Abstract
Vascular endothelial growth factor (VEGF-A) and its family proteins are crucial regulators of blood vessel formation and vascular permeability. Snake venom has recently been shown to be an exogenous source of unique VEGF (known as VEGF-F), and now, two types of VEGF-F with distinct biochemical properties have been reported. Here, we show that VEGF-Fs (venom-type VEGFs) are highly variable in structure and function among species, in contrast to endogenous tissue-type VEGFs (VEGF-As) of snakes. Although the structures of tissue-type VEGFs are highly conserved among venomous snake species and even among all vertebrates, including humans, those of venom-type VEGFs are extensively variegated, especially in the regions around receptor-binding loops and C-terminal putative coreceptor-binding regions, indicating that highly frequent variations are located around functionally key regions of the proteins. Genetic analyses suggest that venom-type VEGF gene may have developed from a tissue-type gene and that the unique sequence of its C-terminal region was generated by an alteration in the translation frame in the corresponding exons. We further verified that a novel venom-type VEGF from Bitis arietans displays unique properties distinct from already known VEGFs. Our results may provide evidence of a novel mechanism causing the generation of multiple snake toxins and also of a new model of molecular evolution.
Vascular endothelial growth factor (VEGF-A)3 and its family proteins are well known angiogenic and lymphangiogenic regulators. The VEGF family has been expanded and currently comprises seven members: VEGF-A, VEGF-B, placenta growth factor (PlGF), VEGF-C, VEGF-D, viral VEGF (also known as VEGF-E), and snake venom VEGF (also known as VEGF-F) (1, 2). These proteins consist of central VEGF homology domains (VHD), which are composed of 92–96 amino acids and share 29–64% identity among the family, and N- and C-terminal extensions. Eight cysteine residues in VHD, which are predicted to form a cystine knot, are strictly conserved among all members. In contrast to the ligands, three receptor tyrosine kinases (RTKs) are known as VEGF receptors: Flt-1 (fms-like tyrosine kinase-1, VEGFR-1), KDR (kinase insert domain-containing receptor, VEGFR-2), and Flt-4 (VEGFR-3). Flt-1 and KDR are mainly distributed on vascular endothelial cells and mediate several major angiogenic activities such as endothelial cell proliferation and migration, whereas Flt-4 is limited to the lymphatic endothelium and involves lymphangiogenesis. VEGF members have been shown to bind these three RTKs with different affinity and selectivity; e.g. VEGF-A binds both Flt-1 and KDR but with 10-fold different affinity, whereas VEGF-B and PlGF are specific to Flt-1. In addition to RTKs, two non-RTK-type receptors, neuropilin-1 (NP-1) and heparin (physiologically heparan sulfate proteoglycan), have also been shown to work as VEGF coreceptors. Some isoforms of VEGF-A, VEGF-B, and PlGF bind NP-1 or heparin/heparan sulfate via their C-terminal regions, resulting in the modulation of RTK-mediated signaling and vessel guidance (3).
For four decades, snake venom proteins/peptides have been used to elucidate the complicated physiology of mammals because of their unique and specific action to target molecules (4). Venom proteins/peptides are generally thought to be developed from endogenous proteins or their domains and often display significant molecular diversity (4). We have previously found snake venom VEGF-Fs named vammin and VR-1 from the venoms of Vipera a. ammodytes and Daboia r. russelli (5). Vammin and VR-1 bind only KDR with high affinity (similar to VEGF-A) but not to other VEGF receptors and show a potent hypotensive effect and stronger enhancement of vascular permeability as compared with human VEGF-A165 (5, 6), which is the predominant isoform of VEGF-A comprising 165 amino acids (7). Vammin and VR-1 are homodimeric proteins similar to other VEGF subtypes and possess short C-terminal positively charged tails that bind heparin (8, 9). Recently, two novel VEGF-Fs, Tf-svVEGF and Pm-VEGF from the venoms of Trimeresurus flavoviridis and Protobothrops mucrosquamatus, respectively, have been shown to bind Flt-1 in preference to KDR, unlike vammin and VR-1 (10, 11). These findings suggest the possibility that VEGF-Fs are functionally diversified similar to other snake venom proteins (2). In the present study, we have demonstrated that the venom-type VEGFs (VEGF-Fs) of snakes are widely distributed in several viper venoms and that their structure and function are extensively variegated among species, in contrast to endogenous tissue-type VEGFs (VEGF-A). Moreover, genomic analyses of venom- and tissue-type VEGFs from T. flavoviridis (Habu snake) strongly suggest that the venom-type VEGF gene developed from a tissue-type gene via a unique mechanism. This is the first report showing that snake venom genes are efficiently diversified to generate multiple toxins separately from the gene encoding the endogenous protein.
