Abstract
Mutation of the inositol polyphosphate 5-phosphatase OCRL1 causes the X-linked disorder oculocerebrorenal syndrome of Lowe, characterized by defects in the brain, kidneys, and eyes. OCRL1 exists as two splice isoforms that differ by a single exon encoding 8 amino acids. The longer protein, termed isoform a, is the only form in brain, whereas both isoforms are present in all other tissues. The significance of OCRL1 splicing is currently unclear. Given its proximity to a clathrin-binding site, we hypothesized that splicing may alter the clathrin binding properties of OCRL1. Here we show that this is indeed the case. OCRL1 isoform a binds clathrin with higher affinity than isoform b and is significantly more enriched in clathrin-coated trafficking intermediates. We also identify a second clathrin-binding site in OCRL1 that contributes to clathrin binding of both isoforms. Association of OCRL1 with clathrin-coated intermediates requires membrane association through interaction with Rab GTPases but not binding to the clathrin adaptor AP2. Expression of OCRL1 isoform a lacking the 5-phosphatase domain impairs transferrin endocytosis, whereas an equivalent version of isoform b does not. Our results suggest that OCRL1 exists as two functional pools, one participating in clathrin-mediated trafficking events such as endocytosis and another that is much less or not involved in this process.
Oculocerebrorenal syndrome of Lowe is a rare X-linked disorder affecting primarily the brain, eyes, and kidneys (1, 2). Lowe syndrome is caused by mutation of OCRL1, a ubiquitously expressed inositol polyphosphate 5-phosphatase that preferentially hydrolyzes phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate (reviewed in Ref. 3). OCRL1 is localized to the trans-Golgi network, early endosomes, plasma membrane ruffles, and clathrin-coated trafficking intermediates (4-8). In addition to a central 5-phosphatase domain, OCRL1 also has C-terminal ASH and RhoGAP-like domains. The ASH domain binds members of the Rab family of small GTPases, which are required for the correct targeting of OCRL1 to the TGN4 and endosomes (9). The RhoGAP-like domain appears to lack catalytic activity and rather serves to bind Rac and Cdc42, which may help anchor OCRL1 to the membrane (8, 10, 11). A recent study also suggested the RhoGAP-like domain may bind ARF1 and ARF6 (11). The C-terminal region of OCRL1 interacts with APPL1, another Rab5 effector that participates in signaling from endocytic membranes (8). In addition, OCRL1 binds directly to the terminal domain of clathrin heavy chain, which occurs via a type I clathrin box with the sequence LIDLE that is present on a loop protruding from the globular RhoGAP-like domain (6-8). Binding studies have indicated that a second clathrin-binding site exists in OCRL1, consistent with its ability to polymerize clathrin in vitro, but it has yet to be identified (6). OCRL1 can also bind via an N-terminal FEDNF motif to the appendage domain of α-adaptin, a subunit of the AP2 plasma membrane clathrin adaptor complex (7).
Together the specificity of OCRL1 5-phosphatase activity, its subcellular localization, and its known interaction partners suggest it could participate in a number of processes, including endocytosis, trafficking at the TGN/endosome interface, signaling from the plasma membrane, endosomes, and/or the TGN or regulation of actin dynamics at these locations (reviewed in Ref. 3; see also Refs. 6, 8, 12). It is also conceivable that OCRL1 could participate in other processes such as cell polarization or housekeeping through removal of ectopic phosphatidylinositol 4,5-bisphosphate and/or phosphatidylinositol 3,4,5-trisphosphate from endomembranes. Although OCRL1 is ubiquitously expressed, the manifestations of Lowe syndrome are restricted to only a few tissues. This may be due to functional compensation by the related 5-phosphatase INPP5B, which shares the same domain organization and substrate specificity as OCRL1 (8, 13, 14). INPP5B also has a similar cellular localization and interaction partner profile as OCRL1, with some differences, the most notable of which is its lack of clathrin binding and absence from clathrin-coated structures (8, 15, 16). Knock-out studies in mice support the idea that OCRL1 and INPP5B can functionally compensate for loss of the other protein (17).
OCRL1 exists as two alternatively spliced forms, termed a and b (18, 19). Isoform a contains an additional exon lacking in isoform b that encodes 8 amino acids adjacent to the LIDLE clathrin box. Isoform a is present in all tissues, whereas isoform b is present in all tissues apart from the brain (19). Isoform a is therefore the only form in brain, but in most other tissues it appears to be the minor form. The significance of OCRL1 splicing is currently unknown. Given the proximity of the alternatively spliced exon to the clathrin box, we hypothesized that it could influence binding of OCRL1 to clathrin. Here we show that this is indeed the case and that it is clathrin binding that determines the amount of OCRL1 that enters clathrin-coated transport intermediates. Isoform a binds clathrin with higher affinity than isoform b and is significantly more enriched in clathrin-coated intermediates. We also identify a second clathrin-binding site in the N terminus of OCRL1 that is important for clathrin binding of both OCRL1 isoforms. Expression of an OCRL1 isoform a construct lacking the 5-phosphatase domain delays transferrin endocytosis, whereas an equivalent version of isoform b does not, consistent with distinct functional roles for the two variants of OCRL1. This is likely of significance in the disease state because the brain, one of the major tissues affected in Lowe syndrome, only expresses isoform a.
EXPERIMENTAL PROCEDURES
Materials and Antibodies—All reagents were from Sigma or Merck unless stated otherwise. Protease inhibitors (mixture set III) were from Calbiochem and used at 1:250. Rabbit KINS anti-OCRL1 and sheep anti-GST antibodies were described previously (9). Mouse anti-clathrin heavy chain X22 for immunofluorescence studies was a kind gift from Prof. Liz Smythe (University of Sheffield, UK). Mouse antibody against clathrin heavy chain for Western blotting was purchased from BD Transduction Laboratories. Mouse anti-transferrin receptor antibody was from Zymed Laboratories Inc.. Sheep anti-golgin-84 and rabbit anti-GM130 antibodies have been described previously (20). Mouse anti-CI-MPR was purchased from Affinity Bioreagents. Rabbit anti-CI-MPR was a kind gift from Prof. Paul Luzio (University of Cambridge, UK). Fluorophore-conjugated (Alexa 594 and Alexa 488) and horseradish peroxidase-conjugated secondary antibodies were purchased from Molecular Probes and Tago Immunologicals, respectively.
