Abstract
The Fas-associated death domain-containing protein (FADD) is an adaptor for relaying apoptotic signals initiated by death receptors such as Fas. Whereas a lack of death receptors has no effect on mouse development, FADD deficiency results in early embryonic lethality, indicating that FADD has additional functions independent of death receptors. We have previously shown that conditional deletion of FADD not only impairs apoptosis but also leads to defective lymphocyte proliferation. The non-apoptotic signaling mediated by FADD remains poorly understood. Earlier studies have suggested that FADD carboxyl terminal serine phosphorylation likely plays a role in FADD-mediated proliferation signaling in T cells. The FADD death domain is presumably only required for apoptotic signaling, as it interacts with death receptors which are dispensable during embryonic development and lymphocyte proliferation. To test this hypothesis, we have performed mutational analyses of the FADD death domain and identified a mutant, R117Q, which lacks binding to Fas and, thus, is incapable of apoptotic signaling in cell lines. Unexpectedly, this death domain point mutation disrupted mouse embryonic development as shown by in vivo functional reconstitution analyses. Interestingly, a second FADD death domain mutant, V121N, retained normal Fas binding and apoptotic signaling ability but also failed to support mouse development. Furthermore, lymphocyte proliferation responses were impaired by V121N. This reverse genetic study has revealed a previously unappreciated role of the FADD death domain, which likely functions as a molecular switch regulating two distinct signals leading to apoptosis and cell proliferation and is critical for embryogenesis, lymphocyte development, and proliferation.
The death domain (DD)3 was initially identified during mutational studies of the pro-apoptotic receptors Fas and TNF-R1, also known as death receptors (DRs) (1, 2). Amino acid replacements targeted at the DD, a stretch of about 80 amino acids within the intracellular sequences of Fas and TNF-R1, abrogate cell death signaling induced by these two DRs. Sequence homology searches led to the identification of additional members of the DR family, including DR3, DR4 (TRAIL-R1), DR5 (TRAIL-R2), and DR6, which also contain an intracellular DD. The physiological function of DRs has been investigated by analyses of humans and animals carrying mutations in the DR genes. Mutations in the DD of Fas lead to lymphoproliferation (lpr) and autoimmune diseases (3, 4) and also result in predisposition to skin and lymphoid malignancy (5–7). TNF-R1-deficient mice are resistant to endotoxic shock but are highly susceptible to pathogens (8, 9). DR3–/– mice have no obvious abnormalities except compromised thymic negative selection (10). DR4/5–/– mice have a normal lymphoid system but show enhanced innate immune responses to viral infection (11). Blocking the binding of TRAIL in mice with soluble DR5 enhances autoimmune inflammation and cell cycle progression in lymphocytes (12). TRAIL–/– mice are susceptible to induced and spontaneous tumors as well as metastasis of engrafted tumors (13–15).
FADD (or Mort1) was initially identified as an adaptor for Fas-induced apoptosis (16–18) and was later shown to be involved in apoptotic pathways initiated by TNF-R1, DR3, and TRAIL-Rs (19–28). The adaptor protein, TRADD, is involved in the TNF-R1 signaling pathway and has been shown to associate with FADD (29). There is a DD-like sequence at the carboxyl proximal region of FADD that interacts with the DDs of DRs and TRADD. FADD contains a second protein interaction structure at the NH2 terminus, called the death effector domain (DED), that binds to the DED present in the pro-domain of caspase 8 (FLICE or MACH) (30, 31). Gene-targeting studies have revealed a novel function of FADD essential for embryonic development (25, 32, 33). FADD–/– and caspase 8–/– mice die midgestation, whereas mice lacking individual DRs have no obvious defects regarding embryonic development. Analyses of conditional mutant mice demonstrated that lymphocytes lacking either FADD or caspase 8 are not only defective in apoptosis induced by Fas but also impaired in proliferative responses induced by the T cell antigen receptor and Toll-like receptors (TLRs) (34–38). Overexpression of a fragment of FADD that contains the DD but lacks the DED blocks both apoptosis and proliferation in T cells (16, 18, 39–41).
During development cell death is required for proper organogenesis and generation of complex multicellular tissues (42, 43). In adults, homeostasis in the lymphoid system and other organs and tissues is controlled in part by induction of cell death (44). The early embryonic lethality phenotype of FADD-deficient mice is intriguing given that mice lacking individual DRs develop normally (8, 9, 11, 45). No embryonic defect or abnormal lymphocyte proliferation responses was reported in double mutant mice containing null alleles of Fas and TNF-R1 (46, 47). Although there is a better understanding of the FADD-mediated apoptosis pathway, very little is known regarding the signaling intermediates and pathways involving FADD in cell proliferation and mouse development. The diverse functions of FADD are likely mediated by its selective interaction with distinct protein partners, including novel ones yet to be identified.
We are interested in dissecting the functional domains in FADD that mediate distinct pathways required for animal development, cell death, and proliferation. In addition to the DD and DED, a functional motif of FADD was proposed to contain a phosphorylated serine residue (Ser-191) at the carboxyl terminus which may play a role in lymphocyte proliferation responses (48). The DD of FADD is clearly essential for apoptosis, as it interacts with the DD of DRs directly or through the TRADD adaptor protein. Structural analyses indicate that the DD of FADD consists of six anti-parallel α-helices, similar to the structure of the DD of Fas (49–52). An initial analysis indicated that a putative Fas binding surface involves six amino acid residues within α-helices two and three of the FADD DD (49). Later, additional residues in α-helices 1, 4, 5, and 6 in the DD of FADD were reportedly involved in mediating binding to Fas (53). To delineate the functional domains of FADD, we have generated mouse FADD DD mutants and performed protein-protein interaction assays in yeast cells and in FADD-deficient (FADD–/–) mouse embryonic fibroblast (MEF) cells. Functional reconstitution analyses were performed by transgenic expression of FADD DD mutants in FADD–/– mice. Taken together, our data from these reverse genetic studies demonstrated that in addition to a role in apoptosis mediated by DRs, the DD of FADD has a non-apoptotic function in vivo which is essential for embryonic development and lymphocyte proliferation.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis and Retrovirus-mediated Gene Expression in FADD–/– Cells—Mutagenesis was performed on a full-length murine FADD cDNA cloned in pSP72 using the QuikChange II kit (Stratagene). The mutant FADD cDNAs were then subcloned into a murine stem cell virus-based CMV-IRES-GFP vector (provided by Dr. W. Sha, University of California at Berkeley). Procedures for the recombinant virus packaging in the 293T cell line and the subsequent mutant protein expression via retroviral transduction in FADD–/– MEFs were performed as previously described (21).
Cell Death Assays—FADD–/– MEFs reconstituted with wild type and mutant FADD were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 μg/ml streptomycin sulfate, and 100 IU/ml penicillin. MEFs were re-seeded into 96-well plates (1.5 × 104 cells per well) in triplicate in Dulbecco's modified Eagle's medium containing 5% fetal calf serum and incubated at 37 °C for 10 h (about 70% confluence). To induce apoptosis, Jo2 anti-Fas antibodies (1 μg/ml, Pharmingen) and cycloheximide (1 μg/ml, Sigma) were added to each well. After 16 h cell death was determined by propidium iodide uptake assays using a flow cytometer (Coulter EPICS XL) as previously described (16). Cell death was induced by a 16-h treatment with mouse recombinant TNFα (100 ng/ml, Genzyme Corp.) and cycloheximide (10 μg/ml) and analyzed by propidium iodide uptake assays.