EXPERIMENTAL PROCEDURES
Cloning, Sequencing, and Genetic Analysis—The cDNAs encoding vammin and VR-1 were cloned using the following sets of degenerate primers for PCR amplification, designed based on the highly conserved amino acid sequences: 5′-GC(any) GT(A/G) TG(C/T) TC(any) (A/G)T(A/G) AA(C/T) TTC AT(any) AC(A/C) TCC AT-3′ for 5′-RACE and 5′-CA(A/G) GA(A/G) (C/T)A(C/T) CC(any) GA(C/T) GA(A/G) AT(not G) (A/T)(C/G)(any) GA(C/T) AT(not G) TT-3′ for 3′-RACE. Then, venom-type VEGFs-specific primer pairs were designed based on the conserved nucleotide sequences: 5′-GCA GCA GCC (A/G)C(C/T) (A/G)CA TCG CAA C-3′ for 5′-RACE and 5′-TTC TGA GCA GCT GTG AAG CCA GGA-3′ for 3′-RACE. The cDNAs encoding VEGF-As of snakes were cloned using the following sets of primers, which are specific to VEGF-As, designed using the conserved nucleotide sequences among several VEGF-As of vertebrate species including Homo sapiens and Bitis gabonica (12): 5′-GG(C/T) CTG CAT TCA CA(G/T) (not A)(C/T)(G/T) (not A)T(A/G) TGC T-3′ for 5′-RACE and 5′-ATG AAC TTT CTG CTC (A/T)CT TGG-3′ for 3′-RACE. The genomic DNA from T. flavoviridis was a gift from Dr. Hideko Atoda. PCR was performed by using specific primers that were designed based on the exon sequences encoding Tf-svVEGF and Tf-VEGF-A (10). Homology searching of the nucleotide sequences was performed by using Genetyx version 7.0. The KA and KS values were calculated using DnaSP software version 4.10.9.
Phylogenetic Analysis—The phylogenetic tree in Fig. 1 was constructed by Genetyx version 7.0 using the unweighted pair group method with arithmetic based on the amino acid sequences. The distance matrix for the alignment sequences was calculated by using the two-parameter method of Kimura as implemented in a computer program (31).
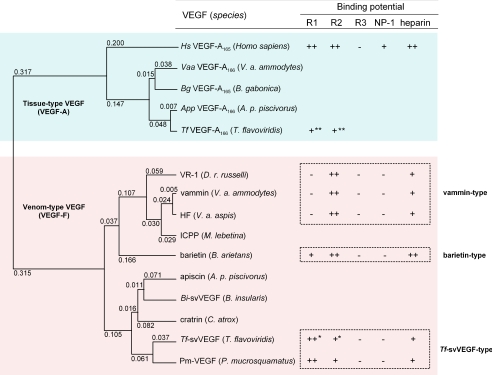
FIGURE 1.
Phylogenetic tree and ligand properties of venom-type VEGFs. The unweighted pair group method with arithmetic phylogenetic tree was built by using amino acid sequences from VHDs of several VEGFs. Tissue-type VEGFs (VEGF-As) are colored in cyan, and venom-type VEGFs (VEGF-Fs) are in magenta. The values at the top of the left corners of the branches indicate the evolutionary distance (expected value of base substitution), calculated by the Genetyx program. The affinities and receptor binding selectivities are referred from the results of Biacore analysis, and the heparin binding abilities are from the eluted NaCl concentrations from heparin affinity column. ++, +, bound; ––, not bound. *, predicted affinity from competitive inhibition assay (10); **, bound but the affinity is not reported (10). R1, Flt-1 (VEGFR-1); R2, KDR (VEGFR-2); R3, Flt-4 (VEGFR-3). Venom-type VEGFs could be classified into three groups based on their structure and receptor binding potentials: vammin-type, Tf-svVEGF-type, and barietin-type. HF, a hypotensive factor from Vipera aspis (29); ICPP, an increasing capillary permeability protein from Macrovipera lebetina (30).
Purification of Barietin from Bitis arietans Venom—Three hundred mg of lyophilized venom of B. arietans (Latoxan, Valence, France) was dissolved in 50 mm Tris-HCl buffer, pH 8.0. After centrifugation to remove a small amount of insoluble particulate, the supernatant was applied onto a Superdex 200-pg gel filtration column with the same buffer. The fractions reacted with anti-vammin antiserum by enzyme-linked immunosorbent assay were pooled and then loaded onto a Q Sepharose high performance column with the same buffer. Barietin could not be retained on the anion-exchange column, and the flow-through fractions that reacted with anti-vammin antiserum were pooled and loaded onto a Hi-Trap heparin column with the same buffer. The column was developed with a linear gradient of NaCl (from 0.2 m up to 0.7 m) (supplemental Fig. S4A). The average molecular mass of purified barietin was determined by MALDI-TOF MS with a Voyager-DE (see supplemental Fig. S4B). The N-terminal and internal peptide sequences were analyzed as described previously (5).