Molecular Biology and Yeast Two-hybrid Experiments—All constructs were made using standard molecular biology techniques. Primer sequences are available on request. GFP- and His/protein S-tagged human OCRL1 isoform b (GenBank™ accession number NP_001578) constructs were described previously (6, 9). DNA encoding full-length isoform a (GenBank™ accession number NP_000267) was cloned from a human liver cDNA library and cloned into pEGFP-C1 (Clontech) for expression of GFP-tagged protein in mammalian cells, pGBKT7 (Stratagene) for yeast two-hybrid experiments, and pBAC2 (Novagen) for expression of His/protein S-tagged recombinant protein in insect cells. OCRL1 isoforms a and b were cloned into a modified version of pcDNA3.1 for N-terminal tagging with mCherry. The OCRL1 LIDIA and LIDLE clathrin box sequences were deleted or point mutated, and the FEDNF α-adaptin-binding site was mutated to AEANF using the QuikChange method (Stratagene). Constructs encoding GST-tagged clathrin terminal domain (residues 1-579), α-adaptin appendage domain, and Rab and Rac GTPases have been described previously (6, 9, 16). An oligonucleotide encoding residues 403-413 of human APPL1 was ligated into the BamHI and EcoRI sites of pGEX4T-2 for preparation of recombinant protein in Escherichia coli. All constructs were verified by DNA sequencing using the ABI Prism Big Dye Terminator Cycle Sequencing kit (Applied Biosystems). Plasmid for expression of mCherry-tagged wild-type Rab5 in mammalian cells was kindly provided by Prof. Philip Woodman (University of Manchester, UK). Yeast two-hybrid analysis was performed according to the Clontech manual as described previously (6). The pGADT7-clathrin terminal domain was a kind gift from Prof. Harald Stenmark (Norwegian Radium Hospital, Oslo, Norway). The pGADT7-α-adaptin appendage domain has been described previously (16).
Cell Culture, Transfection, and Transferrin Uptake—Adherent HeLa and NRK cells were grown at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium containing 10% fetal calf serum. HeLa and NRK cells were transiently transfected with FuGENE 6 (Roche Diagnostics) and JetPEI (Polyplus), respectively, according to the manufacturer's instructions and incubated for 16-20 h before analysis. Transferrin uptake was performed by washing cells three times with warm uptake medium (L-15 containing 2 mg/ml bovine serum albumin) before adding warm uptake medium containing 5 μg/ml Texas Red- or Alexa 594-conjugated transferrin. Cells were incubated for 2, 5, 15, or 30 min at 37 °C and fixed directly into 3% paraformaldehyde at room temperature. Quantitation of uptake was performed using ImageJ by measuring the mean fluorescence intensity of each cell. Semi-quantitative analysis was also performed by comparing fluorescence intensity of transfected cells to neighboring nontransfected cells.
Immunofluorescence Microscopy—Cells were fixed in 3% paraformaldehyde in phosphate-buffered saline, and immunofluorescence microscopy was performed as described previously (6).
Preparation of Cell Extracts and Pulldown Experiments—Recombinant GST-tagged proteins were expressed in E. coli and purified using standard techniques. Extracts were prepared from HeLa cells expressing GFP-tagged OCRL1 by washing the cells twice in ice-cold phosphate-buffered saline followed by extraction in HNMT (20 mm Hepes, pH 7.4, 0.1 m NaCl, 5 mm MgCl2, 0.25% Triton X-100) containing protease inhibitors (1 ml of HNMT/10-cm dish) for 15 min on ice. Extracts were clarified before use by spinning at 15,000 rpm in a microcentrifuge for 10 min. Binding was performed by incubating 250 μl of extract with 20-100 μg of GST-tagged bait protein bound to GSH-Sepharose for 5 h at 4 °C. Binding with purified proteins was performed by incubating 0.5 μg of His/S-tagged OCRL1 in 100 μl of HNMT containing 5 μg of bovine serum albumin with 2 μg of GST-tagged bait bound to 10 μl of GSH-Sepharose for 3 h at 4 °C. Binding with in vitro translated proteins was performed by incubating 5 μl of in vitro translated protein (made using the Promega coupled TnT kit according to the manufacturer's instructions) with 2 μg of GST-tagged bait bound to 10 μl of GSH-Sepharose in 100 μl of HNMT for 3 h at 4 °C. After washing three times with HNMT, proteins were eluted by boiling in SDS sample buffer and analyzed by Western blotting with appropriate antibodies or by autoradiography. Binding experiments were performed 2-4 times per experiment. Representative examples of each experiment are shown in the figures.
Preparation of Clathrin-coated Vesicles—Clathrin-coated vesicles were isolated from HeLa cells expressing GFP-tagged OCRL1 according to Ref. 21. Cells from a confluent 10-cm dish were washed twice with ice-cold phosphate-buffered saline and scraped into 1 ml of buffer C (0.1 m MES, pH 6.5, 0.2 mm EGTA, 0.5 mm MgCl2) containing protease inhibitors before passing 22 times through a ball-bearing homogenizer (8.01-mm ball inside 8.02-mm barrel). The homogenate was centrifuged at 3,900 × g in a Beckman TLS55, and the low speed supernatant was incubated with 50 μg/ml RNase A for 30 min on ice. The membranes were pelleted by spinning at 50,000 rpm for 30 min in a Beckman TLA55 rotor and resuspended in 300 μl of buffer C by passing 10-15 times through a 25-gauge needle. The resuspended pellet (high speed pellet) was mixed with an equal volume of 12.5% Ficoll, 12.5% sucrose (in buffer C) and centrifuged for 25 min at 20,000 rpm in a TLA55 rotor. The supernatant was diluted with 4 volumes of buffer C, and the clathrin-coated vesicles were pelleted by spinning at 50,000 rpm for 30 min in a TLA55 rotor. The clathrin-coated vesicles were resuspended in a total volume of 30 μl of buffer C, snap-frozen in liquid nitrogen, and stored at -80 °C until use.
