Abstract
Toll-like receptors (TLRs) play essential roles in the activation of innate immune responses against microbial infections. TLRs and downstream adaptor molecules contain a conserved cytoplasmic TIR domain. TIRAP is a TIR domain-containing adaptor protein that recruits the signaling adaptor MyD88 to a subset of TLRs. Many pathogenic microorganisms subvert TLR signaling pathways to suppress host immune responses to benefit their survival and persistence. Brucella encodes a TIR domain-containing protein (TcpB) that inhibits TLR2- and TLR4-mediated NF-κB activation. Sequence analysis indicated a moderate level of similarity between TcpB and the TLR adaptor molecule TIRAP. We found that TcpB could efficiently block TIRAP-induced NF-κB activation. Subsequent studies revealed that by analogy to TIRAP, TcpB interacts with phosphoinositides through its N-terminal domain and colocalizes with the plasma membrane and components of the cytoskeleton. Our findings suggest that TcpB targets the TIRAP-mediated pathway to subvert TLR signaling. In vivo mouse studies indicated that TcpB-deficient Brucella is defective in systemic spread at the early stages of infection.
Toll-like receptors (TLRs)2 are crucial components of the innate immune system that recognize conserved microbial components and trigger antimicrobial responses (1, 2). The TLR family is characterized by an extracellular domain containing leucine-rich repeats and a cytoplasmic domain, TIR (3). The leucine-rich repeat domain is involved in the recognition of microbial components, whereas the TIR domain creates a signaling platform to recruit adaptor proteins that ultimately activate NF-κB and cytokine response pathways (4, 5). The specificity of TLR-mediated signaling pathways is dictated by the recruitment of a TIR domain-containing adaptor protein to the cytoplasmic domain of TLRs (6). There are five adaptor proteins (MyD88, TIRAP, TRIF, TRAM, and SARM) known to have TIR domains (7–10). MyD88 is the universal adaptor protein recruited by all TLRs except TLR3 (11). TIRAP has been identified as an adaptor for the MyD88-dependent pathway from TLR2 and TLR4 receptors (12–14). TIRAP is primarily localized to the plasma membrane using its phosphatidylinositol 4,5-bisphosphate-binding domain and functions as a sorting adaptor to deliver MyD88 to activated TLR2 and TLR4 (15). TRIF is involved in TLR3 and TLR4 signaling, and TRAM functions in the TLR4-mediated activation of IRF3 (16–18). The fifth adaptor, SARM, negatively regulates TRIF-dependent TLR signaling (9).
Pathogens have evolved many strategies to evade innate immune responses by interfering with TLR signaling to create a replication-permissive environment inside the host (19–22). Brucella species are highly infectious intracellular pathogens causing brucellosis in mammals, including humans (23). Although Brucella does not encode classical virulence factors such as exotoxins or endotoxins, it initiates chronic infection by surviving and replicating inside professional and nonprofessional phagocytic host cells (24, 25). TcpB is a TIR domain-containing protein encoded by chromosome 1 of three Brucella species, viz. Brucella melitensis, Brucella abortus, and Brucella ovis. TcpB, also termed Btp1, impairs TLR2- and TLR4-mediated NF-κB activation and inhibits maturation of dendritic cells (21, 22). Our studies revealed that TcpB mimics properties of the TLR adaptor protein TIRAP. TcpB interacted with phosphoinositides (PtdIns) and colocalized with the plasma membrane and cytoskeleton. Furthermore, TcpB could efficiently block TIRAP-induced NF-κB activation. TcpB likely interferes with the primary function of TIRAP by mimicking its properties to subvert TLR2 and TLR4 signaling.
EXPERIMENTAL PROCEDURES
Overexpression and Purification of TcpB—The gateway-adapted pDEST-15 destination vector (Invitrogen) was used to generate glutathione S-transferase (GST)-tagged TcpB, TcpBTIR, and TcpBΔTIR. GST-tagged TcpBm1, TcpBm2, and TcpBm3 were generated by site-directed mutagenesis on the parental plasmid pGST-TcpB. The proteins were expressed in l-arabinose-inducible BL21-AI cells (Invitrogen) and affinity-purified using immobilized GST resin (Invitrogen).
Site-directed Mutagenesis—Alanine-scanning mutants were generated using the GeneTailor site-directed mutagenesis system (Invitrogen) according to the manufacturer's instructions. The plasmids were sequenced to confirm the desired mutations.
