Abstract
KshAB (3-Ketosteroid 9α-hydroxylase) is a two-component Rieske oxygenase (RO) in the cholesterol catabolic pathway of Mycobacterium tuberculosis. Although the enzyme has been implicated in pathogenesis, it has largely been characterized by bioinformatics and molecular genetics. Purified KshB, the reductase component, was a monomeric protein containing a plant-type [2Fe-2S] cluster and FAD. KshA, the oxygenase, was a homotrimer containing a Rieske [2Fe-2S] cluster and mononuclear ferrous iron. Of two potential substrates, reconstituted KshAB had twice the specificity for 1,4-androstadiene-3,17-dione as for 4-androstene-3,17-dione. The transformation of both substrates was well coupled to the consumption of O2. Nevertheless, the reactivity of KshAB with O2 was low in the presence of 1,4-androstadiene-3,17-dione, with a kcat/KmO2 of 2450 ± 80 m–1 s–1. The crystallographic structure of KshA, determined to 2.3Å, revealed an overall fold and a head-to-tail subunit arrangement typical of ROs. The central fold of the catalytic domain lacks all insertions found in characterized ROs, consistent with a minimal and perhaps archetypical RO catalytic domain. The structure of KshA is further distinguished by a C-terminal helix, which stabilizes subunit interactions in the functional trimer. Finally, the substrate-binding pocket extends farther into KshA than in other ROs, consistent with the large steroid substrate, and the funnel accessing the active site is differently orientated. This study provides a solid basis for further studies of a key steroid-transforming enzyme of biotechnological and medical importance.
Mycobacterium tuberculosis, arguably the world's most successful pathogen, infects one-third of the human population and has again become a global threat due in part to the emergence of extensively drug-resistant strains (XDR-TB) that are virtually untreatable with current medicines (1). Despite this alarming development, a surprising amount of the pathogen's physiology remains unknown. One recently discovered aspect of the physiology of M. tuberculosis is its cholesterol catabolic pathway (2). Studies of mutants in cholesterol uptake (3) and degradation (4) in various animal models have indicated that cholesterol catabolism is most important during the chronic phase of infection, although the latter study also provided evidence that it occurs from an early stage and contributes to dissemination of the pathogen in the host. Further study of cholesterol catabolism and the pathway enzymes are required to elucidate the precise role of cholesterol catabolism in infection.
The cholesterol catabolic pathway of M. tuberculosis involves degradation of the branched alkyl side chain and the four-ringed steroid nucleus, as occurs in Rhodococcus jostii RHA1, a nonpathogenic, mycolic acid-producing actinomycete (2), although it is unclear whether the order of this degradation is obligatory. Side-chain degradation proceeds via a β-oxidative type process. Degradation of the steroid nucleus is initiated by 3β-hydroxysteroid dehydrogenase, resulting in the formation of 4-cholestene-3-one (5). The successive actions of KstD (3-ketosteroid-1Δ-dehydrogenase), which catalyzes the 1,2-desaturation of ring A, and KshAB (3-ketosteroid 9α-hydroxylase) (Fig. 1) lead to the opening of ring B with concomitant aromatization of ring A (6–9). Ring A is subsequently hydroxylated to yield a catechol and then subject to meta-cleavage (2, 4). Interestingly, M. tuberculosis appears to catabolize ring A completely to CO2 while incorporating the side chain carbon into its lipid pool (3). Finally, the metabolic fate of rings C and D is unclear in M. tuberculosis. Consistent with the role of cholesterol catabolism in pathogenesis, a transposon disruption mutant of kshA strongly attenuated the growth of the pathogen in interferon-γ-activated macrophages, conditions that mimic the immune response (10).
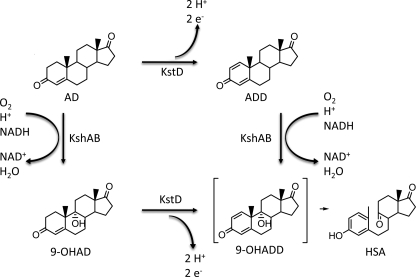
FIGURE 1.
Reactions catalyzed by KshAB in the cholesterol catabolic pathway. Cholesterol is transformed to AD via previous enzymatic steps. KshA catalyzes the 9α-hydroxylation of AD and ADD, yielding 9-OHAD and 9-OHADD, respectively. KstD catalyzes the desaturation of ring A of AD and 9-OHAD to form ADD and 9-OHADD, respectively; the physiological electron acceptor of KstD has yet to be identified. 9-OHADD undergoes a nonenzymatic ring cleavage and aromatization to form HSA.
KshAB of M. tuberculosis is predicted to be a Rieske-type oxygenase (RO)6 comprising a reductase (Rv3571; kshB) and an oxygenase (Rv3526; kshA) (2). Sequence analyses indicate that KshB contains a plant-type [2Fe2S] cluster and a flavin prosthetic group, whereas KshA contains a Rieske-type [2Fe2S] cluster, coordinated by two histidine and two cysteine residues, and a mononuclear iron center, coordinated by two histidines and one aspartate. The latter metallocenter mediates the oxygenation reaction in ROs. Gene disruption studies in Rhodococcus erythropolis SQ1 (6) and Mycobacterium smegmatis (11) have established that KshAB catalyzes the 9α-hydroxylation of 4-androstene-3,17-dione (AD) and 1,4-androstadiene-3,17-dione (ADD) to 9α-hydroxy-4-androstene-3,17-dione (9-OHAD) and 3-hydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (HSA), respectively. However, the substrate specificity of KshAB is unclear, as is the physiological sequence of the reactions catalyzed by this enzyme and KstD.
ROs catalyze a range of reactions in which one or both atoms of O2 are incorporated into an organic substrate in a stereo- and regio-specific manner (12). The reaction cycle requires two reducing equivalents, which originate from NAD(P)H and which are transferred from the reductase to the oxygenase, sometimes via a ferredoxin (12–14). The best characterized ROs, exemplified by naphthalene dioxygenase (NDO9816-4) from Pseudomonas sp. NCIB 9816–4 (15, 16), catalyze the cis-dihydroxylation of aromatic compounds to initiate their aerobic catabolism by bacteria. These ring-hydroxylating dioxygenases have been studied extensively for their potential in bioremediation and as industrial biocatalysts (17, 18). Structural studies have revealed that although the oxygenase component can comprise one or two different subunits, it is always trimeric: α3, α3β3, or(α3)2. In all cases, the α subunit comprises an N-terminal “Rieske” domain harboring the [2Fe-2S] cluster and a larger “catalytic” domain harboring the mononuclear iron (12). The residues coordinating these two metallocenters are conserved. Moreover, the α subunits are arranged head-to-tail within the trimer such that the [2Fe-2S] cluster and the mononuclear iron of adjacent subunits are with within ∼12 Å of each other and comprise the functional unit of the enzyme. An essential aspartate residue bridges the two metallocenters (19) and appears to play a role in preventing the activation of O2 in the absence of substrate (20). Such an uncoupled reaction results in the futile consumption of NADH and production of reactive oxygen species, as has been observed in the presence of poor substrates (21–23). Among better characterized ROs, KshA shares greatest amino acid sequence identity (∼12%) with the oxygenases of 2-oxoquinoline 8-monooxygenase (OMO86) of Pseudomonas putida 86 (20) and carbazole 1,9α-dioxygenase (CARDOJ3) of Janthinobacterium sp. strain J3 (24), for which structural data are available, and phthalate dioxygenase (PDODB01) of Burkholderia cepacia DB01 (25), which has been well characterized kinetically.
We describe herein the characterization of KshAB of M. tuberculosis. Each of the enzyme's two components was heterologously expressed, purified, and used to reconstitute the enzyme's activity. We investigated the substrate specificity of KshAB for two steroid metabolites as well as the enzyme's reactivity with O2. A crystal structure of KshA was solved, enabling structural comparisons with divergent ROs. The data are discussed in terms of the physiological role of KshAB as well as the structure and function of ROs.