RESULTS
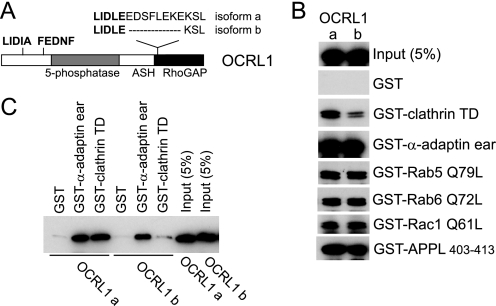
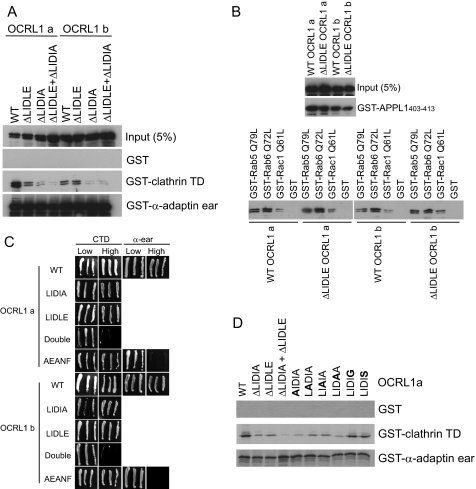
Differential Clathrin Binding of OCRL1 Isoforms—OCRL1 isoform a contains eight amino acids adjacent to the clathrin box LIDLE that are missing in isoform b (Fig. 1A). We hypothesized that this could affect binding of OCRL1 to the terminal domain of clathrin heavy chain. This was tested by expressing GFP-tagged versions of the OCRL1 isoforms in HeLa cells and performing pulldown experiments. As shown in Fig. 1B, GFP-OCRL isoform a bound to clathrin more strongly than isoform b. In contrast, binding to other known OCRL1 binding partners, including the α-adaptin appendage domain, GTP-locked Rab5 and -6 and Rac1, and APPL1, was unaffected. To confirm the effect on clathrin binding was a property of OCRL1 itself and not because of additional interactions, binding was repeated using purified recombinant proteins. Recombinant OCRL1 isoform a bound clathrin significantly better than isoform b, whereas binding to α-adaptin was the same for each isoform (Fig. 1C).
FIGURE 1.
Differential clathrin binding of OCRL1 isoforms. A, schematic view of OCRL1 isoforms a and b. The positions of putative clathrin boxes (LIDIA and LIDLE) and the α-adaptin-binding site (FEDNF) are highlighted in bold. B, GFP-tagged OCRL1 isoforms a and b were expressed in HeLa cells and tested for binding to GST-tagged bait proteins as indicated. Bound proteins were detected by Western blotting with anti-OCRL1 antibodies. C, purified recombinant OCRL1 isoforms a and b were incubated with GST-tagged bait proteins as indicated and bound protein detected by Western blotting with anti-OCRL1 antibodies. TD, terminal domain; α-adaptin ear, α-adaptin appendage domain.
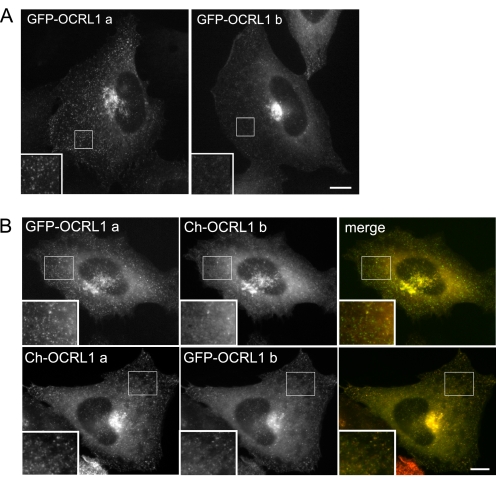
Differential Subcellular Localization of OCRL1 Isoforms—To determine whether the differential clathrin binding of OCRL1 isoforms a and b affects their localization in cells, both isoforms were expressed as GFP-tagged fusions in NRK cells, and their localization was analyzed by fluorescence microscopy. Both proteins predominantly localized to the perinuclear region (Fig. 2A). Double labeling confirmed co-localization with the Golgi apparatus in this region, as expected from previous studies (data not shown) (6, 7). In addition there was punctate staining dispersed throughout the cell, corresponding most likely to endocytic structures and clathrin-coated transport intermediates, as observed previously (see below) (6-8). Strikingly, OCRL1 isoform a was significantly more abundant in the cytoplasmic puncta, and there was less diffuse cytosolic staining compared with isoform b (Fig. 2A). These differences were observed at all expression levels and also seen in HeLa cells (supplemental Fig. S1), indicating they are not a consequence of protein overexpression or dependent upon the cell type used. To further verify these observations, OCRL1 isoforms a and b fused to different fluorescent tags were co-expressed in the same cell. Again, OCRL1 isoform a was more obviously in puncta in the cytoplasm and less diffuse, irrespective of the tag used (Fig. 2B).
FIGURE 2.
Cellular localization of OCRL1 isoforms. A, NRK cells transiently expressing GFP-tagged OCRL1 isoform a or b were fixed and imaged by epifluorescence microscopy. B, NRK cells were co-transfected with GFP or mCherry (Ch)-tagged OCRL1 isoforms as indicated and imaged by epifluorescence microscopy. Bar, 10 μm.
