google Scholar]
- 8.Schneider, D., Skrzypczak, S., Anemuller, S., Schmidt, C. L., Seidler, A., and Rogner, M. (2002) J. Biol. Chem. 277 10949–10954 [DOI] [PubMed] [Google Scholar]
- 9.Shalev-Malul, G., Lieman-Hurwitz, J., Viner-Mozzini, Y., Sukenik, A., Gaathon, A., Lebendiker, M., and Kaplan, A. (2008) Environ. Microbiol. 10 988–999 [DOI] [PubMed] [Google Scholar]
- 10.Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979) J. Gen. Microbiol. 111 1–61 [Google Scholar]
- 11.Zhang, H., Whitelegge, J. P., and Cramer, W. A. (2001) J. Biol. Chem. 276 38159–38165 [DOI] [PubMed] [Google Scholar]
- 12.Hurt, E., and Hauska, G. (1981) Eur. J. Biochem. 117 591–599 [DOI] [PubMed] [Google Scholar]
- 13.Porra, R. J. (1991) Chlorophylls, pp. 31–57, CRC Press Inc., Boca Raton, FL
- 14.Ellefson, W. L., Ulrich, E. A., and Krogmann, D. W. (1980) Methods Enzymol. 69 223–228 [Google Scholar]
- 15.Peck, H. D., Deacon, T. E., and Davidson, J. T. (1965) Biochim. Biophys. Acta 96 429–446 [DOI] [PubMed] [Google Scholar]
- 16.Metzger, S. U., Cramer, W. A., and Whitmarsh, J. (1997) Biochim. Biophys. Acta 1319 233–241 [DOI] [PubMed] [Google Scholar]
- 17.Whitelegge, J. P., Zhang, H., Aguilera, R., Taylor, R. M., and Cramer, W. A. (2002) Mol. Cell Proteomics 1 816–827 [DOI] [PubMed] [Google Scholar]
- 18.Xie, J., Marusich, M. F., Souda, P., Whitelegge, J., and Capaldi, R. A. (2007) FEBS Lett. 581 3545–3549 [DOI] [PubMed] [Google Scholar]
- 19.Zatsman, A. I., Zhang, H., Gunderson, W. A., Cramer, W. A., and Hendrich, M. P. (2006) J. Am. Chem. Soc. 128 14246–14247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGinnis, S., and Madden, T. L. (2004) Nucleic Acids Res. 32 W20–W25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawlings, N. D., Morton, F. R., Kok, C. Y., Kong, J., and Barrett, A. J. (2008) Nucleic Acids Res. 36 D320–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240–255 [DOI] [PubMed] [Google Scholar]
- 24.Bailey, S. (1994) Acta Crystallogr. D Biol. Crystallogr. 50 760–763 [DOI] [PubMed] [Google Scholar]
- 25.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. A 47 110–119 [DOI] [PubMed] [Google Scholar]
- 26.Tomitani, A. (2006) Front. Res. Earth Evol. 2 1–5 [Google Scholar]
- 27.Boronowsky, U., Wenk, S., Schneider, D., Jager, C., and Rogner, M. (2001) Biochim. Biophys. Acta 1506 55–66 [DOI] [PubMed] [Google Scholar]
- 28.Schneider, D., Volkmer, T., and Rogner, M. (2007) Res. Microbiol. 158 45–50 [DOI] [PubMed] [Google Scholar]
- 29.Zhang, H., and Cramer, W. A. (2005) J. Struct. Funct. Genomics 6 219–223 [DOI] [PubMed] [Google Scholar]
- 30.Krinner, M., Hauska, G., Hurt, E., and Lockau, W. (1982) Biochim. Biophys. Acta 681 110–117 [Google Scholar]
- 31.Golden, J. W., and Yoon, H. S. (2003) Curr. Opin. Microbiol. 6 557–563 [DOI] [PubMed] [Google Scholar]
- 32.Hurley, J. K., Morales, R., Martinez-Julvez, M., Brodie, T. B., Medina, M., Gomez-Moreno, C., and Tollin, G. (2002) Biochim. Biophys. Acta 1554 5–21 [DOI] [PubMed] [Google Scholar]
- 33.Elhai, J., and Wolk, C. P. (1988) Methods Enzymol. 167 747–754 [DOI] [PubMed] [Google Scholar]
- 34.Kaneko, T., Nakamura, Y., Wolk, C. P., Kuritz, T., Sasamoto, S., Watanabe, A., Iriguchi, M., Ishikawa, A., Kawashima, K., Kimura, T., Kishida, Y., Kohara, M., Matsumoto, M., Matsuno, A., Muraki, A., Nakazaki, N., Shimpo, S., Sugimoto, M., Takazawa, M., Yamada, M., Yasuda, M., and Tabata, S. (2001) DNA Res. 8 205–213 [DOI] [PubMed] [Google Scholar]