MATERIALS AND METHODS
Chemicals and Reagents—ADD was purchased from Steraloids, Inc. (Newport, RI). AD was purchased from Sigma. 2,3-Dihydroxybiphenyl was a gift from Dr. Victor Snieckus. Restriction enzymes and the Expand high fidelity PCR system were purchased from New England Biolabs (Ipswich, MA) and Roche Applied Science (Laval, Quebec, Canada), respectively. Oligonucleotides for amplifying kshA and kshB were purchased from the Nucleic Acid Protein Service Unit at the University of British Columbia. For kshA, the sequences were 5′-GCAATAGCATATGAGTACCGACACGAGTGGGGTCG-3′ and 5′-TCTAAGCTTTTGCTCGGCGGGCACGTCGT-3′. For kshB, the sequences were 5′-CGGAAGGCATATGACCGAGGCAATT-3′ and 5′-GACAAGCTTCTACTCGTCGTAGGTCACT-3′. Jeffamine M600 was purchased from Hampton Research (La Jolla, CA). All other reagents were of HPLC or analytical grade. Water for buffers was purified using a Barnstead Nanopure Diamond™ system (Dubuque, Iowa) to a resistance of at least 18 megaohms.
DNA Manipulation and Plasmid Construction—DNA was propagated, digested, ligated, and transformed using standard protocols (26). DNA plasmids were purified as described previously (27) and were transformed into Escherichia coli by electroporation using a MicroPulser from Bio-Rad with Bio-Rad 0.1-cm GenePulser cuvettes. The kshA (Rv3526) and kshB (Rv3571) genes were amplified from M. tuberculosis H37Rv genomic DNA using polymerase chain reactions containing 0.2 μg of template DNA, 0.9 units of the Expand High Fidelity PCR System polymerase, a 50 μm concentration of each dNTP, and 75 pmol of each oligonucleotide in a volume of 25 μl. Reactions were subject to 25 temperature cycles using a Stratagene Robocycler Gradient 96 instrument (La Jolla, CA) as follows: 95 °C for 45 s, 45 °C for 45 s, and 72 °C for 90 s. The kshA and kshB amplicons were each digested with NdeI and HindIII and ligated into pET41b to yield pETKA1 and pETKB3, respectively. The kshA gene had its stop codon removed such that KshA was produced with a C-terminal His8 tag. Nucleotide sequences were confirmed by the Nucleic Acid Protein Service Unit at the University of British Columbia.
Bacterial Strains and Growth—KshA and KshB were heterologously produced in E. coli BL21(DE3) using pETKA1 and pETKB3. Production of the two components was improved by producing them in cells that also contained pPAISC-1, a vector containing the iron sulfur cluster genes, as described previously (28). Cells were grown in Luria broth supplemented with 25 μg/ml kanamycin, 7.5 μg/ml tetracycline, and an HCl-solubilized solution of minerals (29). One liter of medium inoculated with 10 ml of an overnight culture was incubated at 25 °C and 37 °C for KshA and KshB, respectively. When the OD reached 0.5, isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.5 mm, and cultures were incubated for a further 18 h before harvesting by centrifugation. Pellets were washed twice with 20 mm sodium phosphate, pH 8.0, containing 10% glycerol and frozen at –80 °C until use.
Protein Purification—Chromatography was performed using anÄKTA Explorer (Amersham Biosciences) unless otherwise stated. KshA was O2-labile and was therefore purified anaerobically by interfacing the system to a Labmaster model 100 glove box (M. Braun, Inc., Peabody, MA) operated at <5 ppm O2, as described previously (29). KshB was stable in air-saturated buffer and was therefore purified aerobically.
To purify KshA, cells from 4 liters of culture were resuspended in 30 ml of 20 mm sodium phosphate, pH 8.0, containing 10% glycerol and disrupted by passing the suspension four times through an Emulsiflex-05 homogenizer (Avestin, Ottawa, Canada) operated at 10,000 p.s.i. Ferrous ammonium sulfate (FAS) was added to a final concentration of 0.25 mm after the first pass. The cell debris was removed by ultracentrifugation (10,000 × g for 45 min). The clear supernatant fluid (∼30 ml) was decanted and filtered through a 0.45-μm filter. This raw extract was loaded onto a gravity-operated Ni2+-nitrilotriacetic acid-agarose (Qiagen) column (1.8 × 4 cm) mounted in the glove box and equilibrated with 20 mm sodium phosphate buffer, pH 8.0, containing 10% glycerol. The protein was eluted using an imidazole step gradient according to the instructions of the manufacturer. The brown fraction eluted using 20 mm sodium phosphate buffer containing 150 mm imidazole, pH 8.0, and was exchanged into 25 mm HEPES, pH 7.5, containing 5% glycerol (buffer A) and concentrated to ∼7 ml using a stirred cell concentrator equipped with a YM30 membrane (Amicon, Oakville, Ontario). The solution was brought to 50 mm NaCl. Human α-thrombin (HTI, Essex Junction, VT) was then added to a molar ratio of 1:410 thrombin/KshA, and the solution was incubated at room temperature overnight. The sample was loaded onto a 1 × 10-cm column of Source™15Q (GE Healthcare) resin equilibrated with buffer A containing 1 mm dithiothreitol and 0.25 mm FAS. The enzyme was eluted with a step gradient of NaCl, with KshA eluting at 0.15 m NaCl. Brown-colored fractions were combined, exchanged into buffer A with 1 mm dithiothreitol and 0.25 mm FAS, concentrated to 20–25 mg/ml, and flash frozen as beads in liquid N2.
To purify KshB, cells from 2 liters of culture were resuspended in buffer A, lysed, and filtered as described above, without adding ferrous ammonium sulfate to the lysate. The raw extract was loaded onto a 1 × 10-cm column Source™15Q resin equilibrated with buffer A. KshB was eluted with a linear gradient of 220–360 mm NaCl in 112 ml of buffer A. Orange-colored fractions were combined, exchanged into buffer A, concentrated to 20–25 mg/ml, and flash frozen in liquid N2. Purified KshA and KshB were stored at –80 °C. 2,3-Dihydroxybiphenyl dehydrogenase was prepared as previously described (29).
Analytical Methods—SDS-PAGE was performed using a Bio-Rad MiniPROTEAN III apparatus with a 12% resolving gel. Gels were stained with Coomassie Blue according to standard protocols. Protein concentrations were determined using the Micro BCA™ protein assay kit (Pierce) using bovine serum albumin as a standard. KshA concentrations were routinely determined using ε280 = 142 mm–1 cm–1 and ε324 = 23.2 mm–1 cm–1. Acid-labile sulfur content of samples was determined colorimetrically using the N,N-dimethyl-paraphenylene diamine assay (30). Iron content was determined using the Ferene S assay (31) adapted for the 96-well plate format. Briefly, 80-μl standards containing 5–75 μm FeCl2 and protein samples containing 1–2 nmol of iron were incubated for 10 min with 10 μl of 12 n HCl. Ten μl of 80% trichloroacetic acid was then added, and protein precipitate was removed by centrifugation. Supernatants were added to 20 μl of 45% sodium acetate in a 96-well plate, to which was then added 100 μl of Ferene S reagent (0.75 mm Ferene S, 10 mm l-ascorbic acid, 45% sodium acetate). Absorbances were read at 562 nm using a VMax kinetic microplate reader (Molecular Devices, Sunnyvale, CA). Prior to these analyses, KshA samples were exchanged into 0.1 m potassium phosphate at pH 7.0 by gel filtration chromatography to remove interfering substances. Gas chromatography-coupled mass spectrometry was performed using an HP 6890 series GC system fitted with an HP-5MS 30 m × 250 μm column (Hewlett-Packard, Palo Alto, CA) and an HP 5973 mass-selective detector.