Lipid Binding Assays—Phosphoinositide phosphate (PIP) strips (Echelon Biosciences) were incubated in blocking buffer (10 mm Tris, pH 8.0, 150 mm NaCl, 0.1% Tween 20, and 0.1% ovalbumin) for 1 h and probed with 500 ng of the indicated GST-TcpB fusion proteins for 3 h at 25 °C in the presence of anti-GST monoclonal antibody (Novagen). The strips were then washed with blocking buffer and probed with horseradish peroxidase-conjugated anti-mouse IgG (Pierce) for 30 min in blocking buffer. The bound protein was detected using SuperSignal West Pico chemiluminescent substrate (Pierce). To perform liposome pulldown assays, 20 μl of 1 mm PolyPIPosomes were mixed with 1 μg of the indicated GST-1674 fusion protein in binding buffer (50 mm Tris, pH 8.0, 150 mm NaCl, and 0.05% Nonidet P-40) and rotated for 30 min at 25 °C. The liposomes were pelleted by centrifugation at 13,000 rpm for 10 min and washed three times with binding buffer. The pellets were resuspended in binding buffer and resolved by SDS-PAGE. The bound protein was detected by Western blot analysis.
Cell Culture and Transfections—Human embryonic kidney (HEK) or HeLa cells were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Sigma) and grown in a 37 °C humidified atmosphere of 5% CO2. Transfections were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
Immunostaining and Confocal Microscopy—Transfected cells were cultured on poly-d-lysine-coated glass bottom culture dishes (MatTek Corp.) overnight. Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline for 10 min, incubated with 50 mm NH4Cl for 10 min, and permeabilized in 0.1% Triton X-100 in phosphate-buffered saline. To stain hemagglutinin (HA)- or c-Myc-tagged TcpB, fluorescein isothiocyanate-labeled mouse anti-HA (Sigma) or fluorescein isothiocyanate-labeled monoclonal anti-c-Myc antibody, respectively, was used. To stain microtubule and actin filaments, Cy3-labeled monoclonal mouse anti-β-tubulin antibody (Sigma) and rhodamine-conjugated phalloidin (Invitrogen), respectively, were used. Cells were examined using a Bio-Rad Radiance 2100 MP Rainbow confocal/multiphoton microscope. Sections (0.2–0.5 μm) were captured using a Nikon camera, and z-stacks were projected using Laser Sharp 2000 software. To disrupt the microtubules, cells were treated with 30 μm nocodazole for 30 min after transfection.
Sequence Analysis—Sequence analyses were performed using the FASTA, SSEARCH, and EMBOSS-Align program. Homology modeling was performed based on the crystal structure of human TLR1 using the Geno3D program.
Luciferase Reporter Assay—To analyze the ability of TcpB to block TIRAP-induced NF-κB activation, HEK293 cells were cotransfected with pHA-TIRAP (5 ng) with various amounts of pHA-TcpB (50, 100, and 250 ng), pNF-κB-Luc (50 ng; Stratagene), and pRL-TK (10 ng; Promega). The total amount of DNA was made constant by adding empty vector (pCMV-HA). To compare the NF-κB suppression activity of wild-type and mutant TcpB, HEK-TLR2 cells were cotransfected with 100 ng of pHA-TcpB, pHA-TcpBm3, or pHA-TcpBG158A with the reporter plasmids mentioned above. HEK-TLR2 cells (24 h post-transfection) were induced with the TLR2 ligand FSL-1 (InvivoGen), and luciferase activity was assayed after 12 h using the Dual-Luciferase reporter assay system (Promega).
Lentivirus Transduction of J774 Cells and Tumor Necrosis Factor (TNF) Enzyme-linked Immunosorbent Assay—The gateway-adapted pLenti6/V5 vector (Invitrogen) was used to generate pLenti-TcpB and pLenti-YFP. Viral packaging was performed with the ViraPower lentiviral expression kit (Invitrogen), and transduction of J774 cells was carried out according to the manufacturer's instructions. 48 h post-transduction, cells were induced with FSL-1 for 10 h. The culture supernatants were collected, and TNF levels were estimated using a mouse TNF ELISA kit (BD Biosciences).
Construction of TcpB-deficient B. melitensis and Infection—A virulent, bioluminescent version of B. melitensis (GR023) was used to construct the tcpB mutant as described (26). Experiments with live B. melitensis were performed in Biosafety level 3 containment facilities in accordance with Center for Disease Control and National Institute of Health guidelines. The tcpB gene was deleted by homologous recombination using a pZEroTcpB plasmid. This mutant strain was used for cell and mouse infection. For cell infection, RAW264.7 and HeLa cell lines were plated overnight in 6-well plates and then infected with a multiplicity of infection of 100 bacteria/cell. The numbers of intracellular viable bacteria were determined at different times after inoculation. For in vivo studies, 6–7-week-old female IRF-1–/– mice were infected in the peritoneum with 1 × 107 colony-forming units of B. melitensis GR023 and the tcpB mutant. For imaging, mice were anesthetized with isoflurane, and bioluminescence was recorded after a 10-min exposure using a CCD camera (Xenogen). The images were analyzed using Living Image 3.0 software (Xenogen).