Immunoprecipitation and Immunoblotting—FADD–/– MEFs (3 × 107) expressing either the wild type or mutant FADD were detached from culture plates by trypsinization and were resuspended in 5 ml of Dulbecco's modified Eagle's medium containing 10% fetal calf serum. These cells were incubated with Jo2 anti-Fas antibodies (10 μg, Pharmingen) for 15 min at 37 °C. Cells were then washed with ice-cold PBS and lysed for 1 h at 4 °C in a lysis buffer containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm dithiothreitol, 1 mm EDTA, 10 mm β-glycerophosphate, 1 mm sodium vanadate, 0.1 mm NaF, 1% Nonidet P-40, 0.2 mm phenylmethylsulfonyl fluoride, and a protease inhibitor mixture (Roche Applied Science). After centrifugal removal of cell debris, additional anti-Fas antibodies were added (1 μg) to the supernatant, and protein complexes were precipitated using protein A-agarose beads (Pierce) by rotational mixing at 4 °C for 3 h. Beads coupled with protein complex were washed four times with ice-cold lysis buffer. Proteins were eluted by boiling in a standard sample buffer for 3 min and fractionated by 12% SDS-PAGE. FADD and caspase 8 associated with Fas were detected with polyclonal anti-FADD antibodies or monoclonal anti-caspase 8 antibodies (1G12, Alexis) by Western blotting procedure.
Genomic Reconstitution of FADD–/– mice—The 12-kb FADD:GFP wild type fusion minigene and the FADD–/– FADD:GFP mice were generated in a previous study (35). To generate the R117Q:GFP and V121N:GFP constructs, site-directed mutagenesis was performed on a 1-kb XhoI fragment containing exon 2 of FADD. The R117Q and V121N mutant fragments were used to replace the 1-kb XhoI fragment on the wild type FADD:GFP minigene construct. The resulting R117Q:GFP and V121N:GFP constructs were injected into wild type C57BL/6 mouse embryos to produce FADD+/+ R117Q:GFP, and FADD+/+ V121N:GFP mice. These mice were then crossed with heterozygous FADD knock-out (FADD+/–) mice, and the resulting FADD+/– R117Q:GFP and FADD+/– V121N:GFP mice were backcrossed with FADD+/– mice to obtain FADD–/– R117Q:GFP and FADD–/– V121N:GFP mice. Offspring were genotyped by Southern blots using genomic DNA isolated from tails. All animal studies were approved by Institutional Animal Care and Use Committee at Thomas Jefferson University.
Embryo Study—Timed mating was performed on crosses between heterozygous FADD knock-out (FADD+/–) mice and FADD+/– R117Q:GFP or FADD+/– V121N:GFP mice. Embryos were isolated at embryonic days 10.5–15.5. Gross morphology was recorded using a stereomicroscope. Genotyping was performed by Southern blotting and PCR using genomic DNA extracted from embryos. Transgene expression was examined by Western blotting using whole cell extract prepared from embryonic tissues.
Fetal Liver Cell Adoptive Transfer—Timed mating was set up as described above. At embryonic days 12.5–15.5, pregnant mice were dissected to surgically remove the uterus. Embryos were isolated using a dissecting microscope. After surgical removal, the fetal liver was disintegrated by pipetting in 1 ml of PBS, and cells were filtered through 50 μm mesh. After centrifugation, cells were resuspended in freezing medium (7% DMSO in fetal bovine serum) and frozen in liquid nitrogen. Genomic DNA was extracted from embryo tissues and used for PCR genotyping as described (35). Proteins were extracted from embryo tissues by 6 passages through a 23-gauge needle in radioimmune precipitation assay buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% deoxycholate, 0.1% SDS, 1 mm EDTA, 1% Nonidet P-40 (Calbiochem), with 1 mm phenylmethylsulfonyl fluoride (Sigma-Aldrich), 0.7 μg/ml pepstatin (Roche Applied Science), and a protease inhibitor mixture (Roche Applied Science). The extracts were subjected to SDS/PAGE, and Western blotting was performed using anti-FADD polyclonal antibodies. After genotypes were determined by PCR or the FADD protein expression was confirmed by Western blotting, fetal liver cells were thawed by incubating at 37 °C for 3 min, immediately re-suspended in PBS, and washed once with PBS. One million fetal liver cells were resuspended in 200 μl of PBS and transferred by tail vein or retroorbital injection into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (NOG, 005557; The Jackson Laboratory) after 200 RAD irradiation as shown previously (54). The resulting chimeras were analyzed 8–14 weeks post-transfer. Lymphocyte proliferation and apoptosis assays were performed as previously described (34, 35).
RESULTS
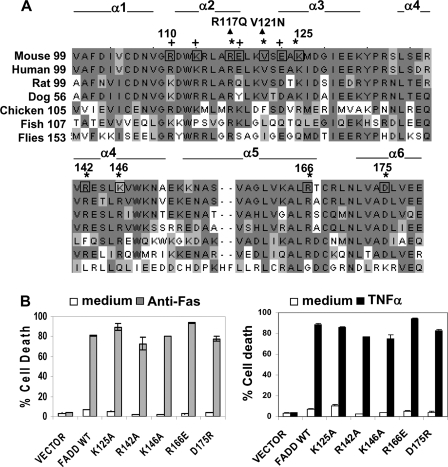
Mutational Analysis of the Death Domain of FADD—We initially sought to identify FADD mutants with selective loss of function to help define the functional domains of FADD. The DD of FADD had previously been targeted for site-directed mutagenesis, as it mediates binding to Fas or TRADD. Cross-species sequence alignments of the FADD DD revealed amino acid residues that are highly conserved from flies to humans (Fig. 1A). It has been indicated that several residues localized within α-helices 2 and 3 of the DD of FADD, as highlighted in Fig. 1A, are involved in mediating FADD binding to Fas and TRADD (49). We have performed yeast two hybrid assays and functional reconstitution analyses using FADD-deficient (FADD–/–) MEF cells and found that mutations at one particular residue, Arg-117, disrupted FADD-Fas and FADD-TRADD interactions and resulted in defective apoptosis induced by Fas or TNFα (21). An expanded Fas binding surface in the DD of FADD was proposed, suggesting that residues including Lys-125, Arg-142, Lys-146, Arg-166, and Asp175 in α-helices 3, 4, 5, and 6 of the DD of FADD were involved in binding of FADD to Fas (Fig. 1A) (53). Therefore, in the current study we performed additional site-directed mutagenesis of the DD of FADD, substituting each of the abovementioned residues with the indicated residues (Fig. 1, A and B). In functional reconstitution assays, the resulting full-length K125A, R142A, K146A, R166E, and D175R FADD mutants were expressed in FADD–/– MEFs lacking the endogenous FADD to determine whether their ability in mediating apoptosis induced by Fas and TNF was affected. To avoid potential side effects caused by transient overexpression, we employed retrovirus-mediated gene transfer to stably express the wild type and the mutant FADD proteins. The wild type FADD WT cDNA was expressed stably in FADD–/– MEFs lacking the endogenous FADD and used as a control (Fig. 1B). FADD–/– MEFs containing the cloning vector were used as the FADD-negative control.
FIGURE 1.