Kinetic Analysis—Enzyme activities were measured by following oxygen consumption using a Clarke-type electrode interfaced to a computer (model 5301; Yellow Springs Instruments, Yellow Springs, OH). The electrode was standardized using 2,3-dihydroxybiphenyl and 2,3-dihydroxybiphenyl dehydrogenase, and initial velocities were calculated from progress curves essentially as described previously (29). The standard activity assay was performed in a total volume of 1.34 ml of air-saturated 0.1 m potassium phosphate, pH 7.0, containing 430 μm NADH, 380 μm ADD, 0.8 μm KshB, and 0.4 μm KshA. The reaction was initiated by adding KshA after equilibration of all other components for 30 s. Reaction velocities were corrected for oxygen consumption observed prior to KshA addition. Stock solutions were prepared fresh daily. KshA was thawed, exchanged into 0.1 m potassium phosphate, pH 7.0, anaerobically using gel filtration chromatography, and stored in a sealed vial on ice. Aliquots were withdrawn as required using gas-tight syringes. One unit of enzyme activity is defined as the amount of enzyme required to consume 1 μmol of substrate/min under the standard assay conditions.
Apparent steady-state kinetic parameters for AD and ADD were determined by measuring rates of O2 consumption in the presence of various concentrations of substrate. Determinations of apparent steady-state kinetic parameters for O2 were measured in the presence of 380 μm ADD. In these experiments, the reaction buffer was equilibrated for 20 min by vigorous bubbling with mixtures of N2 and O2 prior to the reaction, and the reaction cuvette was continually flushed with the same gas mixture during the reaction. Kinetic parameters were evaluated by fitting the Michaelis-Menten equation to the data using the least-squares fitting and dynamic weighting options of LEONORA (32).
HPLC Analysis—Samples were analyzed using a Waters 2695 Separations HPLC module equipped with a Waters 2996 photodiode array detector and a 250 × 4.60-mm C18 Prodigy 10u ODS-Prep column (Phenominex, Torrance, CA). Flavins were identified using the method of Faeder and Siegel (33). Briefly, 5 μl of 400 μm KshB was diluted to 500 μl using 0.1 m potassium phosphate, pH 7.7, containing 0.1 mm EDTA in a light-shielded tube. The mixture was heated at 100 °C for 3 min, rapidly cooled on ice, and then centrifuged at 10,000 × g for 30 min at 4 °C. The supernatant was passed through a 0.2-μm filter, and 100 μl was injected onto the HPLC column equilibrated with aqueous 0.5% phosphoric acid and operated at a flow rate of 1 ml/min. Flavins were eluted using a gradient of 0–100% methanol in 60 ml. Solutions of FMN and FAD were used as standards.
Determination of enzyme coupling using ADD as a substrate was conducted in air-saturated 0.1 m potassium phosphate, pH 7.0, with 1 mm phenylalanine in the presence of 1.3 μm KshA and concentrations of all other components corresponding to those of the standard assay. Coupling experiments using AD as a substrate were performed as above with buffer equilibrated with 80% oxygen. 1 mm phenylalanine was not found to change the rate of reaction with either substrate (results not shown). After 5 min, reactions were quenched by diluting 200 μl of the reaction mixture in 200 μl of methanol. One hundred μl of this mixture was injected onto the column equilibrated with 30% methanol in 0.5% aqueous phosphoric acid. The column was operated at a flow rate of 1 ml/min, and the sample was eluted with a step gradient of methanol, with phenylalanine eluting with 30% methanol and ADD and AD eluting at 80% methanol. A standard curve of the peak area ratios of substrate and phenylalanine was used to quantify the substrate. Oxygen consumption was monitored using the oxygen electrode.
Crystallization—Crystals of KshA were grown aerobically at room temperature (18 °C) using the sitting drop method. Drops contained a 1:1 ratio of 290–400 μm KshA in 25 mm HEPES, pH 7.0, with 1 mm dithiothreitol and 0.25 mm FAS and crystallization solution containing 1.1 m sodium malonate, 0.1 m HEPES, pH 7.0, and 0.5% (v/v) Jeffamine M600. Single dark brown crystals appeared in 3–5 weeks and grew to their full size (200 × 300 × 50 μm) in ∼3 months. Prior to data collection, KshA crystals were serially transferred between solutions of mother liquor supplemented with increasing amounts of ethylene glycol (5–20%) and flash frozen in liquid N2.
X-ray Data Collection and Structure Determination—X-ray data collections were performed under cryogenic conditions using an in-house rotating anode x-ray generator (CuKa radiation λ = 1.542 Å) and at the Canadian Light Source (Beamline CMCF1, λ = 1.000 Å). Data were processed using XDS (34) (Table 1). Substructure solution and initial phasing were performed using the PHENIX program suite (35). The wavelength used (λ = 1.542 Å) and the redundant nature of the data allowed for the location of three iron and six sulfur sites using the HYSS (hybrid substructure search) heavy atom search routine, accounting for one molecule in the asymmetric unit. Initial single wavelength anomalous diffraction phasing was performed using PHASER (36) (as implemented in PHENIX), enabling the calculation of a readily interpretable electron density map. The initial electron density map was then subjected to various cycles of density modification coupled to gradual phase extension and initial backbone tracing using the program Resolve (37). The resulting partial model was iteratively rebuilt using COOT (38), and the structure was refined using REFMAC (39) as well as, for simulated annealing, CNS (40). Electron density maps were calculated using the FFT function of the CCP4 suite (41). The completed model was then refined using the higher resolution data acquired at the Canadian Light Source.
TABLE 1.
Apparent steady-state kinetic parameters of KshAB of M. tuberculosis
Experiments were performed using 0.1 m potassium phosphate, pH 7.0, at 25 °C. Values reported in parentheses represent S.E.
| Substrate | kcat | Km | kcat/Km |
|---|---|---|---|
| s-1 | μm | m-1s-1 | |
| ADD | 0.80 (0.05) | 110 (20) | 7600 (700) |
| AD | 0.07 (0.01) | 24 (16) | 3000 (2000) |
| O2a | >7 | >1200 | 2450 (80) |
Determined in the presence of 380 μm ADD.
Structural Comparisons, Alignments, and Phylogenetic Analysis—Superpositions were calculated using the SSM algorithm in COOT (38). Z scores were calculated using DALI (42). Multiple sequence alignments were generated using STRAP (43) and ClustalX (44). Phylogenetic analyses were performed using PHYLIP (45).
Docking Simulations—ADD was designed using PRODRG (46), energy was minimized using the Dreiding force field (47), and PM6 semiempirical charges were calculated using MOPAC2007 (48). After removing water molecules from the KshA model, docking simulations were performed using PATCHDOCK (49) and Autodock version 4.0 (50) with active site residues held rigid and the initial torsion angles of ADD randomly set. Structural figures and graphics were rendered using PyMOL (51).
RESULTS
Purification of KshAB Components—Purification of KshB from E. coli (pETKB3) by anion exchange chromatography yielded 25 mg of KshB from 1 liter of culture. This material contained 1.7 ± 0.1 mol of iron and 2.0 ± 0.5 mol of sulfur per KshB monomer as determined by the Ferene-S and N,N-dimethyl-paraphenylene diamine assays, respectively. The flavin of KshB eluted from a C18 column with a retention time and spectrum consistent with FAD. The spectrum of oxidized KshB had maxima at 274, 349, and 457 nm (supplemental Fig. 1), consistent with its flavin and [FeS] cluster content. The preparation had an R-value (A274/A349) of 4.6 and an ε349 of 12.0 mm–1 cm–1.
Anaerobic purification of KshA from E. coli (pETKA1) yielded 5 mg of KshA per liter of culture. Over 95% of the protein in the preparation was KshA, as determined by SDS-PAGE analysis. The preparation contained 8.2 ± 0.7 mol of iron and 2.4 ± 0.4 mol of sulfur per mol of KshA protomer. Incubation of KshA for 10 min with 2 mm EDTA, followed by desalting, lowered the iron content to 3.9 ± 0.3 mol/mol of KshA without affecting the specific activity of the latter (results not shown). The spectrum of oxidized KshA had maxima at 324 and 455 nm (supplemental Fig. 1) characteristic of Rieske-type [2Fe-2S] clusters. The preparation had an R-value (A280/A324) of 5.8 and a specific activity of 0.76 ± 0.07 units/mg in the standard assay.