Cytokine Analysis of IRF-1–/– Mice Infected with Wild-type and Knock-out TcpB Brucella—IRF-1–/– mice were infected in the peritoneum with 1 × 107 colony-forming units of B. melitensis GR023 and the tcpB mutant. Spleens were collected after 24 h, and total RNA was isolated. Cytokine RNA levels were quantified using PCR array (SA Biosciences) according to the manufacturer's instructions.
RESULTS
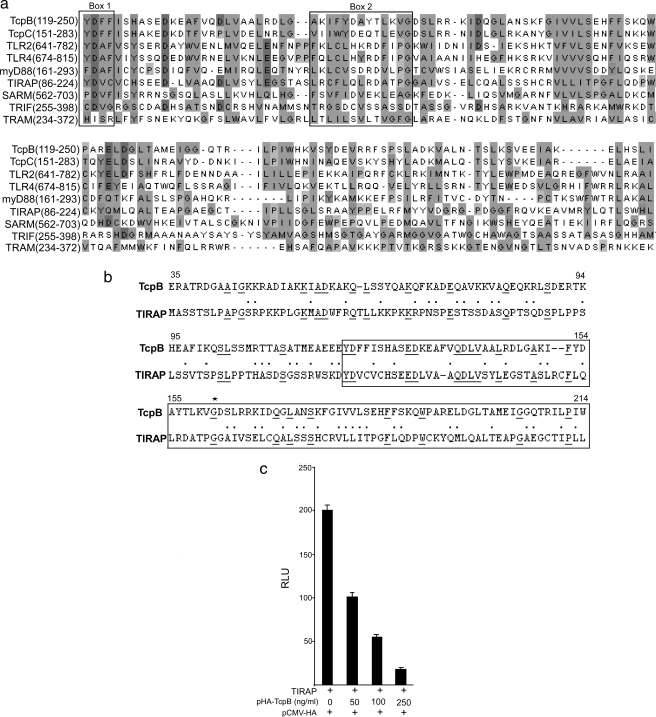
TcpB Shares Sequence Similarity with TIRAP and Inhibits TIRAP-induced NF-κB Activation—The TIR domain region of TcpB displays considerable similarity to other TIR domain-containing proteins of the TLR signaling pathway (Fig. 1a). A sequence similarity search of TcpB against the human complete proteome data base identified a TLR adaptor protein, TIRAP, that shares 20% identity and 53% similarity with TcpB (Fig. 1b). Both TIRAP and TcpB share similar TLR specificity for activation and suppression, respectively. Therefore, we investigated whether TcpB could inhibit TIRAP-mediated NF-κB activation. To determine this, we overexpressed TIRAP in HEK293 cells and analyzed the effect of TcpB on NF-κB activation. The assay indicated that TcpB could efficiently suppress TIRAP-induced NF-κB activation in a dose-dependent manner (Fig. 1c).
FIGURE 1.
a, multiple sequence alignment of TIR domains of TcpB, TcpC, TLR2, TLR4, and five adaptor proteins. Signature motifs (Box 1 and Box 2) of TIR domains are marked. b, pairwise alignment of TcpB (amino acids 35–214) and TIRAP (amino acids 1–180). Identical amino acid sequences are underlined, and similar amino acid residues are represented by a dot. c, TcpB inhibits TIRAP-induced NF-κB activation. HEK293 cells were cotransfected with plasmids encoding TIRAP (5 ng) with increasing concentrations of TcpB (50, 100, and 250 ng), pNF-κB-Luc reporter plasmid (50 ng), and pRL-TK (10 ng). The total amount of DNA was made constant by adding empty vector. Luciferase activity was measured 24 h post-transfection using the Dual-Luciferase reporter assay system. RLU, relative light units.
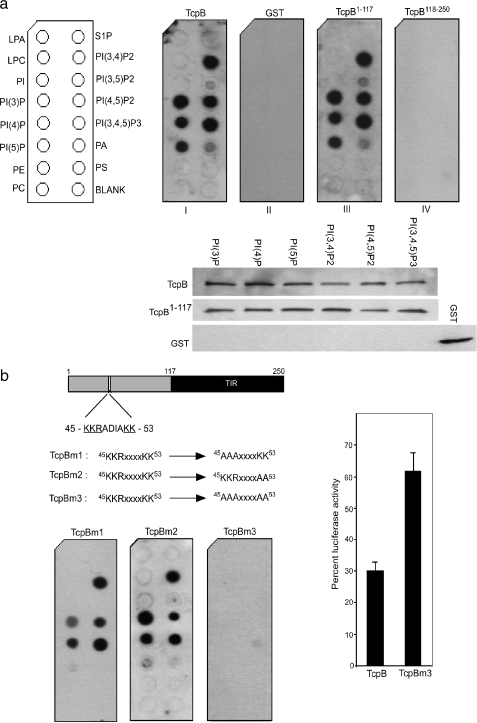
TcpB Mimics Phosphoinositide Binding Properties of TIRAP—To determine whether TcpB mimics the phosphoinositide binding property of TIRAP, we performed in vitro protein-lipid binding assays. TcpB was overexpressed in Escherichia coli as a GST fusion and purified. Nitrocellulose membranes spotted with various purified phospholipids were incubated with the GST-TcpB fusion protein. Binding was detected on the washed membrane using anti-GST antibody to GST-TcpB. Protein overlay assays revealed the interactions of TcpB with PtdIns-3-P, PtdIns-4-P, PtdIns-5-P, PtdIns-3,4-P2, PtdIns-4,5-P2, and PtdIns-3,4,5-P3 (Fig. 2a). We verified the results of protein overlay assays by liposome pulldown. GST-TcpB fusion protein was incubated with PolyPIPosomes. The liposome-protein complexes were pelleted by centrifugation and analyzed by Western blotting (Fig. 2a, lower panel).