Biacore Analysis—Kinetics measurements were performed with a Biacore 3000 SPR biosensor (Uppsala, Sweden). Four recombinant extracellular domains of VEGF receptors/Fc chimeras (R&D Systems, Minneapolis, MN) were immobilized onto the carboxymethylated dextran biosensor surface CM5 by the amine coupling method. Each protein prepared in concentrations of 1–30 nm with HEPES-buffered saline containing EDTA and surfactant P20 (10 mm HEPES, pH 7.4, containing 150 mm NaCl, 3 mm EDTA, and 0.005% surfactant P20) was injected into the flow cells at a flow rate of 20 μl/min. Comparison between sensorgrams was carried out by subtracting the responses in the control flow cell. All kinetic parameters were determined by nonlinear regression analysis using the BIAevaluation version 3.2 software provided by the manufacturer.
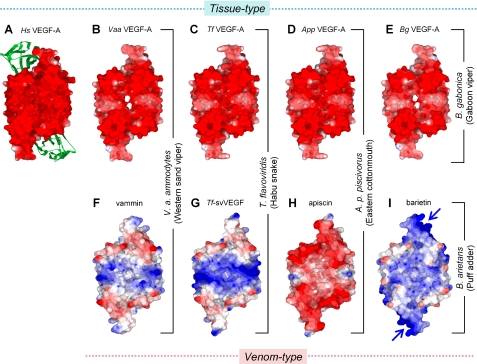
Structural Homology Modeling—The tertiary structures of tissue-type VEGFs of snakes (Vaa-VEGF-A, Tf-VEGF-A, App-VEGF-A, and Bg-VEGF-A) and venom-type VEGFs (Tf-svVEGF, apiscin, and barietin) were constructed by homology modeling based on the crystal structures of Hs-VEGF-A (13) and vammin (9), respectively (see Fig. 5). All structures were constructed with color levels (red, –4.0; white, 0; blue, 4.0) by Swiss PDB Viewer, version 3.7, and drawn by POV-Ray, version 3.6.
FIGURE 5.
Surface electric potential models of tissue- and venom-type VEGFs. A–I, surface electric potential models of tissue-type (A, Hs-VEGF-A; B, Vaa-VEGF-A; C, Tf-VEGF-A, D, App-VEGF-A; E, Bg-VEGF-A) and venom-type (F, vammin; G, Tf-svVEGF; H, apiscin; and I, barietin). Tissue- and venom-type VEGFs were constructed by homology modeling based on the crystal structures of VHDs of Hs-VEGF-A (Glu-13 to Lys-107, panel A) and vammin (Glu-1 to Arg-96, panel F), respectively (9, 23). The green ribbon shows domain 2 of Flt-1 complexed with Hs-VEGF-A (13). The predicted heparin-binding site of barietin is marked by blue arrows. The models were drawn with color levels (red, –4.0; white, 0; blue, 4.0) using the Swiss PDB Viewer. Note that similar surface electric models could be constructed when crystal structures of either vammin or Hs-VEGF-A were used as the template.
RESULTS
Multiple Structures of Venom-type VEGFs—To explore the genetic distribution and diversification of venom-type VEGFs (VEGF-Fs), we screened for venom gland transcripts from 15 venomous snake species using reverse transcription-PCR. Specific amplification was found in the cDNAs from 10 Viperidae snakes but not from four Elapidae snakes and one Colubridae snake. Amplification fragments from V. a. ammodytes and D. r. russelli cDNAs encode vammin and VR-1, respectively. We sequenced three of eight amplified fragments and identified three cDNAs encoding novel venom-type VEGF-like proteins (named barietin, apiscin, and cratrin, from B. arietans, Agkistrodon piscivorus piscivorus, and Crotalus atrox, respectively). Barietin, apiscin, and cratrin cDNAs encode proteins comprising 124–150 amino acids, which show ∼50% identity with human VEGF-A165 (Hs-VEGF-A165) (supplemental Table SI and supplemental Fig. S1). Eight cysteine residues in the VHD, which are predicted to form a cystine knot, are completely conserved in these proteins, whereas the C-terminal region that corresponds to the heparin- and NP-1-binding site of VEGF-A165 (14, 15) is relatively shorter than in VEGF-As and does not include any cysteine residues unlike VEGF-A165 (supplemental Fig. S1B). We next screened venom gland transcripts using different primer sets, which were designed based on the highly conserved sequences among VEGF-As, including several vertebrate species. We identified several transcripts encoding endogenous VEGF-A-like (tissue-type) proteins not only in Viperidae snakes (Vaa-VEGF-A166 and App-VEGF-A166 from V. a. ammodytes and A. p. piscivorus, respectively) but also in Elapidae (Pseudechis australis) and Colubridae (Rhabdophis t. tigrinus) snakes. Unlike the venom-type VEGF-like transcripts described above, all cysteine residues of these tissue-type VEGFs are identical to those of Hs-VEGF-A165, including the C-terminal coreceptor-binding region (supplemental Fig. S1A). From these results, we conclude that Viperidae snakes specifically develop venom-type VEGFs (VEGF-Fs) as toxins separately from endogenous tissue-type VEGFs.