Reconstitution of Activity—Under the conditions of the standard oxygraph assay, the rate of O2 consumption was directly proportional to KshA concentrations from 0.1 to 4.0 μm. KshA was not saturated with KshB, even at a 10-fold excess of KshB over the KshA protomer. Optimal signal/noise ratios were obtained using 0.4 μm KshA and 0.8 μm KshB, concentrations used in subsequent assays. A variant of KshB fused to a C-terminal polyhistidine tag was also tested but resulted in 50% lower specific activity despite containing comparable levels of iron and sulfur as wild-type KshB. Finally, assays conducted over a pH range from 6.5 to 8.0 revealed that although KshAB activity was greater at lower pH, the signal/noise ratio was also significantly lower. Accordingly, assays were performed at pH 7.0. Reaction Products and Coupling—Under the standard assay conditions, KshAB transformed ADD to a single product whose GC retention time and mass spectrum were identical to those of HSA (supplemental Fig. 2). Moreover, HPLC and oxygraph analyses performed on a methanolquenched reaction revealed that 1.02 ± 0.07 mol of O2 were consumed per mol of ADD consumed. Finally, when 650 units of catalase were added to the reaction at a time point corresponding to the methanol quench, no O2 production was detected, indicating that no H2O2 was produced during ADD transformation.
Since the transformation of AD proceeded much more slowly (see below), coupling studies of this reaction were conducted in buffer equilibrated with 80% O2, which increased the reaction rate ∼3-fold. In this reaction, 1.1 ± 0.1 mol of O2 were consumed per mol of AD consumed. Moreover, using catalase, no H2O2 production was detected during the transformation of AD. Taken together, these results indicate that the KshAB-catalyzed transformations of both AD and ADD are well coupled to O2 consumption.
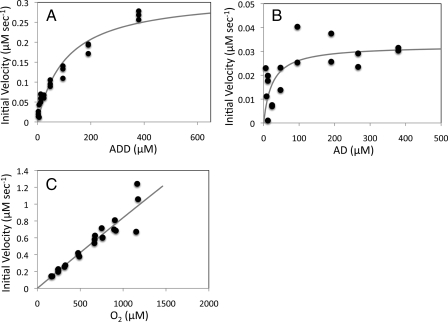
Steady-state Kinetic Analysis—In air-saturated buffer, the initial rates of O2 utilization by KshAB at concentrations of ADD ranging from 3.0 to 380 μm displayed Michaelis-Menten kinetics (Fig. 2A) with the estimated apparent parameters indicated in Table 1. Under the same conditions and over the same range of concentrations of AD, KshAB also appeared to exhibit Michaelis-Menten kinetics (Fig. 2B). However, below 50 μm AD, initial rates of O2 consumption were too low to afford reliable estimates of Km and kcat/Km (Table 1). Nevertheless, the apparent kcat/Km value for ADD was consistently twice that for AD using different preparations of protein and reaction pH.
FIGURE 2.
Steady-state kinetic analyses of KshAB-catalyzed transformations. Shown is the dependence of the initial velocity of O2 consumption on ADD (A) and AD (B) concentrations in air-saturated buffer with fitted parameters Km = 110 ± 20 μm and 24 ± 16 μm, and Vmax = 0.32 ± 0.02 μm s–1 and 0.032 ± 0.006 μm s–1, respectively. C, dependence of the initial velocity of O2 consumption on O2 concentration in the presence of 380 μm ADD. The best fit of the Michaelis-Menten equation to the data using the least squares dynamic weighting options of LEONORA is represented as a solid line.
In the presence of 380 μm ADD, the initial rates of O2 utilization by KshAB depended linearly on the concentration of O2 up to the maximal attainable in this assay, ∼1.2 mm (Fig. 2C), suggesting that the enzyme's apparent KmO2 greatly exceeds this value. Fitting the Michaelis-Menten equation to the data yielded a value for kcat/KmO2 of 2450 ± 80 m–1 s–1. However, the estimates of KmO2 (46 ± 120 nm) and kcat (110 ± 290 s–1 were very poor. Reaction rates with O2 in the presence of AD were slower than those observed in the presence of ADD over a range of O2 concentrations. Therefore, steady state parameters for O2 utilization in the presence of AD were not determined.
Crystal Diffraction Experiments—A dark brown plate-like KshA crystal diffracting over a resolution range of 61 to 2.5 Å on a CuKa rotating anode x-ray source was used for iron/sulfur single wavelength anomalous diffraction structure solution. Data sets from a similar KshA crystal were then obtained using synchrotron light, over a resolution range of 35 to 2.3 Å. The crystals of KshA belonged to the hexagonal space group P321 (Table 2), whose asymmetric unit contained one KshA protomer. Scaling of diffraction data resulted in an Rsym of 0.081. Refinement of the molecular model yielded a structure with Rwork and Rfree values of 0.19 and 0.23, respectively.
TABLE 2.
Crystallographic properties, x-ray diffraction data, phasing, and refinement statistics for KshA
| KshA (Fe/S SAD)a | KshA | |
|---|---|---|
| Diffraction data | ||
| X-ray source | Cu-Kα | Canadian Light |
| Source CMCF1 | ||
| Wavelength (Å) | 1.542 | 1.000 |
| Space group | P321 | P321 |
| Unit cell (Å) | a = b = 116.1, c = 80.8 | a = b = 116.2, c = 81.0 |
| Resolution range (Å) | 61-2.5 | 50-2.3 |
| Highest shell (Å) | 2.5-2.6 | 2.3-2.36 |
| Total observations | 86,930 (1,820) | 204,950 (14,713) |
| Unique reflections | 19,620 (827) | 53,315 (3,841) |
| I/σI | 4.4 (2.2) | 14.6 (3.1) |
| Rsym (%)b | 7.0 (39.1) | 8.1 (43.5) |
| Completeness (%) | 98.6 (97.4) | 98.0 (95.1) |
| Phasing | ||
| Resolution range (Å) | 20-2.6 | |
| No. of used sites | 3 Fe–6 S | |
| Figure of merit | 0.26 | |
| Figure of merit after density modification | 0.62 | |
| Refined model | ||
| Resolution range (Å) | 20-2.3 | |
| No. of reflections | 26,909 | |
| Rfactor/Rfree (%)c | 19/23 | |
| No. of atoms | ||
| Total | 3100 | |
| Protein | 2916 | |
| Solvent | 176 | |
| Mean B values (Å2) | ||
| Protein | 42.2 | |
| Nonheme iron | 36.2 | |
| [2Fe-2S] | 32.1 | |
| rmsd | ||
| Bond lengths (Å) | 0.021 | |
| Bond angles (degrees) | 2.42 |
Iron/sulfur single wavelength anomalous diffraction.
Rsym = ΣhΣi I(hkl) – 〈I(hkl)〉/ΣhΣiI(hkl).
Rwork = Σ∥Fo| – |Fc∥/Σ|Fo|. Rfree is the Rwork value for 5% of the reflections excluded from the refinement. Data for the highest resolution shell are given in parentheses.