Further mutational analyses of the DD of FADD. A, cross-species sequence alignment of the DD of FADD. Conserved resides are shaded. The residues that were mutagenized in previous (+) (21), and current studies (*) are indicated. Six α-helices (α1–α6) of the DD are defined according to previous studies (49, 50). The R117Q and V121N mutations indicated by arrowheads were tested in mice in the current study. B, the DD mutations K125A, R142A, K146A, R166E, and D175R were generated by site-directed mutagenesis, and the resulting full-length mouse FADD mutants were stably expressed in FADD–/– MEFs that lack the endogenous FADD. FADD–/– MEFs stably transfected with the retroviral vector, murine stem cell virus-CMV-IRES-GFP, or with the viral construct containing WT FADD were used as controls. These reconstituted MEFs were treated with anti-Fas antibodies (left) or TNFα (right) to induce apoptosis. Cell death was determined by propidium iodide staining and flow cytometric analysis at 16 h after stimulation. Untreated cells (medium, unfilled bars) were used as controls.
To induce apoptosis, reconstituted MEFs were treated with an agonistic anti-Fas antibody. As shown in Fig. 1B, FADD–/– control MEFs infected with the retroviral vector were highly resistant to this treatment, whereas FADD WT control MEFs underwent extensive death (∼80%) at 16 h after stimulation with anti-Fas antibodies. Interestingly, MEFs expressing K125A, R142A, K146A, R166E, and D175R mutants of FADD can be killed as efficiently as the FADD WT control by anti-Fas antibodies (left, Fig. 1B). When treated with TNFα, there was 80–90% cell death in MEFs reconstituted with each of the five mutants of FADD, which is similar to cell death induced in the FADD WT control (right, Fig. 1B). These results indicate that the K125A, R142A, K146A, R166E, and D175R mutations of the DD of FADD have no major effect on apoptosis induced by Fas or TNF.
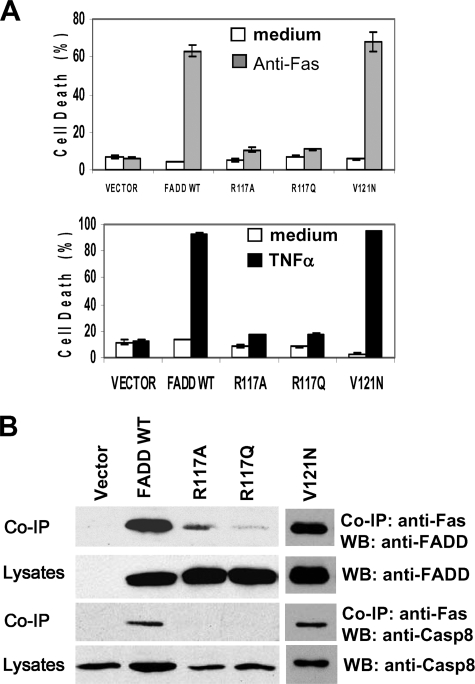
We previously showed that the R117A mutant of FADD is not capable of binding to Fas or TRADD and cannot mediate apoptosis induced by Fas or TNFα (21). In this study a second mutation was introduced into FADD to replace Arg-117 with glutamine, which has a side chain structure similar to that of arginine. The resulting R117Q mutant was stably expressed in FADD–/– MEFs. Like the R117A mutant, R117Q is not functional in mediating apoptosis induced by Fas or TNFα (Fig. 2A). Apoptotic signal-specific protein interaction was determined by co-immunoprecipitation analyses. The Fas binding ability of FADD was diminished dramatically by the R117A or R117Q mutations (Fig. 2B). As expected, signal-specific recruitment of caspase 8 was also disrupted by the R117A and R117Q mutations.
FIGURE 2.
Mutations targeting Arg-117 in the DD disrupted the apoptosis function of FADD. A, Arg-117 was replaced with glutamine, which has a side chain structure similar to that of arginine. The resulting R117Q FADD mutant was stably expressed in FADD–/– MEFs, and apoptosis was induced by anti-Fas antibodies (top) or TNFα (bottom). Similar to MEFs reconstituted with the previously described R117A mutant (21), MEFs expressing the R117Q mutant are highly resistant to apoptosis induced by Fas or TNFα. The Faslpr-like mutant of FADD, V121N, was used as a control, which is functional in apoptosis signaling. B, to detect signal-specific protein interactions, reconstituted MEFs were stimulated with anti-Fas antibodies, and Fas was immunoprecipitated (IP). After SDS/PAGE and blotting, the presence or absence of WT, R117A, R117Q, and V121N mutant FADD proteins as well as Caspase (Casp)8 in the coimmunoprecipitation complex was detected by Western blotting (WB) with anti-FADD or anti-caspase 8 antibodies.
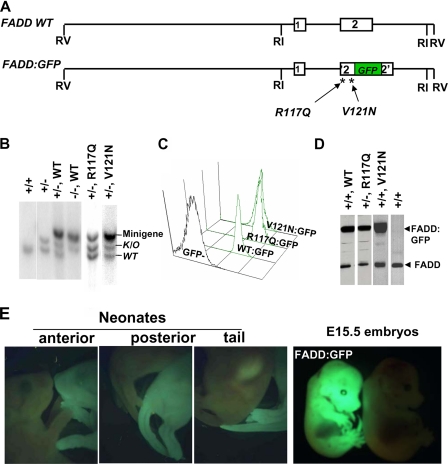
Expression of the FADD DD Mutant Proteins in Mice—To perform in vivo analyses of FADD mutants, we employed a 12-kb minigene containing the wild type mouse FADD gene exon 1 coding for the DED and exon 2 coding for the DD of FADD (FADD WT; Fig. 3A). To facilitate the detection of protein expression, a green fluorescent protein (GFP) tag was fused in-frame at the carboxyl terminus of FADD to generate the FADD:GFP minigene (designated WT:GFP hereafter), as shown previously (35) (Fig. 3A). The R117Q mutation was then introduced to produce the R117Q:GFP mutant minigene (Fig. 3A). This construct was injected into wild type C57BL/6 (B6) mouse embryos, and multiple lines of FADD+/+ R117Q:GFP transgenic mice were produced. A point mutation (lprcs) in the DD of Fas results in a loss of apoptotic function of Fas due to a replacement of isoleucine 125 with asparagine (I125N) (55). It has been suggested that an lprcs-like mutation, V121N, at the junction of α-helices 2 and 3 of FADD might disrupt binding of FADD to Fas (Fig. 1A) (49). In functional reconstitution analyses using FADD–/– MEFs and FADD–/– mice, we found that the V121N mutant protein can bind to Fas and is functional in mediating apoptosis (Fig. 2A) (21). However, V121N appeared to lack a function required for embryonic development, as FADD–/– V121N mice were not viable. It was unclear whether this is due to a lack of expression of the V121N mutant protein in embryos. Therefore, in the current study we fused GFP to V121N as described above for the R117Q construct to facilitate monitoring the FADD mutant protein expression in embryos (Fig. 3A). The resulting V121N:GFP minigene was injected into B6 mouse embryos, and several lines of FADD+/+ V121N:GFP mice were generated. The previously described FADD+/+ WT:GFP transgenic mice were used as controls (35). Genotyping was performed by Southern blotting analyses using mouse tail tissue DNA. The presence of wild type and mutant FADD:GFP fusion minigenes were detected as a 5.4-kb DNA band, distinguishable from the endogenous (WT, 4.7 kb) and the knock-out (K/O, 5.1 kb) alleles of FADD (Fig. 3B).
FIGURE 3.