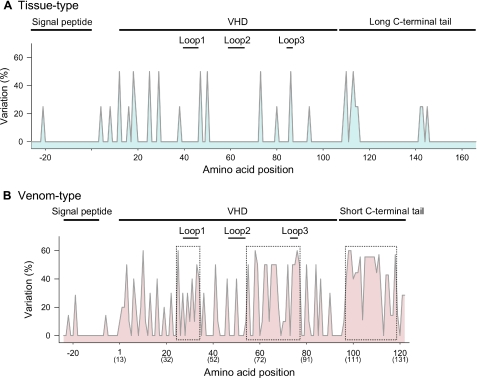
To further understand the development of venom-type VEGFs, we generated a phylogenetic tree based on their VHD sequences (Fig. 1). Venom-type VEGFs (VEGF-Fs) branch separately from tissue-type VEGFs (Fig. 1), and group nearly according to the present venomous snake taxonomy (available through the NCBI Entrez Taxonomy Browser (Serpentes)). Supplemental Table SI shows the sequence identity in VHDs among tissue- and venom-type VEGFs at both the nucleotide and the amino acid levels. Tissue-type VEGFs display high nucleotide and amino acid identities with each other (>∼90%), whereas the amino acids of venom-type VEGFs have far lower identity (<60%) as compared with the nucleotide level (>80%) (supplemental Table SI, colored area). Fig. 2 shows the variation frequency of amino acid positions of tissue- and venom-type VEGFs among snake species. Although significant variation frequency is only observed in a few places in tissue-type VEGFs (Fig. 2A), venom-type VEGFs are highly variegated (Fig. 2B). Variable residues among venom-type VEGFs are particularly observed around receptor-binding loops 1 and 3 and the C-terminal putative coreceptor-binding region (Fig. 2B, dotted boxes), whereas other areas such as the signal peptide region are uniformly conserved. In other words, variations are more pronounced in functionally key regions of mature proteins. This shows that venomous snakes have diversified venom-type VEGFs (VEGF-Fs), especially in functionally key regions, and therefore suggests that their evolutionary process has been distinct from tissue-type VEGFs (VEGF-As).
FIGURE 2.
Variation frequency of amino acid residues of tissue- and venom-type VEGFs among snake species. The variable rate of displaced residues (y axis) was calculated by the following formula: (1 – number of most conserved residues at the position/number of residues at the position) × 100. Highly variable regions of venom-type VEGFs are boxed by dotted lines (B). The amino acid numbers at the bottom (x axis) correspond to supplemental Fig. S1. The parenthesized numbers at the bottom (x axis) in panel B correspond to the residue number of Hs-VEGF-A165.
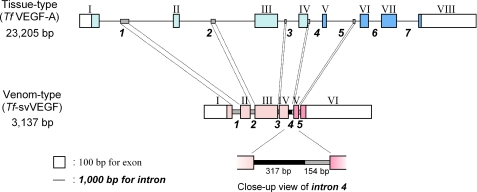
Gene Structures of Tissue- and Venom-type VEGFs—To clarify the genetic development of venom-type VEGFs, we determined the complete gene sequences encoding tissue-type and venom-type VEGFs from T. flavoviridis (Habu snake). The tissue-type VEGF (Tf-VEGF-A) gene is composed of 23,205 bp with eight exons in a structure similar to Hs-VEGF-A, whereas the venom-type VEGF (Tf-svVEGF) gene is composed of 3,137 bp with six exons (Fig. 3, supplemental Fig. S2A and S2B, and supplemental Table SIIA). In both genes, the signal peptide and N-terminal extension regions are encoded in exons I and II, VHD is encoded in exons III and IV, and the C-terminal regions of tissue- and venom-type VEGFs are encoded in exons V–VIII and V–VI, respectively (Fig. 3). The introns of venom-type VEGF gene are significantly shorter than those of the tissue-type gene (385 and 3,762 bp on average, respectively) (supplemental Table SIIA). It is generally known that immature mRNAs with shorter introns are more efficiently possessed to mature forms than those with longer introns, suggesting that the shorter introns of the venom-type gene may be effective for high level expression of venom proteins in the venom glands. In comparing the nucleotide sequences of both genes, high identities were found not only in the exons (45–58%) but also the introns (43–46%) (supplemental Table SIIB and supplemental Fig. S2C), although a 317-bp sequence nonhomologous to the tissue-type VEGF gene was found in intron 4 of the venom-type VEGF gene (Fig. 3 and supplemental Fig. S2B). Interestingly, despite no amino acid sequence homology in the C-terminal regions (encoded in exons V and VI), the nucleotide sequences in exons V and VI of both genes demonstrated ∼50% identity, similar to other exons (supplemental Table SIIB). A sequence alignment of both genes revealed that exons V and VI (C-terminal coreceptor-binding regions) of both genes are translated in frames distinct from each other, resulting in no sequence identity at the amino acid level (Fig. 3 and supplemental Fig. S3). These data suggest that the venom-type VEGF gene developed from an endogenous tissue-type gene but that the unique sequence of the C-terminal region of venom-type VEGF may have been generated by an alteration in the translation frame of tissue-type gene during its evolution.