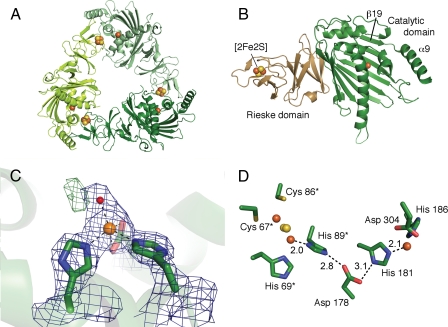
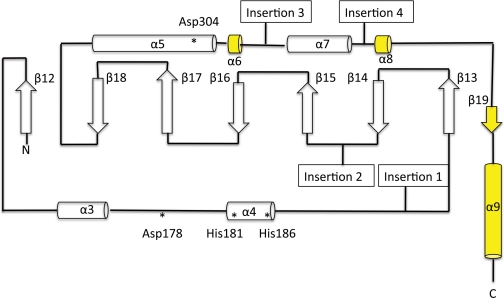
Overall Structure—The refined structure of KshA includes residues 14–374, three iron ions, two acid-labile sulfur atoms, and 176 water molecules (Table 2 and Fig. 3). The model does not include the N- and C-terminal residues (1–13 and 375–386) as well as residues 283–284, for which electron density was not visible. A Ramachandran plot confirms that 99% of the residues have favored configurations. The KshA protomer and its quaternary structure are similar to those of nine structurally characterized ROs (see the legend to supplemental Fig. 4 for Protein Data Bank accession numbers). Thus, KshA comprises an N-terminal Rieske domain harboring the [2Fe-2S] cluster followed by a larger catalytic domain harboring the non-heme mononuclear iron (Fig. 3A). Three symmetry-related subunits interact in a head-to-tail fashion to form the functional enzyme (Fig. 3B). The KshA trimer measures 90 Å at its greatest width, with a conical central pore of 20–30 Å in diameter. Superpositions revealed that the KshA protomer is more similar structurally to the α3 ROs (Cα rmsd values of 2.8 Å for each of CARDOJ3 and OMO86 over 250 and 258 αC atoms, respectively) than to the α subunits of α3β3 ROs (Cα rmsd values of 3.5–3.7 Å over 240–256 αC atoms). A structure-based sequence alignment (supplemental Fig. 3) revealed that the degree of sequence identity between KshA and the α3 ROs is nevertheless very low (11–13%). Moreover, phylogenetic analyses indicated that KshA forms a distinct subfamily of ROs (supplemental Fig. 4).
FIGURE 3.
Crystal structure of KshA from M. tuberculosis. A, ribbon representation of the KshA trimer viewed along the 3-fold symmetry axis. Monomers are colored in different shades of green. Iron ions and acid-labile sulfur atoms are shown as orange and yellow spheres, respectively. B, ribbon representation of the KshA monomer. The Rieske domain is labeled and represented in light brown, and the catalytic domain is labeled and shown in green. The location and position of the β-strand β19 and terminal helix α9 are indicated. C, mononuclear iron coordination sphere. The ribbon representation of KshA is shown as a semitransparent background. The residues ligating the iron are shown as stick representations. The mononuclear iron and solvent molecule S1 are shown as orange and red spheres, respectively. 2Fo – Fc electron density (blue mesh) is shown at 1.7 σ. Residual Fo – Fc density green mesh is shown at 3.5 σ. D, active site and adjacent Rieske center of KshA. The distance between atoms involved in proposed electron transfer is indicated. Residues from the adjacent molecule are indicated with an asterisk. Metal centers are shown as spheres, and side chains are labeled and shown as stick diagrams.
The Rieske domain of KshA (residues 24–153) is highly similar to those of CARDOJ3 and OMO86, with rmsd values of 1.9 and 2.0 Å over 119 and 121 αC atoms, respectively. Briefly, this domain consists of three anti-parallel β-sheets (S1–S3), the third of which harbors the Rieske cluster ligands in loops between strands β4 and β5 and strands β6 and β7, respectively. One iron ion of the cluster is coordinated by Cys67 and Cys86, and the second is coordinated by His69 and His89 (Table 3). As in the other two α3 ROs, the Rieske domain of KshA differs from those of the α3β3 enzymes most significantly in the segment encompassing residues 75 and 80 between sheets S2 and S3; in the latter ROs, this segment is 5 residues longer and interacts with the β subunit.
TABLE 3.
Ligand-iron-ligand distances and angles for the KshA nonheme iron
| Defining atom | Value |
|---|---|
| Metal-ligand distances (Å) | |
| Fe-Nε2, His181 | 2.2 |
| Fe-Nε2, His186 | 2.1 |
| Fe-Oδ1, Asp304 | 2.2 |
| Fe-Oδ2, Asp304 | 2.5 |
| Fe-S1 | 2.1 |
| Metal-ligand angles (degrees) | |
| 181-Fe-186 | 102 |
| 181-Fe-Oδ1, Asp304 | 143 |
| 181-Fe-Oδ2, Asp304 | 92 |
| 186-Fe-Oδ1, Asp304 | 92 |
| 186-Fe-Oδ2, Asp304 | 87 |
| Oδ1, Asp304 –Fe-Oδ2, Asp304 | 54 |
| S1-Fe-181 | 101 |
| S1-Fe-186 | 123 |
| S1-Fe-Oδ1, Asp304 | 98 |
| S1-Fe-Oδ2, Asp304 | 141 |
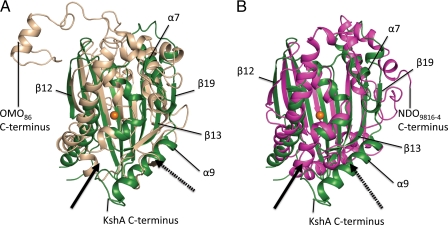
The catalytic domain of KshA (residues 154–374) shares the TBP-like fold of other ROs. In KshA, this domain consists of a eight-stranded antiparallel β-sheet (S4; strands β12-β19) flanked by a series of α-helices on the face distal to the Rieske domain. However, greater structural diversity occurs in this domain than in the Rieske domain (Fig. 4), with rmsd values of 2.8 and 2.9 Å over 135 Cα and Z scores 9.6 and 10.1 for CARDOJ3 and OMO86, respectively. Indeed, the core fold of the KshA catalytic domain contains none of the inserts observed in the other ROs (Fig. 5), consistent with the relatively short length of the enzyme. As compared with the two α3 ROs, the catalytic domain of KshA is missing two secondary structures: 1) a β-strand between α-helix α4 and strand β13 and 2) a β-hairpin and β-strand between strands β14 and β15 (Fig. 5). In CARDOJ3 and OMO86, these two insertions form a structural motif that extends from one corner of the core β sheet to the Rieske domain of an adjacent subunit. As compared with the structurally characterized α3β3 ROs, the catalytic domain of KshA is missing helical structures between β14 and β15.
FIGURE 4.
Structural superposition of KshA (green) with catalytic domains of OMO-O86 (beige, rmsd = 2.8 Å) (A) and NDO-O9816-4 (magenta, rmsd = 3.0 Å) (B). Directionality of the active site channel of KshA is indicated with a solid arrow; that of OMO-O86 and NDO-O9816–4 is indicated with a dashed arrow. Selected secondary structural features of KshA are labeled. C termini are indicated. rmsd values were calculated for superimposed catalytic domains using the SSM algorithm as implemented in COOT.
FIGURE 5.
The secondary structure of KshA. α-Helices and β-strands are represented to scale by cylinders and arrows, respectively. β-Strands and α-helices are numbered. Elements not conserved in all structurally characterized ROs are shown in yellow. The positions of mononuclear iron ligands and Asp178 are indicated with asterisks. Points of inserted elements found in previous structures are indicated as follows. Insertion 1, α-helix in α3β3 ROs or α-helix and 1–2 β-strands in α3 ROs; Insertion 2, 1–2 α-helices in α3β3ROs or β-hairpin and β-strand α3 ROs; Insertion 3, β-strand pairing with N terminus and 1–2 β-strands on central sheet in α3β3 ROs; Insertion 4, C-terminal α-helices in α3 ROs.