The physiological impact of FADD DD mutations was determined by functional reconstitution of FADD knock-out (FADD–/–) mice. A, diagrams of the wild type FADD (FADD WT) and FADD:GFP fusion minigenes containing exon 1 and 2. The R117Q and V121N mutations were introduced into FADD:GFP, and the resulting mutant minigenes were injected to B6 embryos to generated transgenic mice. B, genotyping by Southern blotting detected the presence of the endogenous (WT; 4.7 kb) and knock-out (K/O; 5.1 kb) FADD alleles as well as WT and mutant FADD fusion minigenes (5.4 kb). C, WT and mutant FADD:GFP fusion proteins expressed from minigenes were detected by flow cytometry as a single GFP-positive peak in a cytometric histogram. D, Western blotting was performed to detect the WT and mutant FADD proteins (26 kDa) and those fused to GFP (56 kDa). E, ubiquitous expression of the WT and mutant FADD:GFP in whole E15.5 embryos and neonates was visualized by fluorescent stereomicroscopy.
The expression of FADD WT:GFP, R117Q:GFP, and V121N: GFP fusion proteins in FADD+/+ mice were analyzed by flow cytometry. As shown in Fig. 3C, splenocytes and lymph node cells from minigene-containing mice uniformly express wild type and mutant FADD:GFP, as indicated by a single GFP+ peak in the flow cytometric histograms. In Western blot analyses of splenocytes and lymph node cells, the wild type and mutant FADD:GFP proteins were detected as a 56-kDa band, distinct from the 27-kDa endogenous FADD protein (Fig. 3D). The FADD:GFP fusion protein is ubiquitously expressed in embryos and newborn mice, which was readily visualized using a fluorescent stereomicroscopy (Fig. 3E). In vitro overexpression of FADD by transient transfection reportedly resulted in cell death (18). The FADD+/+ FADD WT and FADD+/+ WT:GFP mice are viable and have no abnormality. These results suggest that elevated FADD expression beyond the endogenous level of FADD and the presence of GFP have no major effect on embryonic and postnatal development in mice (data not shown). Mice expressing the mutant FADD proteins (FADD+/+ V121N, FADD+/+ V121N:GFP, and FADD+/+ R117Q:GFP) also have no apparent defects, indicating that the mutant proteins do not interfere with the function of the endogenous FADD. The data to follow are all obtained from mice carrying wild type or mutant FADD fused to GFP. To simplify gene and protein designations, GFP may be omitted hereafter when possible.
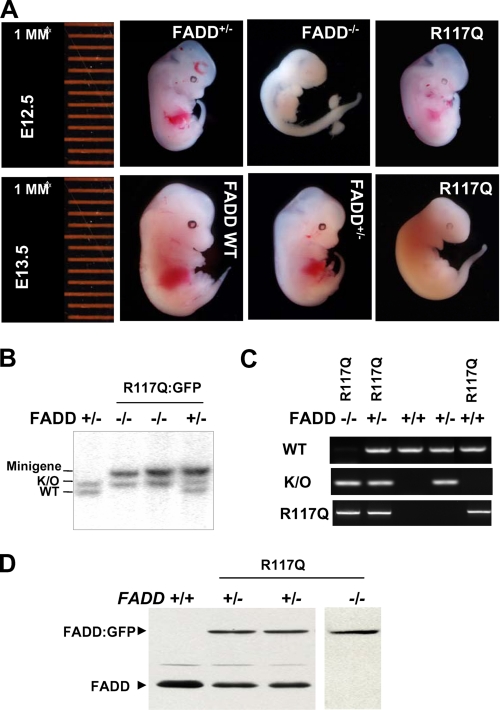
Functional Reconstitution of FADD–/– Mice—The FADD knockout mice (FADD–/–) are embryonic lethal, and this defect can be corrected completely by the presence of the FADD WT minigene in FADD–/– mice (32, 35). The reconstituted FADD–/– WT mice are viable and do not have apparent defects. To determine the physiological impact of the R117Q mutation, we crossed the R117Q mutant minigene into FADD–/– mice. This was achieved by crossing FADD+/+ R117Q mice with FADD+/– mice. The resulting FADD+/– R117Q mice were then crossed with FADD+/– mice, and the offspring were genotyped by Southern blotting and PCR (Fig. 3B and data not shown). Unexpectedly, analyses of multiple lines of mice failed to detect the presence of the FADD–/– R117Q genotype among the 215 postnatal mice genotyped. This result suggests that FADD–/– R117Q died before birth and that the R117Q mutation disrupted a function of FADD essential for embryonic development. Similar analyses were performed to further determine the effect of the V121N mutation by using mice carrying V121N:GFP. The FADD–/– V121N genotype was not present in postnatal mice. These results indicate that the R117Q and V121N mutations of FADD DD result in embryonic lethality.
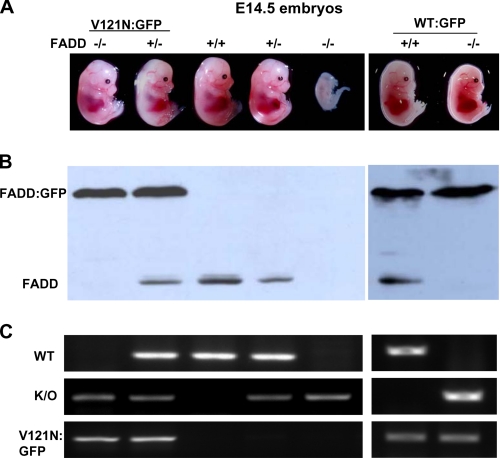
The FADD DD Mutants Failed to Support Normal Ontogenesis—The FADD–/– embryos are present with the expected Mendelian genetic ratio before embryonic day (E) 9.5 and gradually decrease in number thereafter. The few FADD–/– embryos isolated at E12.5 have obvious defects as indicated by the smaller size and a lack of the fetal liver and erythropoiesis. (Fig. 4A). To determine whether the R117Q mutant protein retains a partial function, FADD+/– mice were mated with FADD+/– R117Q mice for timed pregnancy analyses. Southern blotting and PCR were performed to determine the genotypes of embryos of various stages (Fig. 4, B and C). The FADD–/– R117Q embryos were present at day 12.5 (Fig. 4, A and B). The E12.5 R117Q mutant appeared to be normal and similar in size to control FADD WT embryos and contained the developing fetal liver (Fig. 4A). However, the mutant embryos exhibited a slower heart rate compared with control embryos. At E13.5 the R117Q mutant embryos were smaller in size than control FADD WT embryos and had no visible fetal liver or erythropoiesis (Fig. 4A). Therefore, it is evident that the R117Q mutant embryos die between E12.5 and E13.5. To ensure that R117Q mutant protein is expressed during embryogenesis, Western blotting analysis was performed using embryonic tissues. As shown in Fig. 4 D, embryos of the FADD+/– R117Q genotype contain both the endogenous FADD (27 kDa) protein and the fusion protein (56 kDa), whereas embryos of the FADD–/– R117Q genotype express only the fusion protein. Timed pregnancy analyses were also performed with V121N mutant mice. Embryos were isolated and genotyped by Western blotting and PCR (Fig. 5). At E14.5, the FADD–/– V121N embryos appeared normal (Fig. 5A). However, no live FADD–/– V121N embryos were detected after E16.5. These results confirmed that the DD of FADD is essential for mouse development as mutations at either Arg-117 or Val-121 blocked mouse development.
FIGURE 4.
The R117Q mutation resulted in defective embryonic development. A, FADD+/– mice were crossed with FADD+/– R117Q:GFP mice. Pregnancy was timed, and embryos of various developmental stages were isolated for stereomicroscopic analysis. Genotypes of embryos were determined by Southern blotting (B) and PCR (C), proteins were extracted from whole embryos, and wild type and mutant FADD:GFP proteins expression in embryos were confirmed by Western blotting (D). K/O, knock out.