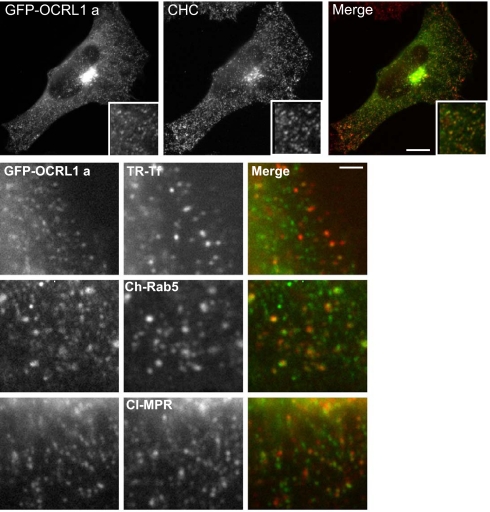
To ascertain whether the OCRL1 isoform a-containing puncta correspond to trafficking intermediates, double labeling with various markers was performed. As expected, there was extensive overlap with clathrin, consistent with significant enrichment of OCRL1 isoform a in clathrin-coated transport intermediates (Fig. 3, top row; see also Fig. 7). We also observed partial overlap of GFP-OCRL1 isoform a with internalized transferrin and overexpressed Rab5, consistent with localization to endocytic structures (Fig. 3). Some of puncta contained CI-MPR, a cargo receptor found in clathrin-coated vesicles shuttling between the TGN and endosomes. Thus, OCRL1 isoform a is abundant in clathrin-coated transport intermediates within the endocytic pathway and those that traffic between the TGN and endosomes. Although previous studies have localized OCRL1 isoform b to similar structures (6, 7), its abundance there is markedly reduced compared with that of isoform a.
FIGURE 3.
Co-localization of OCRL1 isoform a with clathrin. NRK cells transiently expressing GFP-tagged OCRL1 isoform a (green) were fixed and labeled with antibodies to clathrin heavy chain (CHC, top row, red)) or the cation-independent mannose 6-phosphate receptor (CI-MPR, bottom row, red). HeLa cells expressing GFP-OCRL1 a were incubated at 37 °C for 2 min with Texas Red (TR)-transferrin (Tf) prior to fixation (2nd row). NRK cells co-expressed GFP-tagged OCRL1 isoform a and mCherry (Ch)-Rab 5 wild-type (3rd row). All cells were imaged by epifluorescence microscopy. Bars, 10 μm top, 2 μm bottom.
FIGURE 7.
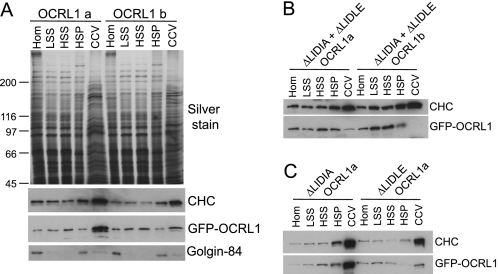
Association of OCRL1 isoforms with clathrin-coated vesicles. Clathrin-coated vesicles were partially purified from HeLa cells expressing the indicated GFP-OCRL1 constructs. A, equal amounts of total protein of homogenate (Hom), low speed supernatant (LSS), high speed supernatant (HSS), membrane fraction (HSP), and clathrin-coated vesicle (CCV) fractions isolated from cells expressing GFP-OCRL1 isoform a or b were analyzed by silver staining or Western blotting with antibodies to clathrin heavy chain (CHC), OCRL1 (to detect GFP-OCRL1), or Golgin-84. B and C, equal total protein amounts of fractions isolated from cells expressing the indicated clathrin box mutants were blotted for clathrin heavy chain and GFP-tagged OCRL1 as indicated.
Identification of a Second Clathrin-binding Site in OCRL1—We previously found that deletion of the LIDLE clathrin box in OCRL1 isoform b did not abrogate clathrin binding indicating the presence of a second clathrin-binding site in the protein (6). Yeast two-hybrid experiments suggested this may reside within the ASH domain, but this region lacks known clathrin-binding motifs (6). We therefore inspected the OCRL1 sequence further and identified an LIDIA sequence within the N-terminal region of mammalian OCRL1 (amino acids 73-77 in human OCRL1) that is similar to the type I clathrin box consensus (L(L/I)(D/E/N)(L/F)(D/E)). Interestingly, in zebrafish OCRL1 the corresponding sequence is LIDID, which matches the consensus perfectly. We therefore deleted this sequence in both human OCRL1 isoforms either alone or in conjunction with LIDLE and monitored binding to clathrin in pulldown assays. As shown in Fig. 4A, deletion of LIDLE had little effect on clathrin binding of OCRL1 isoform b, consistent with our previous findings. In contrast deletion of LIDIA resulted in a dramatic decrease in clathrin binding of isoform b. Deletion of both LIDLE and LIDIA gave a similar result. Deletion of LIDLE from OCRL1 isoform a reduced clathrin binding to a level comparable with that of isoform b, whereas deletion of LIDIA resulted in an even greater decrease in binding. Deletion of both sequences reduced binding to very low levels. None of the mutations affected binding to the α-adaptin appendage domain. Similarly, deletion of LIDLE from OCRL1 isoforms a and b had no effect on binding to APPL1 or GTP-locked Rab5, Rab6, or Rac1 (Fig. 4B).
FIGURE 4.
Protein interactions of OCRL1 clathrin box mutants. A and B, GFP-tagged OCRL1 isoform a or b wild type (WT) or the indicated clathrin box mutants were expressed in HeLa cells and tested for binding to GST-tagged bait proteins as indicated. Bound proteins were detected by Western blotting with anti-OCRL1 antibodies. C, OCRL1 isoform a or b or the indicated mutants were tested for interaction with clathrin terminal domain (CTD) or the α-adaptin appendage domain (α-ear) in the yeast two-hybrid system. Growth on high selection indicates interaction. D, in vitro synthesized wild-type OCRL1 isoform a or the indicated mutants were tested for binding to the indicated GST-tagged bait proteins. Bound OCRL1 was detected by autoradiography. Substituted residues are indicated in bold.