FIGURE 2.
TcpB is a PtdIns-binding protein. a: left panel, identity of each lipid spot; right panels, PIP strip incubated with GST-TcpB (I), GST alone (II), TcpB1–117 (III), and TcpB118–250 (IV). Full-length TcpB or TcpB1–117 was capable of binding to PtdIns but not GST or TcpB118–250. Lower panels, confirmation of the TcpB-PtdIns interaction by liposome pulldown assay. The bound protein was detected by Western blot analysis using the anti-GST antibody. LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PI, phosphatidylinositol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine 1-phosphate; PA, phosphatidic acid; PS, phosphatidylserine. b: analysis of PtdIns binding of TcpB mutants. Upper panel, schematic representation of the cationic motif in TcpB and the mutant constructs; lower panels, mutants incubated with a PIP strip to analyze PtdIns binding. TcpBm1 and TcpBm2 displayed a reduced affinity, whereas TcpBm3 was defective in PtdIns binding. Right panel, luciferase reporter assay to determine the NF-κB suppression activity of TcpBm3. HEK-TLR2 cells were transfected with HA-TcpB (100 ng) or HA-TcpBm3 (100 ng) along with the pNF-κB-Luc reporter plasmid (50 ng) and pRL-TK plasmid (10 ng). The total amount of DNA was made constant by adding empty vector (pCMV-HA). 24 h post-transfection, the cells were induced with the TLR2 ligand FSL-1 (250 ng/ml), and luciferase activity was assayed after 12 h. Luciferase activity was expressed as % luciferase activity relative to mock-transfected cells (pCMV-HA).
To determine the amino acid sequence requirements for PtdIns binding, we expressed the N-terminal (amino acids 1–117) and C-terminal (amino acids 118–250) fragments of TcpB in E. coli as GST fusions, purified them, and performed protein-lipid interaction assays. The assays revealed that the N-terminal amino acids 1–117 of TcpB were sufficient for PIP binding, and the lipid interaction was independent of the C-terminal TIR domain-containing region (Fig. 2a). An analysis of the sequences in the lipid-binding region of TcpB indicated a basic amino acid motif (46KKRxxxxKK54). We constructed alanine-scanning mutants to determine the role of this motif in the TcpB-lipid interaction. TcpBm1 (KKR to AAA) and TcpBm2 (KK to AA) were capable of interacting with lipids with a reduced affinity, whereas mutation of the entire motif (TcpBm3) severely abolished lipid binding (Fig. 2b).
TcpB suppresses NF-κB activation as well as TNF secretion induced by a TLR2 ligand, FSL-1 (supplemental Fig. S1). To determine the effect of mutation in the lipid-binding region of TcpB on NF-κB suppression activity, we transfected HEK-TLR2 cells with various concentrations of TcpB or TcpBm3. Transfected cells were stimulated with FSL-1, and NF-κB activation was quantified. The assay indicated that the activity of TcpBm3 was considerably decreased compared with wild-type TcpB (Fig. 2b, right panel). The minimal NF-κB suppression of TcpBm3 is presumably due to the residual PtdIns binding activity of TcpBm3.