FIGURE 5.
The V121N mutation led to defective embryonic development. A, FADD+/– mice were crossed with FADD+/– V121N:GFP mice. Pregnancy was timed, and embryos of various developmental stages were isolated for stereomicroscopic analysis. The FADD protein expression was determined by Western blotting (B), and embryos were genotyped by PCR (C). K/O, knock out.
The V121N Mutation of FADD Affected Lymphocyte Development—Because of the embryonic lethality phenotype, the impact of R117Q and V121N mutations on lymphocyte development and proliferation responses could not readily be analyzed. Therefore, we performed fetal liver adoptive transfer experiments using NOG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice as hosts. NOG mice lack T cells, B cells, or functional NK cells and are deficient in cytokine signaling, which allows significantly improved engraftment (56). Timed pregnancies were set up, and E12.5 embryos were isolated and genotyped as shown in Fig. 4. The fetal liver was isolated from R117Q mutant embryos and injected into host mice. Although control fetal livers cells isolated from FADD+/– embryos, which contain one allele of the endogenous FADD successfully reconstituted the lymphoid system in NOG hosts, R117Q mutant fetal liver cells failed to do so (data not shown). This indicates that the R117Q mutation resulted in a loss of a FADD function essential for lymphopoiesis.
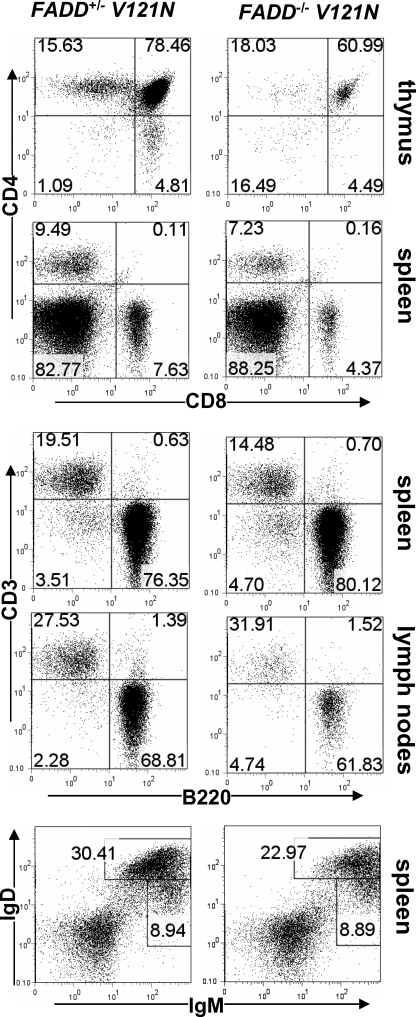
To determine the effect of the V121N mutation on lymphocyte development and functions, adoptive transfer of fetal liver cells was performed with E14.5 embryos. Twelve weeks after adoptive transfer, recipients were sacrificed, and the thymus, spleen, lymph nodes, peripheral blood, and bone marrow were isolated. CD4 and CD8 staining and subsequent flow cytometric analyses revealed that CD4+CD8+ double-positive and CD4+ or CD8+ single-positive T cells were present in the mutant thymus (Fig. 6). However, a 10-fold decrease in total thymocyte numbers was observed in mice reconstituted with FADD–/– V121N fetal liver cells when compared with mice reconstituted with control FADD+/– V121N fetal liver cells (data not shown). Peripheral CD4+ and CD8+ T cells were present in the spleen and lymph nodes of V121N chimeras (Fig. 6 and data not shown). However, the total T cell numbers in the V121N mutant spleen were less than half those in the control spleen (data not shown). This indicates the V121N mutation resulted in reduced generation of T cells from fetal liver precursors. The peripheral T cell and B cell ratios were analyzed by staining spleen and lymph node cells for CD3 and B220. As shown in Fig. 6, the ratios between CD3+ T cells and B220+ B cells were not affected by the V121N mutation. To further analyze peripheral B cells, splenocytes were stained for surface IgD and IgM. The percentages of immature IgM+IgDlo and mature IgD+IgM+ B cells in FADD–/– V121N spleens were not dramatically reduced as compared with those in control FADD+/– V121N mouse spleens (Fig. 6). However, the total cell numbers of B cells were reduced 2-fold in FADD–/– V121N mice, as compared with control FADD+/– V121N mice (data not shown). These results indicate that the V121N mutation leads to compromised generation of peripheral B cells.
FIGURE 6.
The effect of the V121N mutation on peripheral lymphocyte production. Fetal liver cells isolated from FADD+/– V121N control (left) and FADD–/– V121N mutant (right) embryos were adoptively transferred to NOG hosts. Twelve weeks post-transfer, thymic, splenic, and lymph nodes T and B cells were analyzed by staining for CD3, CD4, CD8, B220, IgM, and IgD. Numbers indicate the percentages of each population.
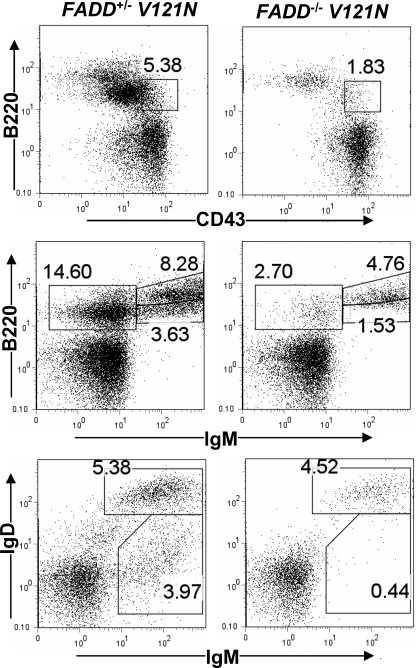
To determine the effect of the V121N mutation on early B cell development, we performed flow cytometric analyses of bone marrow B lineage populations by staining for CD43, B220, IgM, and IgD. In V121N mutant mouse bone marrow, we consistently detected a decrease in the percentages of pro-B cells (B220loCD43+), combined pro- and pre-B cells (B220lo IgM–), immature B cells (B220lo IgM+), and re-circulating mature B cells (B220+IgM+; Fig. 7). Double staining for IgD and IgM also confirmed that the V121N mutation lead to reduced numbers of immature B cells (IgM+IgD–) and re-circulating mature B cells (IgM+IgD+) in the bone marrow (Fig. 7).
FIGURE 7.
Defects in bone marrow B linage cells due to the V121N mutation. Similar to Fig. 6, bone marrow cells were isolated from NOG hosts transferred with FADD+/– V121N control and FADD–/– V121N mutant fetal liver cells. After staining for CD43, B220, IgM, and IgD, bone marrow cells were analyzed by flow cytometry.