To further corroborate these results, binding was also analyzed in the yeast two-hybrid system. Binding to clathrin still occurred when either clathrin-binding site in OCRL1 isoforms a or b was deleted, whereas deletion of both sites completely abolished the interaction (Fig. 4C). Taken together these results indicate that LIDIA is a major clathrin-binding site in both OCRL1 isoforms, and that the difference in clathrin binding between isoforms a and b is because of the LIDLE sequence, which in isoform b contributes little to the interaction with clathrin.
Given that LIDIA does not exactly match the consensus for a type I clathrin box, containing an alanine at position 5 instead of an acidic residue, we performed mutagenesis of this motif to determine the residues important for clathrin binding. As expected, binding of in vitro synthesized OCRL1 isoform a to clathrin was reduced by separate deletion of LIDIA or LIDLE, and almost completely abolished by deletion of both motifs together (Fig. 4C). Individually mutating the first four residues in LIDIA to alanine reduced clathrin binding to a level similar to deletion of the entire motif, indicating that these residues are critical for clathrin binding (Fig. 4C). In contrast, mutagenesis of the terminal alanine to glycine or serine had little effect on clathrin binding, indicating that this residue is not important for binding of OCRL1 via LIDIA to the clathrin terminal domain.
We also tested the effect of mutating the FEDNF α-adaptin-binding motif in either OCRL1 isoform. Mutation of FEDNF to AEANF had little effect on clathrin binding to OCRL1 isoform a in yeast two-hybrid or pulldown assays (Figs. 4C and 5A). Binding of the AEANF mutant isoform b to clathrin in the pulldown assay was slightly reduced compared with wild type (Fig. 5A), suggesting that a minor proportion of the binding may be indirect through association with AP2; this could explain the residual binding seen with the double clathrin box depletions in Fig. 4A. In contrast, mutation of FEDNF resulted in a major decrease in binding of OCRL1 isoform a and a complete loss of isoform b binding to α-adaptin (Fig. 5A). We believe the remaining interaction of isoform a with α-adaptin is indirect via clathrin to which isoform a binds more strongly than isoform b. Consistent with this idea, binding of the AEANF mutants to α-adaptin is completely abolished when assessed in the yeast two-hybrid system (Fig. 4C). We conclude from these experiments that the FEDNF motif is the sole α-adaptin-binding site in OCRL1.
FIGURE 5.
Protein interactions of OCRL1 α-adaptin- and Rab-binding mutants. A and B, GFP-tagged OCRL1 isoform a or b wild-type (WT) or the indicated α-adaptin- (A, AEANF) or Rab-binding (B, G664D) mutants were expressed in HeLa cells and tested for binding to GST-tagged bait proteins as indicated. Bound proteins were detected by Western blotting with anti-OCRL1 antibodies. TD, terminal domain.
We previously showed that mutation of Gly-664 to Asp inhibits binding of OCRL1 isoform b to Rab GTPases (9), and a similar result was reported for isoform a (22). Here, we directly compare the effects of G664D mutation on binding of each isoform to the known OCRL1 interaction partners. G664D mutation had little effect on binding to clathrin, α-adaptin, Rac1, or APPL1, whereas binding to Rab5 and -6 was almost completely abolished (Fig. 5B). The effects of G664D mutation were the same for both OCRL1 isoforms.
Role of Clathrin Boxes in Localization of OCRL1—To examine the role of the individual clathrin-binding sites in determining OCRL1, localization mutants of each isoform lacking either or both clathrin boxes were expressed in cells, and their distribution was analyzed by fluorescence microscopy. None of the mutations affected targeting of OCRL1 to the Golgi apparatus, in agreement with our previous finding that clathrin is not required for Golgi targeting (6) (Fig. 6A). Mutation of the clathrin boxes did, however, affect the amount of OCRL1 present in cytoplasmic punta. In isoform a, deletion of LIDIA partially reduced the amount of OCRL1 in puncta, but the effect was relatively minor (Fig. 6A). In contrast, deletion of LIDLE dramatically reduced the OCRL1 levels in puncta, resulting in a distribution similar to that seen with wild-type isoform b. Deletion of both clathrin boxes resulted in very few OCRL1-positive puncta. In contrast, mutation of the FEDNF α-adaptin-binding site had no discernible effect upon OCRL1 distribution. In isoform b, deletion of LIDIA reduced the amount of OCRL1 in puncta, whereas LIDLE deletion had no effect, and deletion of both clathrin boxes resulted in few OCRL1-positive puncta (Fig. 6A). The G664D Rab binding-deficient mutants of both OCRL1 isoforms a and b were diffuse in the cytoplasm, indicating that recruitment not only to the Golgi apparatus and endosomes but also to clathrin-coated intermediates requires interaction with Rab GTPases (Fig. 6B).
FIGURE 6.
Cellular localization of OCRL1 mutants. GFP-tagged OCRL1 isoform a or b wild-type (WT) or the indicated clathrin box or α-adaptin-binding (A) or Rab-binding (B) mutants were expressed in HeLa cells as indicated and analyzed by epifluorescence microscopy. Bar, 10 μm.
We also analyzed the enrichment of OCRL1 isoforms in clathrin-coated vesicles that were partially purified from HeLa cells. As shown in Fig. 7A, GFP-OCRL1 is dramatically enriched in the vesicle fraction, whereas isoform b is present at much lower levels. As expected the Golgi membrane protein Golgin-84 is depleted from the vesicles, whereas clathrin is highly enriched. Enrichment of OCRL1 in the vesicle fraction is dependent upon clathrin binding, because deletion of both clathrin boxes results in a near complete loss of isoforms a and b from the vesicles (Fig. 7B). To determine which of the clathrin boxes are responsible for enrichment of isoform a in vesicles, mutants lacking either motif were analyzed. As shown in Fig. 7C, deletion of LIDIA had little effect upon enrichment in vesicles, whereas deletion of LIDLE dramatically reduced the levels of OCRL1 in the vesicle fraction. The levels of the isoform a LIDLE deletion mutant in clathrin-coated vesicles was similar to that of wild-type isoform b, consistent with the idea that it is LIDLE that is responsible for the differences in association with clathrin-coated intermediates between the two OCRL1 isoforms.