FIGURE 3.
Genomic structures of tissue-type (Tf-VEGF-A) and venom-type (Tf-svVEGF) genes. Exons and introns are depicted as boxes and lines, respectively. The numbers of exons and introns are shown in Roman and Arabic numerals, respectively. Exons coding open reading frame are colored with cyan (tissue-type) and magenta (venom-type); the C-terminal tail coding exons are in the darker colors. Similar sequences in introns are shown as shaded bars, and an additive sequence in intron 4 of the venom-type gene is shown as a bold bar (close-up view).
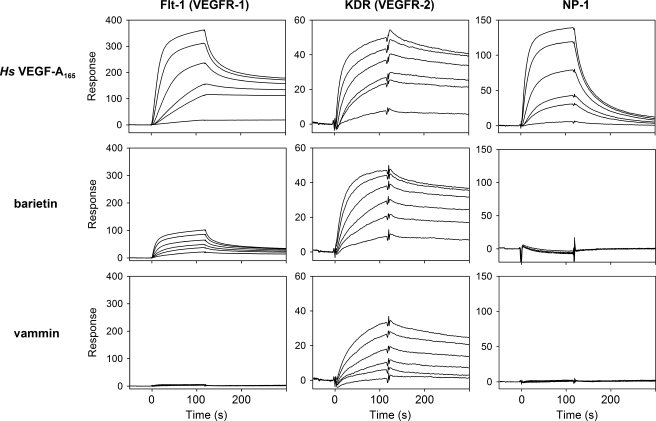
Identification of a Venom-type VEGF with Novel Receptor Selectivity—Among the above mentioned newly cloned venom-type VEGF-like proteins, barietin from B. arietans was predicted to display unique properties; the receptor-binding loop 1 region of barietin is rich in basic amino acid residues rather than acidic residues seen in other known VEGFs. We isolated barietin from the venom of B. arietans by three chromatography steps: gel filtration, anion-exchange, and heparin affinity chromatography (supplemental Fig. S4A). Barietin is a 22-kDa protein under nonreducing conditions and an 11-kDa protein under reducing conditions according to SDS-PAGE (supplemental Fig. S4A), indicating that barietin is a dimeric protein similar to other VEGFs. The N-terminal sequence of purified barietin is EVRPF starting at amino acid 25 from the initial methionine, similar to vammin and VR-1 (supplemental Fig. S1B). The putative mature barietin might be composed of 126 amino acids with a predicted molecular mass of ∼28.2 kDa as homodimer. The average molecular mass of purified barietin was 22,071.6 ± 13.6 as determined by MALDI-TOF MS analysis (supplemental Fig. S4C), an unexpected 6.1 kDa smaller than the molecular mass predicted from its nucleotide sequence. These data suggest that the C-terminal portion barietin is cleaved during maturation. To clarify this, we next performed enzymatic digestion of alkylated mature protein and determined the peptide sequence of barietin. As a result of peptide sequencing and MALDI-TOF MS analyses, the C terminus of mature barietin was found at Ser-97, meaning it almost completely lacks its C-terminal tail (supplemental Fig. S4. (Because it does not include a Lys residue on its C-terminal end in Lys-C digestion, peptide K2 was predicted as the C-terminal peptide. The calculated molecular weight of barietin comprising 97 amino acids is 22,092 as a dimer, which is nearly identical to the determined molecular mass of purified protein.) Because the C-terminal regions of some VEGF isoforms have been shown to interact with heparin/heparan sulfate (supplemental Fig. S1, underline) (14, 16–18), we regarded barietin as having no affinity to heparin; however, barietin was able to bind to a heparin affinity column more tightly than vammin and equally as well as Hs-VEGF-A165 (supplemental Table SIIIA). To obtain detailed biochemical data for barietin, we performed receptor-ligand binding experiments using Biacore. Barietin exhibited a high binding affinity to KDR with a Kd of 4.0 × 10–10 m, an affinity essentially equal to that of vammin and Hs-VEGF-A165. Barietin also bound Flt-1 with 10-fold less affinity than to KDR (Kd = 3.3 × 10–9 m) but did not bind Flt-4 or NP-1 (Fig. 4 and supplemental Table SIIIB). These data indicate that barietin has unique binding properties, unseen in other known VEGFs.
FIGURE 4.