- 35.Mulkidjanian, A. Y., Koonin, E. V., Makarova, K. S., Mekhedov, S. L., Sorokin, A., Wolf, Y. I., Dufresne, A., Partensky, F., Burd, H., Kaznadzey, D., Haselkorn, R., and Galperin, M. Y. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 13126–13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker, F. A., Emrick, D., Rivera, J. E., Hanquet, B. J., and Buttlaire, D. H. (1988) J. Am. Chem. Soc. 110 6234–6240 [DOI] [PubMed] [Google Scholar]
- 37.Leys, D., Mowat, C. G., McLean, K. J., Richmond, A., Chapman, S. K., Walkinshaw, M. D., and Munro, A. W. (2003) J. Biol. Chem. 278 5141–5147 [DOI] [PubMed] [Google Scholar]
- 38.Esser, L., Quinn, B., Li, Y. F., Zhang, M., Elberry, M., Yu, L., Yu, C. A., and Xia, D. (2004) J. Mol. Biol. 341 281–302 [DOI] [PubMed] [Google Scholar]
- 39.Solmaz, S. R., and Hunte, C. (2008) J. Biol. Chem. 283 17542–17549 [DOI] [PubMed] [Google Scholar]
- 40.Esser, L., Elberry, M., Zhou, F., Yu, C. A., Yu, L., and Xia, D. (2008) J. Biol. Chem. 283 2846–2857 [DOI] [PubMed] [Google Scholar]
- 41.Cramer, W. A., Baniulis, D., Yamashita, E., Zhang, H., Zatsman, A. I., and Hendrich, M. P. (2008) in Photosynthetic Protein Complexes (Fromme, P., ed) pp. 155–179, Wiley-VCH, Weinheim
- 42.Baymann, F., Giusti, F., Picot, D., and Nitschke, W. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson, M. E., Finazzi, G., Wang, Q. J., Middleton-Zarka, K. A., Whitmarsh, J., and Kallas, T. (2005) J. Biol. Chem. 280 10395–10402 [DOI] [PubMed] [Google Scholar]
- 44.Mackay, D. T., Botting, C. H., Taylor, G. L., and White, M. F. (2007) Mol. Microbiol. 64 1540–1548 [DOI] [PubMed] [Google Scholar]
- 45.Kouzarides, T. (2000) EMBO J. 19 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Balbas, M. A., Bauer, U. M., Nielsen, S. J., Brehm, A., and Kouzarides, T. (2000) EMBO J. 19 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takemura, R., Okabe, S., Umeyama, T., Kanai, Y., Cowan, N. J., and Hirokawa, N. (1992) J. Cell Sci. 103 953–964 [DOI] [PubMed] [Google Scholar]
- 48.Waltzer, L., and Bienz, M. (1998) Nature 395 521–525 [DOI] [PubMed] [Google Scholar]
- 49.Chen, H., Lin, R. J., Xie, W., Wilpitz, D., and Evans, R. M. (1999) Cell 98 675–686 [DOI] [PubMed] [Google Scholar]
- 50.Dhalluin, C., Carlson, J. E., Zeng, L., He, C., Aggarwal, A. K., and Zhou, M. M. (1999) Nature 399 491–496 [DOI] [PubMed] [Google Scholar]
- 51.Pesaresi, P., Gardner, N. A., Masiero, S., Dietzmann, A., Eichacker, L., Wickner, R., Salamini, F., and Leister, D. (2003) Plant Cell 15 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomez, S. M., Bil', K. Y., Aguilera, R., Nishio, J. N., Faull, K. F., and Whitelegge, J. P. (2003) Mol. Cell Proteomics 2 1068–1085 [DOI] [PubMed] [Google Scholar]
- 53.Michel, H., Hunt, D. F., Shabanowitz, J., and Bennett, J. (1988) J. Biol. Chem. 263 1123–1130 [PubMed] [Google Scholar]
- 54.Michel, H., Griffin, P. R., Shabanowitz, J., Hunt, D. F., and Bennett, J. (1991) J. Biol. Chem. 266 17584–17591 [PubMed] [Google Scholar]
- 55.Starai, V. J., Celic, I., Cole, R. N., Boeke, J. D., and Escalante-Semerena, J. C. (2002) Science 298 2390–2392 [DOI] [PubMed] [Google Scholar]
- 56.Barak, R., Prasad, K., Shainskaya, A., Wolfe, A. J., and Eisenbach, M. (2004) J. Mol. Biol. 342 383–401 [DOI] [PubMed] [Google Scholar]
- 57.Vetting, M. W., Magnet, S., Nieves, E., Roderick, S. L., and Blanchard, J. S. (2004) Chem. Biol. 11 565–573 [DOI] [PubMed] [Google Scholar]