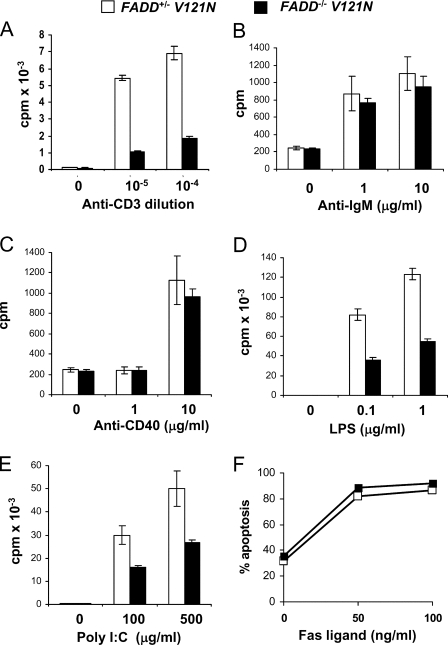
The V121N Mutation Leads to Impaired Proliferation without Affecting Apoptosis in Lymphocytes—Although reduced in number, mature T cells and B cells are present in V121N mutant chimeras. To analyze the effect of the V121N mutation on T cell proliferation responses, peripheral T cells were purified and treated with anti-CD3 antibodies to stimulate the T cell antigen receptor. Proliferation was indicated by the relative amount of [3H]thymidine incorporation. As shown in Fig. 8A, V121N T cells had greatly diminished proliferative responses when compared with control wild type FADD-expressing T cells. Purified peripheral B cells were induced to proliferate by stimulation of the B-cell receptor with anti-IgM antibodies. This assay revealed no significant difference between V121N mutant and control B cells (Fig. 8B). The V121N mutation also did not appear to affect B cell proliferation induced by CD40 stimulation (Fig. 8C). In contrast, lipopolysaccharide-induced proliferation of V121N mutant B cells was impaired as compared with control B cells (Fig. 8D). Similarly, poly(I:C)-induced proliferation was also defective in V121N B cells (Fig. 8E). Poly(I:C) and lipopolysaccharide are ligands for TLR3 and TLR4, respectively. B cells and T cells were stimulated with FasL to induced apoptosis. This response was not impacted by the V121N mutation of FADD (Fig. 8F and data not shown).
FIGURE 8.
The effect of the FADD DD mutation, V121N, on lymphocyte proliferation and apoptosis. Peripheral T and B cells were isolated from NOG hosts transferred with FADD+/– V121N control and FADD–/– V121N mutant fetal liver cells and treated with the stimulants indicated. At 40 h after stimulation, cells were pulsed with [3H]thymidine (A–E). Proliferation was indicated by the amount of incorporated thymidine. At 16 h after stimulation with FasL, apoptosis was determined by propidium staining and flow cytometry (F).
DISCUSSION
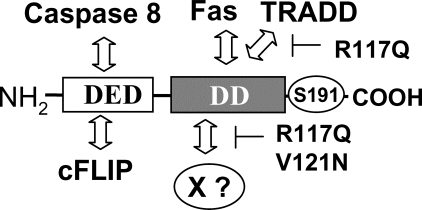
FADD is a common adaptor shared by several DRs including Fas, TNF-R1, and DR4/5 for apoptotic signaling, which plays an essential role in modulating immune responses as well as in tumor surveillance. To help dissect the multiple functions of FADD, we performed site-directed mutagenesis and functional reconstitution to identify domains which mediate distinct signals. To this end the DD of FADD was chosen as the target for mutations, as it has been shown to interact directly with the DDs of Fas and TRADD. Attempts have been made by others to understand the protein interaction interface present in the DD of FADD. Residues including Arg-110, Lys-113, Arg-117, Glu-118, Val-121, Glu-123, Lys-125, Arg-142, Lys-146, Arg-166, and Asp-175 of the DD of FADD were suggested to be essential for binding of FADD to Fas (18, 49, 53). Therefore, mutations targeting these residues presumably render FADD incapable of binding to Fas and TRADD. Among the residues mutagenized in our previous and current studies, Arg-117 appeared to be critical for DR-induced apoptosis, as R117A and R117Q mutations disrupt binding to Fas and TRADD and abolish apoptosis induced by Fas and TNF (Figs. 1 and 2) (21). Therefore, the DD of FADD is critical for mediating apoptosis, and the Arg-117 residue localized within α-helix 2 of the DD of FADD likely represents a point of contact with Fas and TRADD (Fig. 9).
FIGURE 9.
A model for the functional domains of FADD. The DED at the NH2 terminus binds to the DED of caspase 8 and cFLIP. The DD of FADD interacts with the DD of Fas and TRADD, which is blocked by the R117Q mutation. The sequence in the proximity of Arg-117 and V121N may represent an interface contacting a novel protein (X?), which is critical for mouse development. The phosphorylated Ser-191 is indicated.
The function of FADD in embryos is poorly understood. To date it is unclear whether multiple FADD-mediated apoptotic signaling pathways play an indispensable role in mouse embryogenesis. The absence of a single DR has no discernable effect on mouse development (8, 9, 11, 45). Mice lacking both Fas and TNF-R1 have no developmental abnormality (46, 47). Alternatively, a DR-independent function of FADD may be essential for mouse development. Analyses of these possibilities was facilitated by using two FADD DD mutants, R117Q and V121N in functional reconstitution assays in FADD–/– mice (Fig. 3). The FADD:GFP minigene restored normal development in FADD–/– mice lacking the endogenous FADD (35). However, the R117Q mutant only allowed development of FADD–/– embryos to proceed for an additional 2 days (Fig. 4). The R117Q mutant protein is expressed in embryos as indicated by the presence of the fused GFP visualized by fluorescent stereo microscope as well as by Western blotting of embryo extracts (Fig. 4). This result appears to indicate that multiple FADD-dependent apoptotic signaling pathways are required for embryogenesis, which is blocked by the R117Q mutation. However, the Faslpr-like FADD mutant V121N, which retains normal binding to Fas and TRADD and is fully functional in DR-induced apoptosis, was also unable to restore normal embryogenesis in FADD–/– mice (Figs. 2 and 5). Therefore, in total these data argue that the DD of FADD has a dual function that is essential for DR-induced apoptosis as well as for mouse development, which could not be readily uncoupled by point mutations.
A lack of FADD results in developmental blockage at E9 to E10 of gestation. The R117Q mutation results in a nearly complete loss of function mutation, as it leads to death between E12 and E13 and poor development of the fetal liver (Fig. 4). In contrast, the V121N mutant embryos survive up to E15 (Fig. 5), and fetal liver cells were readily isolated, allowing hematopoietic engraftment in immunodeficient NOG host mice. Whereas wild type FADD-expressing fetal liver cells readily reconstituted the lymphoid system of NOG mice, V121N fetal liver progenitors are defective in lymphopoiesis in host mice. This mutation resulted in reduced number of T cells and B cells (Figs. 6 and 7 and data not shown). V121N T cells have reduced proliferation in response to T cell antigen receptor stimulation, and V121N B cells are defective in proliferation induced by the stimulation of TLR3 and TLR4 with double-stranded RNA and lipopolysaccharide, respectively (Fig. 8). These proliferation defects caused by a point mutation in FADD DD are reminiscent of those exhibited by FADD-deficient T cells and B cells (34, 35). However, a lack of FADD or the R117Q mutation resulted in defective apoptosis, whereas the V121N mutation had no effect on lymphocyte apoptosis (Fig. 8F and data not shown). Therefore, using these two hypomorphic mutants of selective loss of function, we demonstrated that the FADD DD has dual functions in DR-induced cell death as well as non-apoptotic signaling essential in lymphocyte development and proliferation.