The catalytic domain of KshA is further distinguished from that of the other ROs by its C terminus (residues 338–374). In KshA, the polypeptide backbone loops tightly back on itself after α7, forms an eighth β strand (β19), and ends with a long terminal helix (α9) (Fig. 3B). The last is situated on the outer surface of the trimer and next to residues 103–106 in the Rieske domain of the adjacent protomer. In the tertiary structure, this helix is located in the same place as the β-hairpin in CARDOJ3 and OMO86 and like this hairpin appears to stabilize the trimer. By contrast, the helical C terminus of the other two α3 ROs does not include the β-strand (β19) observed in KshA and extends into the interior of the trimer after the final conserved α-helix (KshA α7) (Fig. 4). Like KshA, the C-terminal segments of the α3β3 enzymes of known structure also contain one or two strands. However, in contrast to KshA, these β-strands occur before the last conserved helix in the linear peptide sequence (Fig. 5, Insertion 3).
Active Site Channel—The location of the substrate-binding pocket in KshA is similar to that observed in other ROs. However, the orientation of the active site access channel and position of the channel mouth are strikingly different. In KshA, the channel mouth is located between the C-terminal section of helix α5 and the loop between strands β15 and β16. By contrast, in the two α3 ROs, this entrance occurs between the second and third strands of the central β-sheet (β14 and β15 in KshA), whereas in α3β3 enzymes, it occurs in a similar position near the N- and C-terminal segments of strands β13 and β14, respectively. Indeed, the channel leading from the active site iron to the surface in KshA is angled at ∼90° with respect to those of the other ROs (Fig. 4 and supplemental Fig. 5) and is significantly longer in KshA (28 Å) than in OMO86 and CARDOJ3 (18 Å), as measured from the mononuclear iron to the surface of the protein. The different orientation and length of this channel are the result of two structural differences in KshA; strands β13 and β14 are 5–7 residues longer, and secondary structural elements between β14 and β15 are missing. The former effectively occludes the site where the channel mouth is located in the other ROs, whereas the latter allows for the channel observed in KshA. Despite these differences, the width of the channel mouth in KshA (∼9 Å measured at various points along the channel length; e.g. Cζ of Phe182 and Cδ2 of Leu255) is similar to that of OMO86 and CARDOJ3 (∼10 Å).
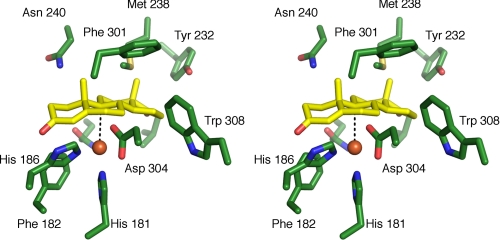
Active Site—The active site mononuclear iron is fully occupied, as indicated by comparison of temperature factor values with those from surrounding atoms. The metal ion is coordinated by His181, His186, Asp304 (bidentate), and an exogenous ligand in a five-coordinate distorted tetragonal geometry (Fig. 3C). Bean-shaped residual density was observed for the exogenous ligand prior to its inclusion in the model. This is similar to what has been observed in structures of other ROs (52–54). When this density was modeled as a single fully occupied solvent species, the temperature factor of the refined ligand (34 Å2) was comparable with those of surrounding protein atoms. However, positive residual density (up to 4 σ) adjacent to this species was still present (Fig. 3C). Alternatively, refinement in this position of a side-on bound molecule of dioxygen starting from a relaxed O–O bond length of 1.45 Å resulted in an O–O bond length of 1.25 Å, significantly shorter than typical for iron-coordinating O2 (1.45 Å). Although positive residual density was no longer present after refinement with dioxygen, we observed significantly higher temperature factors (∼52 Å2). Finally, a tentative refinement with two fully occupied solvent species resulted in a distance between them of 1.9 Å, with positive residual density present whenever occupancy of either or both species was reduced. Overall, the observable density probably reflects the presence of multiple species with partial occupancies, as has been postulated for other ROs (52–54).
To identify the substrate-binding pocket of KshA, ADD was docked using AutoDock. In the two best solutions from these simulations, ADD fit tightly into the pocket extending past the mononuclear iron but was rotated 180° about the short axis such that either ring A or ring D was farthest from the channel mouth (supplemental Fig. 6). The orientation in which ring A was farthest from the channel mouth is potentially biologically relevant, since C9, the atom to be hydroxylated, is located 3.8 Å from the mononuclear iron in an orientation conducive to 9α-hydroxylation. Indeed, its position relative to the catalytic metallocenter is very similar to that of the atom that is hydroxylated in 2-oxoquinoline, C8, in the OMO86·2-oxoquinoline complex (20). The substrate-binding pocket defined by the docking studies extends deeper into the catalytic domain of KshA than what has been observed in other ROs, although the mononuclear irons are similarly positioned. The elongation of the substrate binding site in relation to other RO structures is consistent with the larger size of ADD and AD with respect to the preferred substrates of other ROs. Furthermore, residues identified to interact with the bound substrate (Val176, Gln204, Tyr232, Met238, Asn240, Asn257, Phe301, and Trp308) (Fig. 6) are conserved in known and predicted steroid-transforming ROs (2) but not in other ROs of known structure. The conservation of first shell residues, the tight fit of ADD into the pocket, and the position of the carbon to be hydroxylated with respect to the mononuclear iron strongly suggest that the docking experiment correctly identified the substrate-binding pocket.
FIGURE 6.
Stereo image of ADD docking simulation in the KshA active site. The model shown represents the best ranking solution (predicted free energy of binding =–7.04 kcal/mol) of the top rmsd cluster. The distance between the mononuclear iron and the C9 atom of ADD is 3.8 Å. The shown residues are conserved in KshAs and are labeled, except for Val176 and Gln204. Protein carbons are shown in green, ADD carbons in yellow, oxygen atoms in red, nitrogen in blue, and sulfur in yellow. The mononuclear iron is represented as an orange sphere.
Bridging Aspartate—As in the other ROs, the Rieske cluster and mononuclear iron center of adjacent subunits of KshA interact via a network of hydrogen bonds over the subunit interface. This includes conserved Asp178, whose carboxylate bridges the metallocenters by forming hydrogen bonds with each of the following: the Nε2 atom of His89 (2.8 Å) and the Nδ1 atom of His181 (3.1 Å). In other ROs, this residue has been implicated in catalysis, although its precise role is unclear (20, 55). The conformation of this aspartate is similar in all previous RO structures, but in KshA, the χ1 angle differs by ∼150°, such that the bond between the α- and β-carbons points toward the catalytic domain rather than the Rieske domain of the adjacent subunit (Fig. 3D). However, it is unclear whether the occurrence of this different rotomer in KshA has any mechanistic significance.
DISCUSSION
KshAB from M. tuberculosis was purified, reconstituted in vitro, and kinetically characterized. In addition, the oxygenase component was structurally characterized. Previous attempts to purify a ketosteroid hydroxylase were hampered by the O2 lability of one or more of the components (56). Although the terminal oxygenase of that enzyme, which was also from an actinomycete, was not identified, the enzyme appeared to be a three-component system. In the current study, anaerobic handling of KshA was critical to obtaining preparations that had fully occupied metallocenters.
The ability of KshAB to transform both ADD and AD is consistent with studies based on gene deletion and recombinant strains that have indicated that both compounds are physiological substrates of KshAB from R. erythropolis SQ1 (6, 57, 58). In contrast to what was proposed in R. erythropolis SQ1 based on the moderate toxicity of ADD (58), the clear preference of KshAB from M. tuberculosis for ADD suggests that, at least in the pathogen, KstD precedes KshAB in the steroid catabolic pathway. This conclusion is consistent with KstD2SQ1 possessing 15 times greater specific activity for AD than for 9OHAD (59). However, in the same study, another isozyme, KstD1SQ1, had approximately equal relative specificities for AD and 9-OHAD. Unfortunately, KstDH37Rv from M. tuberculosis H37Rv had undetectable levels of activity with both substrates. However, previous characterization of Δ1-KstD enzymes indicated relatively high levels of activity with AD (7, 8).