Receptor binding properties of tissue- and venom-type VEGFs. The extracellular domains of Flt-1 (left), KDR (center), and NP-1 (right) were immobilized on a CM5 sensor chip. Several concentrations of Hs-VEGF-A165, barietin, and vammin (1, 3, 6, 10, 20, and 30 nm) were then applied. Binding properties are summarized in supplemental Table SIIIB.
DISCUSSION
Here, we have shown that venom-type VEGFs are efficiently diversified in their structure, especially in functionally key regions such as receptor-binding loops and C-terminal putative coreceptor-binding regions (Fig. 2B). Diversification of the genes encoding snake venom proteins/peptides has been shown to be caused by a mechanism called accelerated evolution (19–21). In this mechanism, nucleotides of venom protein/peptide genes are preferentially substituted in their mature protein coding exons rather than introns, untranslated region, and signal peptide coding exons. In addition, the rate of non-synonymous substitutions (amino acid substitutions, KA) are unusually high as compared with that of synonymous (neutral substitutions, KS) (19–21). The KA and KS values for the pairwise combinations among venom-type VEGFs (VEGF-Fs) and tissue-type VEGFs (VEGF-As) are shown in supplemental Table SIV. When the ratio of KA/KS is larger than 1 (ordinary genes are known to be in the range of 0.1–0.2), it could be assumed that the genes have been subjected to positive selection (22). Although the frequency of KA for the VHD-coding sequence of venom-type VEGFs was 7–18%, much greater than that of tissue-type VEGF (0.5–4%), the ratio of KA/KS of venom-type VEGFs (0.65–0.79) is slightly greater than in tissue-type VEGFs (0.09–0.58), indicating that modest accelerated evolution occurred in the VHD-coding region of venom-type VEGFs. Fig. 5 shows the result of homology modeling of VHDs of several tissue- and venom-type VEGFs constructed based on the crystal structures of Hs-VEGF-A (23) and vammin (9). Surface electric potential models of venom-type VEGFs are variable among the snakes, whereas those of tissue-type VEGFs are uniformly positively charged in a manner similar to Hs-VEGF-A (Fig. 5); these data suggest that the functions of venom-type VEGFs may be extensively variegated, whereas those of tissue-type VEGFs would be universally conserved. In fact, here we have identified a novel venom-type VEGF (named barietin) that shows receptor binding selectivity distinct from other known VEGFs; barietin was able to bind KDR with essentially equal affinity to vammin and Hs-VEGF-A165, able to bind Flt-1 to a lesser degree, and also tightly bound heparin (Fig. 4 and supplemental Table SIII). Fig. 1 shows a phylogenetic tree of VEGFs and their receptor and coreceptor binding potentials. In this tree, venom-type VEGFs can be classified into three groups based on their structure and receptor binding potentials: vammin-type (vammin, VR-1, and HF), which selectively bind KDR and heparin (5), Tf-svVEGF-type (Tf-svVEGF and Pm-VEGF), which bind Flt-1 in preference to KDR and heparin (10, 11), and the barietin-type. These data indicate that viper venoms contain at least three groups of structurally and functionally distinct venom-type VEGFs (VEGF-Fs). Viral VEGF from the genome of Orf-viruses is also known as a multiple exogenous member of the VEGF family (24–26); however, viral VEGFs from more than 20 viral strains show no significant variation in their structures (27) and functions (28) as compared with snake venom-type VEGFs. Considering these facts, venom-type VEGF in snakes is the most strikingly diversified VEGF member.
We here showed that the primary structures of venom-type VEGFs display significantly lower identities (<60%) as compared with the identities at the nucleotide level (>80%), whereas the tissue-type VEGF exhibits high identities at both amino acid and nucleotide levels (supplemental Table SI). A sequence alignment shows that nucleotide sequences of venom-type VEGFs are highly conserved among their cDNAs, despite their lower sequence identity at the amino acid level, especially in receptor-binding loops and the C-terminal putative coreceptor-binding region (supplemental Figs. S1 and S5). In contrast to the modest accelerated evolution observed in VHD (supplemental Table SIV), the diversification in the C-terminal region may be caused by a distinct mechanism; several frameshift mutations generated by insertions/deletions can be seen in exons V and VI (supplemental Fig. S5, highlighted in yellow). For example, distinct stop codons are in-frame in the cDNAs of barietin and apiscin as compared with other venom-type VEGF cDNAs (supplemental Fig. S5). From this aspect, we speculate that the diversity of the C-terminal region of venom-type VEGFs is caused by frameshift mutations in the corresponding exons. Genomic sequence analyses of tissue- and venom-type VEGFs of T. flavoviridis (Tf-svVEGF and Tf-VEGF-A) revealed that an additive sequence not seen in tissue-type VEGF genes is found in intron 4 of the venom-type VEGF gene (Fig. 3 and supplemental Fig. S2B), and consequently, the splicing site of the corresponding exons may be altered (supplemental Fig. S3). An Hs-VEGF-A mutant, in which the C-terminal tail is replaced with that of vammin, fully retained vascular permeability enhancement activities, although another VEGF-A mutant with the C-terminal tail of VEGF-B significantly reduced the activity.4 These mutants show similar receptor selectivity to known VEGF receptors; they could bind Flt-1 and KDR with essentially equal affinity to Hs-VEGF-A165 but not Flt-4, NP-1, and heparin.4 These data strongly suggest that the C-terminal region of snake venom VEGF may affect another undefined molecule in addition to known VEGF receptors.