The DD defines a novel protein-protein interaction motif that is present in pro-apoptotic proteins including DRs and the adaptors FADD and TRADD. Additional proteins including MyD88, IRAK4, and NF-κB also contain DD-like domains but are not known to directly participate in DR-induced apoptosis (57, 58). Overexpression of a fragment of FADD that contains the DD but not the DED blocks apoptosis, possibly by competing with the endogenous FADD for binding to Fas (16, 18). However, this dominant negative FADD mutant also inhibits T cell proliferation (39–41), indicating that the DD of FADD binds to additional proteins (Fig. 9). The Arg-117 residue is located toward the carboxyl terminus of α-helix 2, and V121N is within the turn between α-helix 2 and 3 (Figs. 1 and 9). Our results imply that sequences within the proximity of Arg-117 and Val-121 interact with a novel protein(s), which is essential for unique signaling pathways required during development. Perhaps the DD of FADD acts as a bifunctional switch where FADD could determine whether death or proliferative signals are transmitted depending on upstream signals or cues. The hypomorphic FADD R117Q and V121N mutants will facilitate further analysis of the novel pathways mediated by FADD.
Acknowledgments
We thank Stephen Rosenberg, James Martin, and Tara Robinson for critical reading of this manuscript and the Kimmel Cancer Center Transgenic and Flow Cytometry facilities at Thomas Jefferson University for technical support.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA95454 (NCI). This work was also supported by the W. W. Smith Charitable Trust, CORNCERN Foundation, and Thomas Jefferson University Grants 920011 and 910580 (to J. Z.).
Footnotes
The abbreviations used are: DD, death domain; DR, death receptor; TNF, tumor necrosis factor; TRADD, TNF-R-associated death domain; TRAIL, TNF-related apoptosis-inducing ligand; -R, receptor; DED, death effector domain; FADD, Fas-associated death domain-containing protein; PBS, phosphate-buffered saline; kb, kilobase(s); WT, wild type; TLR, Toll-like receptor; MEF, mouse embryonic fibroblast; GFP, green fluorescent protein.
References
- 1.Tartaglia, L. A., Ayres, T. M., Wong, G. H. W., and Goeddel, D. V. (1993) Cell 74 845–853 [DOI] [PubMed] [Google Scholar]
- 2.Itoh, N., and Nagata, S. (1993) J. Biol. Chem. 268 10932–10937 [PubMed] [Google Scholar]
- 3.Nagata, S., and Golstein, P. (1995) Science 267 1449–1455 [DOI] [PubMed] [Google Scholar]
- 4.Fisher, G. H., Rosenberg, F. J., Straus, S. E., Dale, J. K., Middleton, L. A., Lin, A. Y., Strober, W., Lenardo, M. J., and Puck, J. M. (1995) Cell 81 935–946 [DOI] [PubMed] [Google Scholar]
- 5.Hill, L. L., Ouhtit, A., Loughlin, S. M., Kripke, M. L., Ananthaswamy, H. N., and Owen-Schaub, L. B. (1999) Science 285 898–900 [DOI] [PubMed] [Google Scholar]
- 6.Peng, S. L., Robert, M. E., Hayday, A. C., and Craft, J. (1996) J. Exp. Med. 184 1149–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson, W. F., Giese, T., and Fredrickson, T. N. (1998) J. Exp. Med. 187 1825–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothe, J., Lesslauer, W., Lotscher, H., Lang, Y., Koebel, P., Kontgen, F., Althage, A., Zinkernagel, R., Steinmetz, M., and Bluethmann, H. (1993) Nature 364 798–802 [DOI] [PubMed] [Google Scholar]
- 9.Pfeffer, K., Matsuyama, T., Kundig, T. M., Wakeham, A., Kishihara, K., Shahinian, A., Wiegmann, K., Ohashi, P. S., Kronke, M., and Mak, T. W. (1993) Cell 73 457–467 [DOI] [PubMed] [Google Scholar]
- 10.Wang, E. C. Y., Thern, A., Denzel, A., Kitson, J., Farrow, S. N., and Owen, M. J. (2001) Mol. Cell. Biol. 21 3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diehl, G. E., Yue, H. H., Hsieh, K., Kuang, A. A., Ho, M., Morici, L. A., Lenz, L. L., Cado, D., Riley, L. W., and Winoto, A. (2004) Immunity 21 877–889 [DOI] [PubMed] [Google Scholar]
- 12.Song, K. M., Chen, Y. G., Goke, R., Wilmen, A., Seidel, C., Goke, A., Hilliard, B., and Chen, Y. H. (2000) J. Exp. Med. 191 1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smyth, M. J., Cretney, E., Takeda, K., Wiltrout, R. H., Sedger, L. M., Kayagaki, N., Yagita, H., and Okumura, K. (2001) J. Exp. Med. 193 661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cretney, E., Takeda, K., Yagita, H., Glaccum, M., Peschon, J. J., and Smyth, M. J. (2002) J. Immunol. 168 1356–1361 [DOI] [PubMed] [Google Scholar]
- 15.Zerafa, N., Westwood, J. A., Cretney, E., Mitchell, S., Waring, P., Iezzi, M., and Smyth, M. J. (2005) J. Immunol. 175 5586–5590 [DOI] [PubMed] [Google Scholar]
- 16.Zhang, J., and Winoto, A. (1996) Mol. Cell. Biol. 16 2756–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boldin, M. P., Varfolomeev, E. E., Pancer, Z., Mett, I. L., Camonis, J. H., and Wallach, D. (1995) J. Biol. Chem. 270 7795–7798 [DOI] [PubMed] [Google Scholar]
- 18.Chinnaiyan, A. M., O'Rourke, K., Tewari, M., and Dixit, V. M. (1995) Cell 81 505–512 [DOI] [PubMed] [Google Scholar]
- 19.Juo, P., Woo, M. S., Kuo, C. J., Signorelli, P., Biemann, H. P., Hannun, Y. A., and Blenis, J. (1999) Cell Growth Differ. 10 797–804 [PubMed] [Google Scholar]
- 20.Kuang, A. A., Diehl, G. E., Zhang, J., and Winoto, A. (2000) J. Biol. Chem. 275 25065–25068 [DOI] [PubMed] [Google Scholar]
- 21.Imtiyaz, H. Z., Zhang, Y., and Zhang, J. (2005) J. Biol. Chem. 280 31360–31367 [DOI] [PubMed] [Google Scholar]
- 22.Chinnaiyan, A. M., Tepper, C. G., Seldin, M. F., O'Rourke, K., Kischkel, F. C., Hellbardt, S., Krammer, P. H., Peter, M. E., and Dixit, V. M. (1996) J. Biol. Chem. 271 4961–4965 [DOI] [PubMed] [Google Scholar]
- 23.Chinnaiyan, A. M., O'Rourke, K., Yu, G.-L., Lyons, R. H., Garg, M., Duan, D. R., Xing, L., Gentz, R., Ni, J., and Dixit, V. M. (1996) Science 274 990–992 [DOI] [PubMed] [Google Scholar]