The apparent specificity of KshAB for ADD (kcat/Km = 7600 ± 700 m–1 s–1) is 2–3 orders of magnitude less than that of other ROs for their best substrates, including BPDOB356 of Pandoraea pnomenusa B-356 (23, 28), PDODB01 (60), and toluate dioxygenase (TADOmt-2) of P. putida mt-2 (61). It is likely that the relatively low levels of activity observed in KshAB are due to the enzyme's relatively poor reactivity with O2, as discussed below. However, it is possible that the physiological substrate of KshAB is ADD with a different functional group at C-17 resulting from partial side chain degradation. Although metabolite analyses of gene deletion mutants of each of R. jostii RHA1, R. erythropolis SQ1, M. smegmatis, and Mycobacterium bovis BCG have indicated AD and ADD as the most probable substrates for KshAB (6–8, 11, 57, 58), it is unclear whether side chain degradation precedes ring degradation in M. tuberculosis. In analyses of the ΔhsaC mutant of R. jostii RHA1, three catecholic metabolites were identified, possessing ketone, hydroxyl, and propionate groups at C-17, respectively,7 consistent with some simultaneous degradation of the side chain and rings in this strain. Nevertheless, the docking simulations indicate that the binding pocket of KshA would not accommodate isopropionate at C-17 without reorientation of the amino acid side chains lining the binding pocket, consistent with ADD being a good substrate. Another possibility is that the activity of KshAB is regulated in vivo, either allosterically or through covalent modification. In this respect, it would be interesting to investigate the activity of KshAB from a nonpathogenic strain.
The low activity of KshAB appears to be due to the enzyme's poor reactivity with O2. The apparent kcat/KmO2 of KshAB in the presence of ADD (2.5 × 103 m–1 s–1) is around 2 orders of magnitude lower than the corresponding values for BPDOB-356 and TADOmt-2 (220–360 × 103 m–1 s–1) (23, 61). Even more strikingly, although the apparent KmO2 values of BPDOB-356 and TADOmt-2 were 10–28 μm, that of KshAB was too high to be accurately determined, with little saturation occurring even at 1.2 mm O2, the maximum used in these studies. The measured KmO2 of KshAB is also much higher that of HsaC (∼90 μm), a downstream oxygenase in the cholesterol catabolic pathway (4), as well as the concentrations of O2 found in lung tissues of infected and uninfected rabbits (∼2.7–102 μm (62)). It is possible that limiting the rate of KshAB reaction with O2 reduces the accumulation of catechols, which are the substrate of HsaC and which appear to be toxic to M. tuberculosis (4). Alternatively, as noted above, it is possible that the reactivity of KshAB with O2 is modulated in vivo.
The catalytic domain of the KshA appears to represent a minimal and perhaps archetypical RO catalytic domain, helping to define the core fold. Comparison of KshA with the ROs of known structures indicates that the core catalytic domain extends from strand β12 to helix α7, inclusive. Within these limits, the structure of KshA exhibits only those elements that are common to all characterized ROs. The other characterized ROs each have secondary structural elements inserted throughout this core. Furthermore, as summarized in Fig. 5 and described under “Results,” these insertions are specific to the respective α3 and α3β3 ROs characterized to date, both in terms of their location in the polypeptide chain and the identity of the inserted motif. Given the minimal catalytic domain of KshA, this enzyme may be a useful tool to investigate the functional implications of the structural features unique to the other enzymes. Sequence and structural analyses indicate that KshA forms a distinct subfamily of ROs within the α3 enzymes. Moreover, many of the unique structural features of KshA appear to be functionally significant. First, the C-terminal region (residues 337–386) provides a mechanism of trimer stabilization that is distinct from the other two reported in ROs. Second, the deeper substrate-binding pocket of KshA is required to accommodate the ketosteroid, which is larger than the preferred substrates of the structurally characterized ROs. Finally, the differently positioned active site channel of KshA appears to be necessary to allow proper orientation of the substrate within this binding pocket. These features together with the minimal catalytic domain of KshA help explain why KshA diverges so much from the characterized ROs.
Although animal studies have suggested that cholesterol catabolism is not essential to the pathogenesis of M. tuberculosis (3, 4), the uncoupling of oxygen activation from substrate turnover that has been reported in a variety of ROs in the presence of suboptimal substrates (21–23) underlines the potential of KshA as a target for a novel class of therapeutics. Inhibitors that cause uncoupling in KshA would not only block cholesterol catabolism but would also result in the intracellular production of reactive oxygen species and the depletion of energy reserves through the futile depletion of NADH. Further studies of this enzyme should facilitate the design of such inhibitors.
Supplementary Material
Acknowledgments
We thank Christine Florizone and Jie Liu for skilled technical assistance. We also thank Dr. Victor Snieckus for generously providing 2,3-dihydroxybiphenyl. We thank the Canadian Light Source (Saskatoon, Canada) for access to Beamline CMCF1 for x-ray synchrotron data collection.
This work was supported by grants from the Canadian Institute for Health Research (to L. D. E. and N. S.), the British Columbia Lung Association (to L. D. E.), and the Michael Smith Foundation for Health Research (MSFHR) Infrastructure (to N. S.) and Emerging Team programs.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
The atomic coordinates and structure factors (code 2ZYL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Footnotes
The abbreviations used are: RO, Rieske oxygenase; ADD, 1,4-androstadiene-3,17,-dione; AD, 4-androstene-3,17-dione; 9-OHAD, 9-hydroxy-4-androstene-3,17-dione; 9-OHADD, 9-hydroxy-1,4-androstadiene-3,17-dione; HSA, 3-hydroxy-9,10-seconandrost-1,3,5(10)-triene-9, 17-dione; NDO9816-4, naphthalene dioxygenase from Pseudomonas sp. NCIB 9816-4; OMO86, 2-oxoquinoline 8-monooxygenase of P. putida 86; CARDOJ3, carbazole 1,9α-dioxygenase of Janthinobacterium sp. J3; PDODB01, phthalate dioxygenase of B. cepacia DB01; BPDOB-356, biphenyl dioxygenase of P. pnomenusa B-356; TADOmt-2, toluate dioxygenase of P. putida mt-2; FAS, ferrous ammonium sulfate; rmsd, positional root mean square deviation; HPLC, high pressure liquid chromatography.