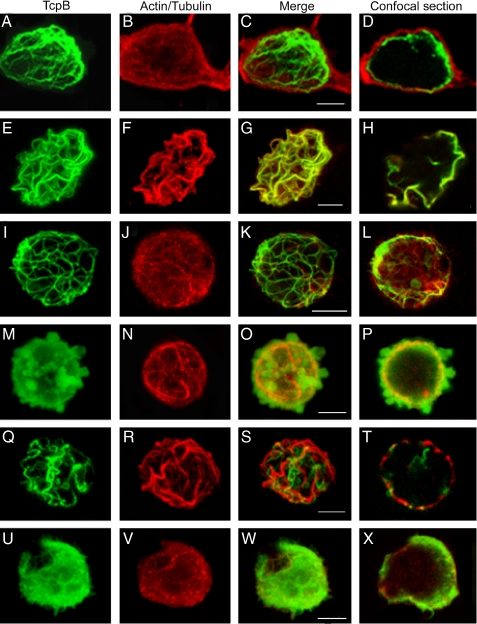
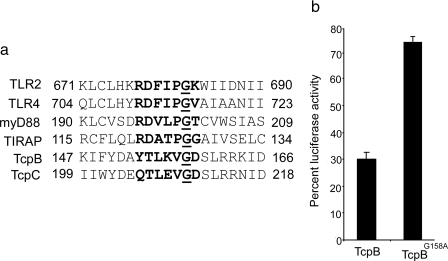
TcpB Colocalizes with Microtubules and Plasma Membrane—To determine the subcellular localization of TcpB, we overexpressed HA-TcpB in HEK cells and stained them with anti-HA monoclonal antibody conjugated with fluorescein isothiocyanate in combination with rhodamine-phalloidin to identify actin filaments or anti-β-tubulin antibody conjugated with Cy3 to identify microtubules. The cells were analyzed by confocal microscopy. TcpB formed tubular networks that appeared to colocalize with microtubules and the plasma membrane (Fig. 3, A–H). Noticeably, TcpB induced severe cell shrinkage, rounding up of the cells, and cytotoxicity. The cytoskeleton appeared thick and condensed. Similar results were obtained with the expression of HA-TcpB or c-Myc-TcpB in HeLa cells (supplemental Figs. S2 and S3). To test the requirement of microtubules for TcpB localization, transfected cells were treated with nocodazole, stained, and analyzed. TcpB-decorated tubules in the cytoplasm appeared partially disrupted after nocodazole treatment (Fig. 3, I–L, and supplemental Fig. S2, D1–D4). To determine the requirement of the TIR domain for characteristic localization of TcpB, we analyzed the expression of HA-TcpBΔTIR (amino acids 1–117) or HA-TcpBTIR (amino acids 118–250) in HEK cells. HA-TcpBΔTIR localized to the plasma membrane and induced dramatic blebs (Fig. 3, M–P, and supplemental Fig. S2, E1–E4), whereas TcpBTIR appeared to form condensed tubules (Fig. 3, Q–T, and supplemental Fig. S2, F1–F4). Plasma membrane localization of HA-TcpBΔTIR confirms the PtdIns binding property of the N-terminal non-TIR region of TcpB. Site-directed mutagenesis was performed in the TIR domain of TcpB to identify the amino acid required for its characteristic localization. Interestingly, mutation of glycine 158 to alanine in the TIR domain of TcpB failed to form tubular networks, and the protein primarily localized to the plasma membrane (Fig. 3, U–X, and supplemental Fig. S2, G1–G4). Sequence analysis indicated that glycine 158 is located in the BB-loop region of TcpB (Fig. 4a). The homology model of the TIR domain of TcpB suggests that glycine 158 is positioned in the loop region (supplemental Fig. S4). Glycine 158 is highly conserved in the BB-loop region (5) of TIR domain-containing proteins, including TIRAP (Fig. 4a). To determine the effect of the glycine → alanine mutation on the activity of TcpB, we performed a NF-κB reporter assay with TcpBG158A. HEK-TLR2 cells were transfected with TcpB or TcpBG158A, and NF-κB activation was quantified after FSL-1 induction. Interestingly, the activity of TcpBG158A was markedly reduced, suggesting the requirement of the intact TIR domain for TcpB-mediated interference with TLR signaling (Fig. 4b).
FIGURE 3.
Subcellular localization of TcpB. HEK cells were transfected with pCMV-HA-TcpB plasmid; stained for HA-TcpB (A and E), actin (B), and tubulin (F); and merged (C and G). D and H are sections of C and G, respectively, to indicate the plasma membrane-bound TcpB. To analyze the requirement of microtubules for TcpB localization, cells were treated with nocodazole and stained for TcpB (I) and tubulin (J) and merged (K), and a section of the projected image (L) is shown. To determine the role of the TIR domain for characteristic localization of TcpB, cells were transfected with HA-TcpBΔTIR (M–P) or HA-TcpBTIR (Q–T), stained for TcpB (M and Q) and tubulin (N and R), merged (O and S), and sectioned (P and T). Images U–X are HEK cells expressing HA-TcpBG158A stained for TcpB (U) or tubulin (V), merged (W), and sectioned (X). Scale bars, 5 μm.
FIGURE 4.
a, alignment of the BB-loop region. Amino acid sequences within the BB-loop region of TLR2, TLR4, MyD88, TIRAP, TcpB, and TcpC were compared. The conserved glycine residue is underlined. b, luciferase reporter assay to determine the NF-κB suppression activity of HA-TcpBG158A. The assay was performed as described in the legend to Fig. 2.