- 24.Chaudhary, P. M., Edby, M., Jasmin, A., Bookwalter, J. M., and Hood, L. (1997) Immunity 7 821–830 [DOI] [PubMed] [Google Scholar]
- 25.Yeh, W.-C., Pompa, J. L., McCurrach, M. E., Shu, H.-B., Elia, A. J., Shahinian, A., Ng, M., Wakeham, A., Khoo, W., Mitchell, K., El-Deiry, W. S., Lowe, S. W., Goeddel, D. V., and Mak, T. W. (1998) Science 279 1954–1958 [DOI] [PubMed] [Google Scholar]
- 26.Bodmer, J.-L., Holler, N., Reynard, S., Vinciguerra, P., Schneider, P., Juo, P., Blenis, J., and Tschopp, J. (2000) Nat. Cell Biol. 2 241–243 [DOI] [PubMed] [Google Scholar]
- 27.Kischkel, F. C., Lawrence, D. A., Chuntharapai, A., Schow, P., Kim, K. J., and Ashkenazi, A. (2000) Immunity 12 611–620 [DOI] [PubMed] [Google Scholar]
- 28.Sprick, M. R., Weigand, M. A., Rieser, E., Rauch, C. T., Juo, P., Blenis, J., Krammer, P. H., and Walczak, H. (2000) Immunity 12 599–609 [DOI] [PubMed] [Google Scholar]
- 29.Hsu, H., Shu, H.-B., Pan, M.-G., and Goeddel, D. V. (1996) Cell 84 299–308 [DOI] [PubMed] [Google Scholar]
- 30.Muzio, M., Chinnaiyan, A. M., Kischkel, F. C., O'Rourke, K., Shevchenko, A., Ni, J., Scaffidi, C., Bretz, J. D., Zhang, M., Gentz, R., Mann, M., Kramer, P. H., Peter, M. E., and Dixit, V. M. (1996) Cell 85 817–827 [DOI] [PubMed] [Google Scholar]
- 31.Boldin, M. P., Goncharov, T. M., Goltsev, Y. V., and Wallach, D. (1996) Cell 85 803–815 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, J., Cado, D., Chen, A., Kabra, N. H., and Winoto, A. (1998) Nature 392 296–300 [DOI] [PubMed] [Google Scholar]
- 33.Varfolomeev, E. E., Schuchmann, M., Luria, V., Chainnilkulchai, N., Beckmann, S. J., Mett, I., Rebrikov, D., Brodianski, V. M., Kemper, O. C., Kollet, O., Lapidot, T., Soffer, D., Sobe, T., Avraham, K. B., Goncharov, T., Holtman, H., Lonai, P., and Wallach, D. (1998) Immunity 9 267–276 [DOI] [PubMed] [Google Scholar]
- 34.Imtiyaz, H. Z., Rosenberg, S., Zhang, Y., Rahman, Z. S., Hou, Y. J., Manser, T., and Zhang, J. (2006) J. Immunol. 176 6852–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Y., Rosenberg, S., Wang, H., Imtiyaz, H. Z., Hou, Y. J., and Zhang, J. (2005) J. Immunol. 175 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salmena, L., Lemmers, B., Hakem, A., Matysiak-Zablocki, E., Murakami, K., Au, P. Y., Berry, D. M., Tamblyn, L., Shehabeldin, A., Migon, E., Wakeham, A., Bouchard, D., Yeh, W. C., McGlade, J. C., Ohashi, P. S., and Hakem, R. (2003) Genes Dev. 17 883–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su, H., Bidere, N., Zheng, L., Cubre, A., Sakai, K., Dale, J., Salmena, L., Hakem, R., Straus, S., and Lenardo, M. (2005) Science 307 1465–1468 [DOI] [PubMed] [Google Scholar]
- 38.Beisner, D. R., Ch'en, I. L., Kolla, R. V., Hoffmann, A., and Hedrick, S. M. (2005) J. Immunol. 175 3469–3473 [DOI] [PubMed] [Google Scholar]
- 39.Walsh, C. M., Wen, B. G., Chinnaiyan, A. M., O'Rourke, K., Dixit, V. M., and Hedrick, S. M. (1998) Immunity 8 439–449 [DOI] [PubMed] [Google Scholar]
- 40.Newton, K., Harris, A. W., Bath, M. L., Smith, K. G. C., and Strasser, A. (1998) EMBO J. 17 706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zornig, M., Hueber, A.-O., and Evan, G. (1998) Curr. Biol. 8 467–470 [DOI] [PubMed] [Google Scholar]
- 42.Meier, P., Finch, A., and Evan, G. (2000) Nature 407 796–801 [DOI] [PubMed] [Google Scholar]
- 43.Vaux, D. L., Haecker, G., and Strasser, A. (1994) Cell 76 777–779 [DOI] [PubMed] [Google Scholar]
- 44.Rathmell, J. C., and Thompson, C. B. (2002) Cell 109 (suppl.) 97–107 [DOI] [PubMed] [Google Scholar]
- 45.Adachi, M., Suematsu, S., Kondo, T., Ogasawara, J., Tanaka, T., Yoshida, N., and Nagata, S. (1995) Nat. Genet. 11 294–300 [DOI] [PubMed] [Google Scholar]
- 46.Zhou, T., Edwards, C. K. r., Yang, P., Wang, Z., Bluethmann, H., and Mountz, J. D. (1996) J. Immunol. 156 2661–2665 [PubMed] [Google Scholar]
- 47.Hildeman, D. A., Mitchell, T., Teague, T. K., Henson, P., Day, B. J., Kappler, J., and Marrack, P. C. (1999) Immunity 10 735–744 [DOI] [PubMed] [Google Scholar]
- 48.Hua, Z. C., Sohn, S. J., Kang, C., Cado, D., and Winoto, A. (2003) Immunity 18 513–521 [DOI] [PubMed] [Google Scholar]
- 49.Jeong, E. J., Bang, S., Lee, T. H., Park, Y. I., Sim, W. S., and Kim, K. S. (1999) J. Biol. Chem. 274 16337–16342 [DOI] [PubMed] [Google Scholar]
- 50.Berglund, H., Olerenshaw, D., Sankar, A., Federwisch, M., McDonald, N. Q., and Driscoll, P. C. (2000) J. Mol. Biol. 302 171–188 [DOI] [PubMed] [Google Scholar]
- 51.Carrington, P. E., Sandu, C., Wei, Y., Hill, J. M., Morisawa, G., Huang, T., Gavathiotis, E., and Werner, M. H. (2006) Mol. Cell 22 599–610 [DOI] [PubMed] [Google Scholar]
- 52.Huang, B., Eberstadt, M., Olejniczak, E. T., Meadows, R. P., and Fesik, S. W. (1996) Nature 384 638–641 [DOI] [PubMed] [Google Scholar]
- 53.Hill, J. M., Morisawa, G., Kim, T., Huang, T., Wei, Y., Wei, Y., and Werner, M. H. (2004) J. Biol. Chem. 279 1474–1481 [DOI] [PubMed] [Google Scholar]
- 54.Kambe, N., Hiramatsu, H., Shimonaka, M., Fujino, H., Nishikomori, R., Heike, T., Ito, M., Kobayashi, K., Ueyama, Y., Matsuyoshi, N., Miyachi, Y., and Nakahata, T. (2004) Blood 103 860–867 [DOI] [PubMed] [Google Scholar]
- 55.Watanabe, F. R., Brannan, C. I., Copeland, N. G., Jenkins, N. A., and Nagata, S. (1992) Nature 356 314–317 [DOI] [PubMed] [Google Scholar]
- 56.Shultz, L. D., Lyons, B. L., Burzenski, L. M., Gott, B., Chen, X., Chaleff, S., Kotb, M., Gillies, S. D., King, M., Mangada, J., Greiner, D. L., and Handgretinger, R. (2005) J. Immunol. 174 6477–6489 [DOI] [PubMed] [Google Scholar]
- 57.Hofmann, K., and Tschopp, J. (1995) FEBS Lett. 371 321–323 [DOI] [PubMed] [Google Scholar]
- 58.Feinstein, E., Kimchi, A., Wallach, D., Boldin, M., and Varfolomeev, E. (1995) Trends Biochem. Sci. 20 342–344 [DOI] [PubMed] [Google Scholar]