K. Yam, C. Dresen, and L. D. Eltis, unpublished observations.
References
- 1.World Health Organization (2008), Anti-tuberculosis Drug Resistance in the World, Report No. 4, pp. 14–21, World Health Organization, Geneva, Switzerland
- 2.van der Geize, R., Yam, K., Heuser, T., Wilbrink, M. H., Hara, H., Anderton, M. C., Sim, E., Dijkhuizen, L., Davies, J. E., Mohn, W. W., and Eltis, L. D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandey, A. K., and Sassetti, C. M. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yam, K., D'Angelo, I., Kalscheuer, R., Zhu, H., Wang, J., Snieckus, V., Ly, L. H., Converse, P. J., Jacobs, W. R., Strynadka, N., and Eltis, L. D. (2009) PLoS Pathog., in press [DOI] [PMC free article] [PubMed]
- 5.Yang, X., Dubnau, E., Smith, I., and Sampson, N. S. (2007) Biochemistry 46 9058–9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Geize, R., Hessels, G. I., van Gerwen, R., van der Meijden, P., and Dijkhuizen, L. (2002) Mol. Microbiol. 45 1007–1018 [DOI] [PubMed] [Google Scholar]
- 7.van der Geize, R., Hessels, G. I., van Gerwen, R., Vrijbloed, J. W., van Der Meijden, P., and Dijkhuizen, L. (2000) Appl. Environ. Microbiol. 66 2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Geize, R., Hessels, G. I., and Dijkhuizen, L. (2002) Microbiology 148 3285–3292 [DOI] [PubMed] [Google Scholar]
- 9.van der Geize, R., and Dijkhuizen, L. (2004) Curr. Opin. Microbiol. 7 255–261 [DOI] [PubMed] [Google Scholar]
- 10.Rengarajan, J., Bloom, B. R., and Rubin, E. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8327–8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andor, A., Jekkel, A., Hopwood, D. A., Jeanplong, F., Ilkoy, E., Konya, A., Kurucz, I., and Ambrus, G. (2006) Appl. Environ. Microbiol. 72 6554–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferraro, D. J., Gakhar, L., and Ramaswamy, S. (2005) Biochem. Biophys. Res. Commun. 338 175–190 [DOI] [PubMed] [Google Scholar]
- 13.Kovaleva, E. G., and Lipscomb, J. D. (2008) Nat. Chem. Biol. 4 186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bugg, T. D., and Ramaswamy, S. (2008) Curr. Opin. Chem. Biol. 12 134–140 [DOI] [PubMed] [Google Scholar]
- 15.Kauppi, B., Lee, K., Carredano, E., Parales, R. E., Gibson, D. T., Eklund, H., and Ramaswamy, S. (1998) Structure 6 571–586 [DOI] [PubMed] [Google Scholar]
- 16.Karlsson, A., Parales, J. V., Parales, R. E., Gibson, D. T., Eklund, H., and Ramaswamy, S. (2003) Science 299 1039–1042 [DOI] [PubMed] [Google Scholar]
- 17.Furukawa, K. (2003) Trends Biotechnol. 21 187–190 [DOI] [PubMed] [Google Scholar]
- 18.Gibson, D. T., and Parales, R. E. (2000) Curr. Opin. Biotechnol. 11 236–243 [DOI] [PubMed] [Google Scholar]
- 19.Parales, R. E., Parales, J. V., and Gibson, D. T. (1999) J. Bacteriol. 181 1831–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martins, B. M., Svetlitchnaia, T., and Dobbek, H. (2005) Structure 13 817–824 [DOI] [PubMed] [Google Scholar]
- 21.Lee, K. (1999) J. Bacteriol. 181 2719–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernhardt, F. H., and Kuthan, H. (1981) Eur. J. Biochem. 120 547–555 [DOI] [PubMed] [Google Scholar]
- 23.Imbeault, N. Y., Powlowski, J. B., Colbert, C. L., Bolin, J. T., and Eltis, L. D. (2000) J. Biol. Chem. 275 12430–12437 [DOI] [PubMed] [Google Scholar]
- 24.Nojiri, H., Ashikawa, Y., Noguchi, H., Nam, J. W., Urata, M., Fujimoto, Z., Uchimura, H., Terada, T., Nakamura, S., Shimizu, K., Yoshida, T., Habe, H., and Omori, T. (2005) J. Mol. Biol. 351 355–370 [DOI] [PubMed] [Google Scholar]
- 25.Tarasev, M., and Ballou, D. P. (2005) Biochemistry 44 6197–6207 [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 27.Pulleyblank, D., Michalak, M., Daisley, S. L., and Glick, R. (1983) Mol. Biol. Rep. 9 191–195 [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Gil, L., Kumar, P., Barriault, D., Bolin, J. T., Sylvestre, M., and Eltis, L. D. (2007) J. Bacteriol. 189 5705–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaillancourt, F. H., Han, S., Fortin, P. D., Bolin, J. T., and Eltis, L. D. (1998) J. Biol. Chem. 273 34887–34895 [DOI] [PubMed] [Google Scholar]
- 30.Chen, J. S., and Mortenson, L. E. (1977) Anal. Biochem. 79 157–165 [DOI] [PubMed] [Google Scholar]
- 31.Zabinski, R., Munck, E., Champion, P. M., and Wood, J. M. (1972) Biochemistry 11 3212–3219 [DOI] [PubMed] [Google Scholar]
- 32.Cornish-Bowden, A. (1994) Analysis of Enzyme Kinetic Data, Oxford University Press, Oxford
- 33.Faeder, E. J., and Siegel, L. M. (1973) Anal. Biochem. 53 332–336 [DOI] [PubMed] [Google Scholar]
- 34.Kabsch, W. (1993) J. Appl. Crystallogr. 26 795–800 [Google Scholar]
- 35.Adams, P. D., Grosse-Kunstleve, R. W., Hung, L. W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K., and Terwilliger, T. C. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1948–1954 [DOI] [PubMed] [Google Scholar]
- 36.McCoy, A. J. (2007) Acta Crystallogr. Sect. D Biol. Crystallogr. 63 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terwilliger, T. C. (2000) Acta Crystallogr. Sect. D Biol. Crystallogr. 56 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240–255 [DOI] [PubMed] [Google Scholar]
- 40.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905–921 [DOI] [PubMed] [Google Scholar]
- 41.(1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760–763 [DOI] [PubMed] [Google Scholar]
- 42.Holm, L., Kaariainen, S., Rosenstrom, P., and Schenkel, A. (2008) Bioinformatics 24 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gille, C., and Frommel, C. (2001) Bioinformatics 17 377–378 [DOI] [PubMed] [Google Scholar]
- 44.Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007) Bioinformatics 23 2947–2948 [DOI] [PubMed] [Google Scholar]
- 45.Felsenstein, J. (1989) Cladistics 5 164–166 [Google Scholar]
- 46.Schuttelkopf, A. W., and van Aalten, D. M. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 1355–1363 [DOI] [PubMed] [Google Scholar]
- 47.Mayo, S. L., Olafson, B. D., and Goddard, W. A. (1990) J. Phys. Chem. 94 8897–8909 [Google Scholar]
- 48.Stewart, J. J. (2007) J. Mol. Model 13 1173–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneidman-Duhovny, D., Inbar, Y., Nussinov, R., and Wolfson, H. J. (2005) Nucleic Acids Res. 33 W363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., Hart, W. E., Belew, R. K., and Olson, A. J. (1998) J. Comp. Chem. 19 1639–1662 [Google Scholar]
- 51.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA
- 52.Ferraro, D. J., Brown, E. N., Yu, C. L., Parales, R. E., Gibson, D. T., and Ramaswamy, S. (2007) BMC Struct. Biol. 7 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Furusawa, Y., Nagarajan, V., Tanokura, M., Masai, E., Fukuda, M., and Senda, T. (2004) J. Mol. Biol. 342 1041–1052 [DOI] [PubMed] [Google Scholar]
- 54.Dong, X., Fushinobu, S., Fukuda, E., Terada, T., Nakamura, S., Shimizu, K., Nojiri, H., Omori, T., Shoun, H., and Wakagi, T. (2005) J. Bacteriol. 187 2483–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarasev, M., Pinto, A., Kim, D., Elliott, S. J., and Ballou, D. P. (2006) Biochemistry 45 10208–10216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strijewski, A. (1982) Eur. J. Biochem. 128 125–135 [DOI] [PubMed] [Google Scholar]
- 57.van der Geize, R., Hessels, G. I., van Gerwen, R., van der Meijden, P., and Dijkhuizen, L. (2001) FEMS Microbiol. Lett. 205 197–202 [DOI] [PubMed] [Google Scholar]
- 58.van der Geize, R., Hessels, G. I., Nienhuis-Kuiper, M., and Dijkhuizen, L. (2008) Appl. Environ. Microbiol. 74 7197–7203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knol, J., Bodewits, K., Hessels, G. I., Dijkhuizen, L., and van der Geize, R. (2008) Biochem. J. 410 339–346 [DOI] [PubMed] [Google Scholar]
- 60.Pinto, A., Tarasev, M., and Ballou, D. P. (2006) Biochemistry 45 9032–9041 [DOI] [PubMed] [Google Scholar]
- 61.Ge, Y., Vaillancourt, F. H., Agar, N. Y., and Eltis, L. D. (2002) J. Bacteriol. 184 4096–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Via, L. E., Lin, P. L., Ray, S. M., Carrillo, J., Allen, S. S., Eum, S. Y., Taylor, K., Klein, E., Manjunatha, U., Gonzales, J., Lee, E. G., Park, S. K., Raleigh, J. A., Cho, S. N., McMurray, D. N., Flynn, J. L., and Barry, C. E., 3rd. (2008) Infect. Immun. 76 2333–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.