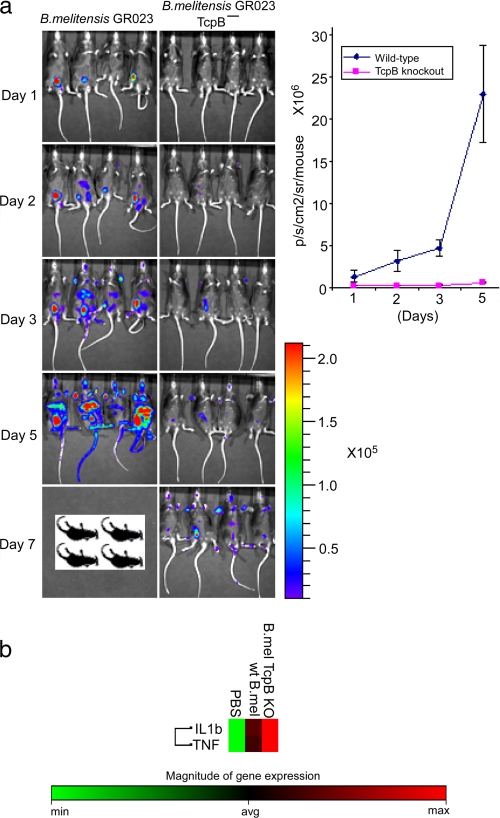
Role of TcpB in the Virulence of B. melitensis—We performed in vitro and in vivo infection studies to compare the intracellular replication and infection dynamics of wild-type and mutant TcpB Brucella. Although a TcpB mutant was not defective in intracellular replication in macrophages or HeLa cells (data not shown), the mutant presented an attenuated phenotype in mice. A bioluminescent version of B. melitensis GR023 (27) was used to generate a TcpB mutant, and the infection dynamics were monitored in real time in IRF-1–/– mice by biophotonic imaging. Infection of mice with wild-type B. melitensis GR023 resulted in systemic spread of bacteria with strong bioluminescence from the areas encompassing liver, spleen, and testis by day 3 and induced death by day 6 (Fig. 5a). In contrast, systemic spread of the TcpB mutant was markedly retarded at the early stages of infection, as only weak bioluminescence was observed until 5 days post-infection. The mutant appeared to gradually overcome the defect and induced the death of infected mice by day 14. We have analyzed the cytokine expression profile of IRF-1–/– mice infected with wild-type and mutant TcpB. Expression of the proinflammatory cytokines TNF and interleukin-β appears to have elevated in TcpB-deficient Brucella compared with the wild type (Fig. 5b).
FIGURE 5.
a, biophotonic imaging of IRF-1–/– mice infected with B. melitensis GR023 or TcpB mutant. The dissemination of bacterial strains was monitored following intraperitoneal inoculation (1 × 107 colony-forming units). Mice were imaged with a 10-min exposure using an in vivo imaging system (Xenogen). The rainbow scale represents approximate photon counts. b, suppression of proinflammatory cytokines by TcpB. IRF-1–/– mice were infected with B. melitensis GR023 or TcpB mutant, and spleens were collected after 24 h. Total RNA was extracted from spleens, and cytokine RNA levels were quantified using PCR array (SA Biosciences). IL1b, interleukin-1β; wt, wild-type; PBS, phosphate-buffered saline; KO, knock-out; min, minimum; avg, average; max, maximum.
DISCUSSION
The subversion of TLR signaling by TIR domain-containing proteins from three pathogenic microorganisms, viz. Salmonella, E. coli, and Brucella, has been characterized (19, 21, 22). All the proteins inhibited NF-κB activation mediated by TLR2 and TLR4. Our sequence analysis revealed that TcpB shares a moderate level of similarity with a TLR adaptor protein, TIRAP. TIRAP acts as a sorting adaptor to recruit MyD88 and mediates downstream signaling of TLR2 and TLR4 receptors (12–15). Interestingly, TIRAP and TcpB share similar TLR specificity to activate and inhibit NF-κB, respectively. TIRAP also plays a major role in dendritic cell maturation (13), whereas TcpB inhibits the maturation of dendritic cells (21). Furthermore, studies have indicated that TIRAP is a critical mediator for host defense against many invasive bacterial pathogens, and its mutation accounts for a high susceptibility to infectious diseases (28–30). On the basis of these observations, we investigated whether TIRAP is the target of TcpB to subvert TLR signaling. TcpB was reported to inhibit NF-κB activation induced by MyD88, IRAK1, and IRAK4 (22). The involvement of MyD88 in TcpB-mediated NF-κB suppression has been reported, but TcpB did not block the NF-κB activation mediated by TLR9, which is also a MyD88-dependent receptor (21). TIRAP functions upstream of MyD88 by delivering MyD88 to membrane-bound TLR2 and TLR4 (15). TcpB is capable of inhibiting TIRAP-induced NF-κB activation, suggesting that TIRAP could be the target of TcpB to inhibit TLR2- and TLR4-mediated NF-κB activation.