The C-terminal regions of Hs-VEGF-A165 (Ala-111 to Arg-165) and vammin (Arg-94 to Arg-110) are shown to act as basic heparin-binding domains (supplemental Fig. S1, underlined) (8, 14). The heparin-binding domain of VEGF-A is highly conserved among vertebrate species including humans (supplemental Fig. S1). Despite its binding ability to heparin, barietin apparently does not possess a C-terminal basic tail-like vammin and other heparin-binding VEGFs (supplemental Fig. S1). From homology modeling of the tertiary structure of barietin, it appears that barietin possess highly basic clusters consisting of six residues (Lys-30, Lys-33, Gln-70, Lys-74, Lys-79, and Lys-81; supplemental Fig. S1B, bold letters) around predicted receptor-binding loops (Fig. 5I, arrows), indicating that barietin may form unique heparin-binding site that are not seen in other VEGFs. To further understand the consequence of the unique heparin-binding region of bariein, we tested the effects of heparin on barietin; however, we could not find any apparent effect of heparin on the biochemical and biological activities of barietin, such as its receptor-biding ability and effect on endothelial cell growth (data not shown). Although we did not check the receptor phosphorylation induced by barietin here, the effect of heparin on receptor phosphorylation should be tested in future.
Snake venom proteins are known to be variegated in their structures, resulting in the acquisition of specific and potent functions unseen in related proteins. In this study, we demonstrated that venom-type VEGFs (VEGF-Fs) are diversified in their structures and functions in contrast to endogenous tissue-type VEGFs. We have also shown that venom-type VEGF genes would have developed from tissue-type VEGF genes via a unique mechanism. We believe that these data suggest the existence of a novel mechanism causing the generation of multiple snake toxins.
Supplementary Material
Acknowledgments
We thank Dr. Hideko Atoda for supplying the genome sample from T. flavoviridis. We also thank Yoshihiro Hasegawa for technical assistance.
This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T. M.).
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBank™/EBI Data Bank with accession number(s) FJ554642, FJ554643, FJ554635, FJ554636, FJ554637, FJ554638, FJ554639, FJ554640, FJ554641.
The on-line version of this article (available at http://www.jbc.org) contains five supplemental figures and four supplemental tables.
Footnotes
The abbreviations used are: VEGF, vascular endothelial growth factor; VEGFR, VEGF receptor; VHD, VEGF homology domain; PlGF, placenta growth factor; KDR, kinase insert domain-containing receptor; RTK, receptor tyrosine kinase; NP-1, neuropilin-1; RACE, rapid amplification of cDNA ends; MALDI-TOF MS, matrix-assisted laser desorption/ionization-time of flight mass spectrometry; Hs, Homo sapiens; Tf, Trimeresurus flavoviridis; Vaa, Vipera ammodytes ammodytes; Bg, Bitis gabonica; App, Agkistrodon piscivorus piscivorus; Pm, Protobothrops mucrosquamatus.