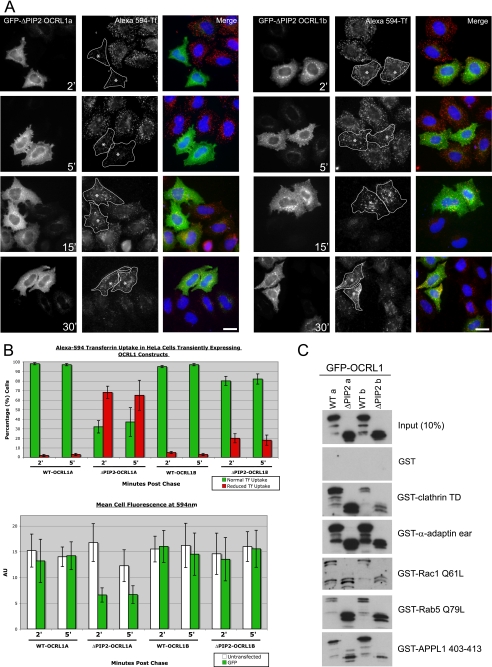
Functional Analysis of OCRL1 Isoforms—We have previously shown that expression of GFP-OCRL1 isoform b lacking the entire catalytic domain perturbs Golgi organization, alters the morphology of early endosomes, and blocks trafficking from endosomes to the TGN (6, 9). To determine whether there may be a functional distinction between the two isoforms of OCRL1, we generated an equivalent 5-phosphatase deletion construct in isoform a and studied effects upon organelle morphology and marker protein distribution. Similar to isoform b, expression of OCRL1 isoform a ΔPIP2 (where ΔPIP2 is deletion mutant lacking 5-phosphatase domain) also resulted in Golgi fragmentation and altered endosome morphology (supplemental Fig. S2 and S3). The morphologically altered endosomes generated by expression of either isoform contained transferrin receptor and cation-independent mannose 6-phosphate receptor (supplemental Fig. S3). However, the GFP-OCRL1 decorated endosomes appeared generally smaller and less distinct with isoform a compared with isoform b, and there was a less marked redistribution of clathrin onto these structures (supplemental Figs. S2 and S3). Importantly, the mutant OCRL1 constructs were expressed at the same level indicating that the observed effects are not because of different expression levels of the proteins (see Fig. 8C). These results suggest there may be a functional distinction between OCRL1 isoforms.
FIGURE 8.
Expression of GFP-OCRL1 isoform a ΔPIP2 impairs transferrin endocytosis. A, GFP-tagged OCRL1 isoform a or b ΔPIP2 deletion mutants were expressed in HeLa cells and Alexa 594-transferrin uptake analyzed at the indicated time points by fluorescence microscopy. Asterisks indicate cells expressing comparable levels of each transfected protein. Bar, 10 μm. B, quantitation of transferrin uptake was monitored in two ways. Top, the level of transferrin uptake in transfected cells relative to neighboring untransfected cells was counted (100 transfected cells counted per construct per time point for each experiment; results are presented as the mean ± S.D. for three experiments). Bottom, the mean transferrin fluorescence per transfected cell was quantitated (10 transfected cells for each construct and time point; results are presented as the mean ± S.D. for three experiments). C, binding of the indicated GFP-tagged OCRL1 constructs to GST-tagged bait proteins was analyzed by Western blotting with antibodies to OCRL1. α-ear, α-adaptin appendage domain.
To investigate this possibility in more detail we studied transferrin uptake in transfected cells, which occurs exclusively by clathrin-mediated endocytosis. Because OCRL1 isoform a is concentrated in clathrin-coated intermediates, including those at the cell periphery, we hypothesized that the dominant ΔPIP2 mutant of isoform a might impair the rate of transferrin uptake, whereas the equivalent version of isoform b would not. As shown in Fig. 8, expression of wild-type GFP-OCRL1 isoform a or b had no effect on transferrin uptake. The ΔPIP2 mutant of isoform b also did not affect transferrin uptake (Fig. 8, A and B). This is consistent with our previous observation that transferrin is efficiently delivered to the morphologically altered endosomes induced by this construct (6). In contrast to isoform b, there was a significant inhibition of transferrin uptake at early time points (2 and 5 min) in cells expressing the ΔPIP2 mutant of isoform a (Fig. 8, A and B). Transferrin did enter the cells at later times indicating that the effect is a delay in uptake rather than a block. Interestingly, for mutants of both isoforms the endocytosed transferrin appeared to accumulate in the enlarged OCRL1-coated endosomes at later times suggesting a possible delay in receptor recycling to the plasma membrane (Fig. 8A).
To address the mechanisms underlying the effects of mutant OCRL1 expression, the binding of the various constructs to known interaction partners was analyzed. Importantly, the expression levels of wild-type and mutant OCRL1 isoforms a and b were similar (Fig. 8C). As seen with the full-length wild-type OCRL1 proteins, binding of the deletion mutant of isoform a to clathrin was to a higher level than that of isoform b. For both isoforms, binding of the deletion mutants to clathrin was similar to that of their wild-type counterparts. The binding to α-adaptin, Rac1, and APPL1 was similar for all constructs, whereas the binding to Rab5 appeared greater for the deletion mutants compared with the wild-type proteins (Fig. 8C). This suggests that altered protein-protein interactions may lead to the observed effects of mutant OCRL1 expression.