Yeast-two hybrid or immunoprecipitation analyses did not indicate an interaction between TcpB and TIRAP. TIRAP is a PtdIns-binding protein and colocalizes with actin at membrane ruffles (15). TcpB mimics the PtdIns binding property of TIRAP, and both proteins interact with PtdIns through a cationic motif at the N terminus. Mutation in the PtdIns-binding domain of TIRAP was reported to have abolished its activity (15). Noticeably, a similar mutation of cationic motifs in TcpB affected NF-κB suppression activity. Therefore, TIRAP and TcpB likely share a similar pathway for NF-κB activation and suppression, respectively. TcpB requires an intact TIR domain for its functional activity, as a point mutation in the BB-loop region of TcpB (TcpBG158A) abolished its NF-κB suppression activity. Similarly, a point mutation in the TIR domain of TIRAP (TIRAPP125H) is reported to abolish the NF-κB activation property of TIRAP (13). TIRAP interacts with TLR2 or TLR4 and facilitates the assembly of the signaling complex. Our analysis did not indicate an interaction between TcpB and TLR2 or TLR4. TcpB and TIR-like proteins from E. coli and Paracoccus were shown to have interacted with MyD88 (22, 31). Because TcpB did not inhibit MyD88-dependent TLR9 signaling, it appears that the PtdIns binding property provides the receptor specificity for TcpB. We speculate that TcpB is targeted to the plasma membrane, where it interferes with the signaling complex formation and blocks downstream activation.
Bacterial TIR-like proteins are important virulent factors that facilitate the survival and persistence of the pathogen. tlpA mutant Salmonella enterica was defective in survival in macrophages and mice (19). Similarly, tcpC of E. coli was reported to be involved in pathogenesis (22). TcpB-deficient Brucella replicated in professional and nonprofessional phagocytes but was attenuated in mice. The slow rate of dissemination and the prolonged survival of infected animals point out that TcpB plays an important role in bacterial virulence, especially at the early stages of the infectious process. In vitro and in vivo studies clearly indicated TcpB-mediated suppression of cytokine levels. Reduced virulence of the TcpB mutant could be attributed to elevated cytokine expression resulting from the inefficient suppression of TLR-mediated innate immune responses.
In conclusion, we have demonstrated that TcpB mimics properties of the TLR adaptor molecule TIRAP and efficiently blocks TIRAP-induced NF-κB activation. TIRAP is required for specific signaling of TLR2 and TLR4, and similar TLR specificity is exhibited by TcpB. Both TIRAP and TcpB contain two distinct domains, an N-terminal PtdIns-binding domain and a C-terminal TIR domain. Point mutations in the PtdIns-binding domain or in the TIR domain abolish function of TIRAP. We have demonstrated that the similar domains are also important for the activity of TcpB. Collectively, our data suggest that TcpB targets the TIRAP-mediated pathway by mimicking TIRAP to inhibit TLR2- and TLR4-mediated signaling.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health/NIAID GLRCE for Biodefense and Emerging Infectious Disease Research Program Grant 1U54-AI-057153 and National Institutes of Health Grant 1R01AI073558. This work was also supported by Binational Agricultural Research and Development Grant US-3829-06 R.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: TLR, toll-like receptor; GST, glutathione S-transferase; HEK, human embryonic kidney; HA, hemagglutinin; TNF, tumor necrosis factor; PIP, phosphoinositide phosphate; PtdIns, phosphatidylinositide/phosphatidylinositol.
References
- 1.Pasare, C., and Medzhitov, R. (2005) Adv. Exp. Med. Biol. 560 11–18 [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783–801 [DOI] [PubMed] [Google Scholar]
- 3.Miggin, S. M., and O'Neill, L. A. (2006) J. Leukocyte Biol. 80 220–226 [DOI] [PubMed] [Google Scholar]
- 4.Takeda, K., Kaisho, T., and Akira, S. (2003) Annu. Rev. Immunol. 21 335–376 [DOI] [PubMed] [Google Scholar]
- 5.Xu, Y., Tao, X., Shen, B., Horng, T., Medzhitov, R., Manley J. L., and Tong, L. (2000) Nature 408 111–115 [DOI] [PubMed] [Google Scholar]
- 6.Akira, S., and Takeda, K. (2004) Nat. Rev. Immunol. 4 499–511 [DOI] [PubMed] [Google Scholar]