Y. Yamazaki, H. Uzuki, and T. Morita, unpublished observation.
References
- 1.Olsson, A. K., Dimberg, A., Kreuger, J., and Claesson-Welsh, L. (2006) Nat. Rev. Mol. Cell Biol. 7 359–371 [DOI] [PubMed] [Google Scholar]
- 2.Yamazaki, Y., and Morita, T. (2006) Mol. Divers. 10 515–527 [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet, P. (2003) Nat. Rev. Genet. 4 710–720 [DOI] [PubMed] [Google Scholar]
- 4.Yamazaki, Y., and Morita, T. (2007) Curr. Pharm. Des. 13 2872–2886 [DOI] [PubMed] [Google Scholar]
- 5.Yamazaki, Y., Takani, K., Atoda, H., and Morita, T. (2003) J. Biol. Chem. 278 51985–51988 [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki, Y., Nakano, Y., Imamura, T., and Morita, T. (2007) Biochem. Biophys. Res. Commun. 355 693–699 [DOI] [PubMed] [Google Scholar]
- 7.Ferrara, N., Gerber, H. P., and LeCouter, J. (2003) Nat. Med. 9 669–676 [DOI] [PubMed] [Google Scholar]
- 8.Yamazaki, Y., Tokunaga, Y., Takani, K., and Morita, T. (2005) Biochemistry 44 8858–8864 [DOI] [PubMed] [Google Scholar]
- 9.Suto, K., Yamazaki, Y., Morita, T., and Mizuno, H. (2005) J. Biol. Chem. 280 2126–2131 [DOI] [PubMed] [Google Scholar]
- 10.Takahashi, H., Hattori, S., Iwamatsu, A., Takizawa, H., and Shibuya, M. (2004) J. Biol. Chem. 279 46304–46314 [DOI] [PubMed] [Google Scholar]
- 11.Chen, Y. L., Tsai, I. H., Hong, T. M., and Tsai, S. H. (2005) Thromb. Haemostasis 93 331–338 [DOI] [PubMed] [Google Scholar]
- 12.Francischetti, I. M., My-Pham, V., Harrison, J., Garfield, M. K., and Ribeiro, J. M. (2004) Gene (Amst.) 337 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiesmann, C., Fuh, G., Christinger, H. W., Eigenbrot, C., Wells, J. A., and de Vos, A. M. (1997) Cell 91 695–704 [DOI] [PubMed] [Google Scholar]
- 14.Keyt, B. A., Berleau, L. T., Nguyen, H. V., Chen, H., Heinsohn, H., Vandlen, R., and Ferrara, N. (1996) J. Biol. Chem. 271 7788–7795 [DOI] [PubMed] [Google Scholar]
- 15.Soker, S., Takashima, S., Miao, H. Q., Neufeld, G., and Klagsbrun, M. (1998) Cell 92 735–745 [DOI] [PubMed] [Google Scholar]
- 16.Makinen, T., Olofsson, B., Karpanen, T., Hellman, U., Soker, S., Klagsbrun, M., Eriksson, U., and Alitalo, K. (1999) J. Biol. Chem. 274 21217–21222 [DOI] [PubMed] [Google Scholar]
- 17.Mamluk, R., Gechtman, Z., Kutcher, M. E., Gasiunas, N., Gallagher, J., and Klagsbrun, M. (2002) J. Biol. Chem. 277 24818–24825 [DOI] [PubMed] [Google Scholar]
- 18.Tokunaga, Y., Yamazaki, Y., and Morita, T. (2006) Biochem. Biophys. Res. Commun. 348 957–962 [DOI] [PubMed] [Google Scholar]
- 19.Ogawa, T., Oda, N., Nakashima, K., Sasaki, H., Hattori, M., Sakaki, Y., Kihara, H., and Ohno, M. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 8557–8561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakashima, K., Ogawa, T., Oda, N., Hattori, M., Sakaki, Y., Kihara, H., and Ohno, M. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 5964–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakashima, K., Nobuhisa, I., Deshimaru, M., Nakai, M., Ogawa, T., Shimohigashi, Y., Fukumaki, Y., Hattori, M., Sakaki, Y., Hattori, S., and Ohno, M. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 5605–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa, T., Chijiwa, T., Oda-Ueda, N., and Ohno, M. (2005) Toxicon 45 1–14 [DOI] [PubMed] [Google Scholar]
- 23.Muller, Y. A., Li, B., Christinger, H. W., Wells, J. A., Cunningham, B. C., and de Vos, A. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 7192–7197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, M., Clauss, M., Lepple-Wienhues, A., Waltenberger, J., Augustin, H. G., Ziche, M., Lanz, C., Buttner, M., Rziha, H. J., and Dehio, C. (1999) EMBO J. 18 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wise, L. M., Veikkola, T., Mercer, A. A., Savory, L. J., Fleming, S. B., Caesar, C., Vitali, A., Makinen, T., Alitalo, K., and Stacker, S. A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3071–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiba, A., Yabana, N., and Shibuya, M. (2003) J. Biol. Chem. 278 13453–13461 [DOI] [PubMed] [Google Scholar]
- 27.Mercer, A. A., Wise, L. M., Scagliarini, A., McInnes, C. J., Buttner, M., Rziha, H. J., McCaughan, C. A., Fleming, S. B., Ueda, N., and Nettleton, P. F. (2002) J. Gen. Virol. 83 2845–2855 [DOI] [PubMed] [Google Scholar]
- 28.Wise, L. M., Ueda, N., Dryden, N. H., Fleming, S. B., Caesar, C., Roufail, S., Achen, M. G., Stacker, S. A., and Mercer, A. A. (2003) J. Biol. Chem. 278 38004–38014 [DOI] [PubMed] [Google Scholar]
- 29.Komori, Y., Nikai, T., Taniguchi, K., Masuda, K., and Sugihara, H. (1999) Biochemistry 38 11796–11803 [DOI] [PubMed] [Google Scholar]
- 30.Gasmi, A., Abidi, F., Srairi, N., Oijatayer, A., Karoui, H., and Elayeb, M. (2000) Biochem. Biophys. Res. Commun. 268 69–72 [DOI] [PubMed] [Google Scholar]
- 31.Kimura, M. (1980) J. Mol. Evol. 16 111–120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.