DISCUSSION
We report here that the two known splice isoforms of OCRL1 differ in their association with clathrin and the extent to which they localize to clathrin-coated trafficking intermediates in cells. The additional eight residues present in isoform a that result in increased clathrin binding lie immediately adjacent to the LIDLE clathrin box and form part of a loop that projects out of the compact RhoGAP-like domain (8). The loop likely allows better access to the terminal domain of clathrin heavy chain, explaining the increased clathrin binding compared with isoform b, in which the LIDLE motif is predicted to lie closer to the surface of the RhoGAP-like domain and be less exposed. Although OCRL1 has a second clathrin-binding site, identified in this study as LIDIA, it is the LIDLE motif that is most important for association with clathrin-coated vesicles.
The role of the LIDIA motif is less clear. It differs from the consensus type I clathrin box by having an alanine at position 5 instead of an acidic residue. However, substitution of this residue has little effect on clathrin binding, indicating that it is not important for this interaction. Similar results have been reported for the tyrosine kinase activated Cdc42-associated kinase and epsin, which both bind clathrin via atypical type I clathrin boxes lacking a terminal acidic residue (23, 24). In both cases binding is to the same site in clathrin as that bound by the consensus type I clathrin box motif, which corresponds to a groove on the surface of the terminal domain β-propeller (25). Thus it is likely that the LIDIA and LIDLE motifs of OCRL1 bind to the same site in clathrin. Both motifs could bind sequentially to the same terminal domain, or more likely to adjacent terminal domains in a clathrin assembly. This would allow high avidity binding in the case of OCRL1 isoform a. In the case of isoform b, LIDIA would allow for a low level of association with clathrin-coated trafficking intermediates. An alternative scenario is that LIDIA promotes association with nonvesicle associated pools of clathrin, such as those forming flat lattices at the plasma membrane or on the surface of endosomes (26, 27).
Although the FEDNF motif of OCRL1 confers α-adaptin binding, its deletion does not appear to affect the amount of OCRL1 in clathrin-coated trafficking intermediates. Presumably, binding to α-adaptin allows sorting of OCRL1 into buds or vesicles containing this adaptor, which in turn could link OCRL1 with various types of internalized receptor. Another possibility is that α-adaptin binding helps orientate OCRL1 with respect to the membrane or the clathrin coat, facilitating its subsequent engagement with lipid or additional binding partners.
The difference in association with clathrin-coated intermediates between the two OCRL1 isoforms suggests they may have different cellular roles. The enrichment of isoform a in clathrin-coated vesicles argues for a role during the trafficking of these intermediates. The recent localization of OCRL1 isoform a to clathrin-coated pits at the plasma membrane suggests a possible role during endocytosis (8). We show here that a dominant negative isoform a ΔPIP2 deletion mutant impairs transferrin endocytosis, providing the first functional evidence for an endocytic role for OCRL1. Interestingly, the equivalent mutant version of isoform b did not affect transferrin uptake, although it did perturb endosome and Golgi morphology. This suggests that two functional pools of OCRL1 exist, one that is intimately associated with clathrin-mediated endocytosis and possibly other clathrin-mediated trafficking steps (isoform a), and another that is much less involved in such events (isoform b). Isoform b may participate in other non-clathrin-mediated trafficking or function in other processes such as signaling, actin dynamics, or serve a housekeeping function to maintain phosphoinositide homeostasis on Golgi and endosome membranes (3).
The mechanisms by which the ΔPIP2 deletion mutants lead to the observed cellular effects are unclear. They are unlikely to be solely due to changes in phosphoinositide metabolism because catalytically inactive point mutant versions of either isoform fail to give the same effects (data not shown). However, we did observe an increase in binding to Rab5 for the deletion mutants compared with their wild-type counterparts, suggesting that changes in protein-protein interactions may be responsible. This alone could be sufficient to induce the observed phenotype, or it may also require changes in phosphoinositide metabolism caused by loss of the catalytic domain.
Interestingly, isoform a is the only form of OCRL1 in the brain, one of the major tissues affected in Lowe syndrome, suggesting that defects in clathrin-mediated trafficking contribute to the Lowe syndrome pathology. In particular, neurons are highly active in endocytosis, both at the pre-synaptic membrane, where the bulk of clathrin participates in synaptic vesicle recycling, and the post-synaptic membrane. Endocytosis is also important for signaling during brain development. The identification of APPL1 as an OCRL1 binding partner suggests OCRL1 may participate in signaling from endocytic structures (8). Interestingly, recent studies have shown that APPL1 is important for TrkA trafficking and signaling in a neuronal cell model, and that it is abundant in the brain during embryogenesis where it is plays a key role in cell survival (28, 29). OCRL1 may therefore participate in these processes in the brain.
Both clathrin-binding sites are present in OCRL1 from vertebrates but absent from orthologues from “lower” organisms, including Drosophila melanogaster, Caenorhabditis elegans, and Dictyostelium discoideum, which also lack the FEDNF α-adaptin-binding motif. This would suggest OCRL1 is not present in clathrin-coated trafficking intermediates in these species, and that its enrichment there is a property unique to vertebrates, although this has yet to be investigated. Presumably the adaptation to bind clathrin reflects an important function for OCRL1 in clathrin-coated structures in vertebrates. It is therefore surprising that INPP5B, which lacks the clathrin and α-adaptin-binding sites and is absent from clathrin-coated structures (8, 16), can compensate for loss of OCRL1 in mouse knock-out studies (17). The reasons for this are currently unclear. Clearly, further work using appropriate model systems will be required to better understand the physiological importance of the OCRL1-clathrin interaction and the mechanisms by which INPP5B can compensate for the loss of OCRL1 in vivo.
Supplementary Material
Acknowledgments
We thank Professors Liz Smythe, Paul Luzio, Philip Woodman, and Harald Stenmark for generously providing reagents as noted above. We are indebted to Matthew Ball and Guanhua Yan for excellent technical assistance. We also thank Dr. Irene Barinaga-Rementeria Ramirez for critically reading the manuscript.