- 7.McGettrick, A. F., and O'Neill, L. A. (2004) Mol. Immunol. 41 577–582 [DOI] [PubMed] [Google Scholar]
- 8.Takeda, K., and Akira, S. (2005) Int. Immunol. 17 1–14 [DOI] [PubMed] [Google Scholar]
- 9.Carty, M., Goodbody, R., Schröder, M., Stack, J., Moynagh, P. N., and Bowie, A. G. (2006) Nat. Immunol. 7 1074–1081 [DOI] [PubMed] [Google Scholar]
- 10.O'Neill, L. A., and Bowie, A. G. (2007) Nat. Rev. Immunol. 7 353–364 [DOI] [PubMed] [Google Scholar]
- 11.Janssens, S., and Beyaert, R. (2002) Trends Biochem. Sci. 27 474–482 [DOI] [PubMed] [Google Scholar]
- 12.Horng, T., Barton, G. M., Flavell, R. A., and Medzhitov, R. (2002) Nature 420 329–333 [DOI] [PubMed] [Google Scholar]
- 13.Horng, T., Barton, G. M., and Medzhitov, R. (2001) Nat. Immunol. 2 835–841 [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto, M., Sato, S., Hemmi, H., Sanjo, H., Uematsu, S., Kaisho, T., Hoshino, K., Takeuchi, O., Kobayashi, M., Fujita, T., Takeda, K., and Akira, S. (2002) Nature 420 324–329 [DOI] [PubMed] [Google Scholar]
- 15.Kagan, J., and Medzhitov, R. (2006) Cell 125 943–955 [DOI] [PubMed] [Google Scholar]
- 16.Oshiumi, H., Matsumoto, M., Funami, K., Akazawa, T., and Seya, T. (2003) Nat. Immunol. 4 161–167 [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto, M., Sato, S., Mori, K., Hoshino, K., Takeuchi, O., Takeda, K., and Akira, S. (2002) J. Immunol. 169 6668–6672 [DOI] [PubMed] [Google Scholar]
- 18.Hoebe, K., Du, X., Georgel, P., Janssen, E., Tabeta, K., Kim, S. O., Goode, J., Lin, P., Mann, N., Mudd, S., Crozat, K., Sovath, S., Han, J., and Beutler, B. (2003) Nature 424 743–748 [DOI] [PubMed] [Google Scholar]
- 19.Newman, R. M., Salunkhe, P., Godzik A., and Reed, J. C. (2006) Infect. Immun. 74 594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowie, A. G. (2007) Clin. Exp. Immunol. 147 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salcedo, S. P., Marchesini, M. I., Lelouard, H., Fugier, E., Jolly, G., Balor, S., Muller, A., Lapaque, N., Demaria, O., Alexopoulou, L., Comerci, D. J., Ugalde, R. A., Pierre, P., and Gorvel, J. P. (2008) PLoS Pathog. 4 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cirl, C., Wieser, A., Yadav, M., Duerr, S., Schubert, S., Fischer, H., Stappert, D., Wantia, N., Rodriguez, N., Wagner, H., Svanborg, C., and Miethke, T. (2008) Nat. Med. 14 399–406 [DOI] [PubMed] [Google Scholar]
- 23.Corbel, M. J. (1997) Emerg. Infect. Dis. 3 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pappas, G., Akritidis, N., Bosilkovski, M., and Tsianos, E. (2005) N. Engl. J. Med. 352 2325–2336 [DOI] [PubMed] [Google Scholar]
- 25.Spera, J. M., Ugalde, J. E., Mucci, J., Comerci, D. J., and Ugalde, R. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16514–16519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rambow-Larsen, A. A., Rajashekara, G., Petersen E., and Splitter, G. (2008) J. Bacteriol. 190 3274–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajashekara, G., Glover, D. A., Krepps, M., and Splitter, G. A. (2005) Cell. Microbiol. 7 1459–1473 [DOI] [PubMed] [Google Scholar]
- 28.Khor, C. C., Chapman, S. J., Vannberg, F. O., Dunne, A., Murphy, C., Ling E. Y., Frodsham, A. J., Walley, A. J., Kyrieleis, O., Khan, A., Aucan, C., Segal, S., Moore, C. E., Knox, K., Campbell, S. J., Lienhardt, C., Scott, A., Aaby, P., Sow, O. Y., Grignani, R. T., Sillah, J., Sirugo, G., Peshu, N., Williams, T. N., Maitland, K., Davies, R. J., Kwiatkowski, D. P., Day, N. P., Yala, D., Crook, D. W., Marsh, K., Berkley, J. A., O'Neill, L. A., and Hill, A. V. (2007) Nat. Genet. 39 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeyaseelan, S., Young, S. K., Yamamoto, M., Arndt, P. G., Akira, S., Kolls, J. K., and Worthen, G. S. (2006) J. Immunol. 177 538–547 [DOI] [PubMed] [Google Scholar]
- 30.Hawn, T. R., Dunstan, S. J., Thwaites, G. E., Simmons, C. P., Thuong, N. T., Lan, N. T., Quy, H. T., Chau, T. T., Hieu, N. T., Rodrigues, S., Janer, M., Zhao, L. P., Hien, T. T., Farrar, J. J., and Aderem, A. (2006) J. Infect. Dis. 194 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Low, L. Y., Mukasa, T., Reed, J. C., and Pascual, J. (2007) Biochem. Biophys. Res. Commun. 356 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.