This work was supported by Medical Research Council Senior Nonclinical Research Fellowship G117/494 and Biotechnology and Biological Sciences Research Council Project Grant 34/C17842 (to M. L.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: TGN, trans-Golgi network; GST, glutathione S-transferase; GFP, green fluorescent protein; NRK, normal rat kidney; MES, 4-morpholineethanesulfonic acid.
References
- 1.Lowe, C. U., Terrey, M., and Mac, L. E. (1952) AMA Am. J. Dis. Child 83 164-184 [DOI] [PubMed] [Google Scholar]
- 2.Nussbaum, R., and Suchy, S. F. (2001) in Metabolic and Molecular Basis of Inherited Diseases (Scriver, C. R., Beauder, A. L., Sly, W. S., and Valle, D., eds) pp. 6257-6266, McGraw-Hill Inc., New York
- 3.Lowe, M. (2005) Traffic 6 711-719 [DOI] [PubMed] [Google Scholar]
- 4.Olivos-Glander, I. M., Janne, P. A., and Nussbaum, R. L. (1995) Am. J. Hum. Genet. 57 817-823 [PMC free article] [PubMed] [Google Scholar]
- 5.Faucherre, A., Desbois, P., Nagano, F., Satre, V., Lunardi, J., Gacon, G., and Dorseuil, O. (2005) Hum. Mol. Genet. 14 1441-1448 [DOI] [PubMed] [Google Scholar]
- 6.Choudhury, R., Diao, A., Zhang, F., Eisenberg, E., Saint-Pol, A., Williams, C., Konstantakopoulos, A., Lucocq, J., Johannes, L., Rabouille, C., Greene, L. E., and Lowe, M. (2005) Mol. Biol. Cell 16 3467-3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ungewickell, A., Ward, M. E., Ungewickell, E., and Majerus, P. W. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 13501-13506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdmann, K. S., Mao, Y., McCrea, H. J., Zoncu, R., Lee, S., Paradise, S., Modregger, J., Biemesderfer, D., Toomre, D., and De Camilli, P. (2007) Dev. Cell 13 377-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyvola, N., Diao, A., McKenzie, E., Skippen, A., Cockcroft, S., and Lowe, M. (2006) EMBO J. 25 3750-3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faucherre, A., Desbois, P., Satre, V., Lunardi, J., Dorseuil, O., and Gacon, G. (2003) Hum. Mol. Genet. 12 2449-2456 [DOI] [PubMed] [Google Scholar]
- 11.Lichter-Konecki, U., Farber, L. W., Cronin, J. S., Suchy, S. F., and Nussbaum, R. L. (2006) Mol. Genet. Metab. 89 121-128 [DOI] [PubMed] [Google Scholar]
- 12.Suchy, S. F., and Nussbaum, R. L. (2002) Am. J. Hum. Genet. 71 1420-1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jefferson, A. B., and Majerus, P. W. (1995) J. Biol. Chem. 270 9370-9377 [DOI] [PubMed] [Google Scholar]
- 14.Schmid, A. C., Wise, H. M., Mitchell, C. A., Nussbaum, R., and Woscholski, R. (2004) FEBS Lett. 576 9-13 [DOI] [PubMed] [Google Scholar]
- 15.Shin, H. W., Hayashi, M., Christoforidis, S., Lacas-Gervais, S., Hoepfner, S., Wenk, M. R., Modregger, J., Uttenweiler-Joseph, S., Wilm, M., Nystuen, A., Frankel, W. N., Solimena, M., De Camilli, P., and Zerial, M. (2005) J. Cell Biol. 170 607-618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams, C., Choudhury, R., McKenzie, E., and Lowe, M. (2007) J. Cell Sci. 120 3941-3951 [DOI] [PubMed] [Google Scholar]
- 17.Janne, P. A., Suchy, S. F., Bernard, D., MacDonald, M., Crawley, J., Grinberg, A., Wynshaw-Boris, A., Westphal, H., and Nussbaum, R. L. (1998) J. Clin. Investig. 101 2042-2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nussbaum, R. L., Orrison, B. M., Janne, P. A., Charnas, L., and Chinault, A. C. (1997) Hum. Genet. 99 145-150 [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P. M., Armour, C. D., Santos, R., Schadt, E. E., Stoughton, R., and Shoemaker, D. D. (2003) Science 302 2141-2144 [DOI] [PubMed] [Google Scholar]
- 20.Diao, A., Rahman, D., Pappin, D. J., Lucocq, J., and Lowe, M. (2003) J. Cell Biol. 160 201-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirst, J., Miller, S. E., Taylor, M. J., von Mollard, G. F., and Robinson, M. S. (2004) Mol. Biol. Cell 15 5593-5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCrea, H. J., Paradise, S., Tomasini, L., Addis, M., Melis, M. A., De Matteis, M. A., and De Camilli, P. (2008) Biochem. Biophys. Res. Commun. 369 493-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drake, M. T., Downs, M. A., and Traub, L. M. (2000) J. Biol. Chem. 275 6479-6489 [DOI] [PubMed] [Google Scholar]
- 24.Teo, M., Tan, L., Lim, L., and Manser, E. (2001) J. Biol. Chem. 276 18392-18398 [DOI] [PubMed] [Google Scholar]
- 25.ter Haar, E., Harrison, S. C., and Kirchhausen, T. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1096-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Signoret, N., Hewlett, L., Wavre, S., Pelchen-Matthews, A., Oppermann, M., and Marsh, M. (2005) Mol. Biol. Cell 16 902-917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sachse, M., Urbe, S., Oorschot, V., Strous, G. J., and Klumperman, J. (2002) Mol. Biol. Cell 13 1313-1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varsano, T., Dong, M. Q., Niesman, I., Gacula, H., Lou, X., Ma, T., Testa, J. R., Yates, J. R., III, and Farquhar, M. G. (2006) Mol. Cell. Biol. 26 8942-8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schenck, A., Goto-Silva, L., Collinet, C., Rhinn, M., Giner, A., Habermann, B., Brand, M., and Zerial, M. (2008) Cell 133 486-497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.