Abstract
In therapeutic antibody preparation, acidic pH conditions are generally used for elution from Protein A affinity column of IgG or for its viral inactivation. Exposing IgG to low pH conditions induces conformational changes, leading to its functional damage or loss, although the mechanisms have not been fully elucidated. In this study using random peptide T7 phage display libraries, we isolated a unique and novel peptide motif that specifically recognized the non-native conformer (acid conformer) of human IgG that was generated by the low pH treatment, but not the native conformer. We examined the generation conditions and biochemical properties of acid conformer using the peptide motif as an affinity ligand. The acid conformer was easily generated at acidic pH (<pH 3.0) and at moderate temperatures (20-40 °C). The conformer was present in a monomeric form functionally maintaining antigen or Fc receptor binding, but showed a tendency to aggregate with a long incubation time at neutral pH (>25 °C). The peptides isolated here could contribute to the elucidation of the mechanisms of antibody dysfunction or aggregation during acid exposure as well as storage of human IgG.
Protein A is widely used as an affinity ligand for the purification of human antibodies because it specifically binds to the Fc region of IgG (1). However, we must consider possible contamination with bacterial endotoxin and Protein A itself in purified antibody preparations for clinical use, because Protein A is derived from bacteria and possesses high anti-genicity (2). Therefore, many investigators have attempted to construct purification systems as alternatives to the Protein A column. By investigating low molecular weight compounds, Li et al. made Protein A mimetics and performed IgG purification from human plasma and murine ascites fluid (3). Fassina et al. used peptide derivatives as affinity ligands, which have broader specificity to IgG than Protein A, and applied them to antibody affinity chromatography (4). Ehrlish and Bailon, and Krook et al. also reported IgG-binding peptides discovered using the filamentous phage display technique (5, 6). Recently, Verdoliva et al. screened a synthetic peptide library and identified an IgG-binding cyclic dimeric peptide that recognized the lower hinge region of IgG (7, 8). Of these earlier attempts, the Fc-III peptide with an intramolecular disulfide bond reported by DeLano et al. in 2000 (9) is a potential candidate that can displace Protein A functions. This is because the Fc-III peptide binds with relatively high affinity (the apparent dissociation constant Kd is 30 nm) to the groove between the CH2 and CH3 domains of human IgG and shares common binding sites with Protein A. In addition, the Fc-III peptide has been used in studies of affinity enhancement (10) and artificial cell-surface antibody receptors (11).
Krumpe et al. recently reported the construction of a random peptide library on a T7 phage display system (12). The library has marked characteristics including reduced bias of amino acids generated by the mixed nucleotides in the displayed peptides and increased peptide diversity, as compared with that of the M13 filamentous peptide library. By using this library, we tried to isolate novel IgG-binding peptide ligands. In the process of this research, we identified several peptide sequences sharing high consensus motifs with extremely high binding specificity to human IgG. Unexpectedly, however, this peptide motif did not recognize the normal conformation of human IgG. To identify the target species for our peptides, we examined the binding of our peptides to human IgG treated with a purification process and found that our peptides targeted particular conformational species, which was induced by acid treatment of human IgG. We refer to this alternative conformer as an “acid conformer.”
Acidic pH conditions are not only used for elution of IgG from the Protein A column, but are also used as a method for eradicating virus contamination (2, 13). It has been reported that, when antibodies are exposed to acidic pH conditions, a conformer with properties that are different to normal IgG is generated (14). Although the effects of acid treatment on antibody structure has been studied using mouse or rabbit IgG (15, 16), the properties of the acid conformer are not fully understood. This is the first report describing the generation conditions and biophysical characteristics of the human IgG acid conformer that was identified using a specific affinity ligand. Our data will aid understanding of the causes and mechanisms of dysfunction and aggregation of IgG that occur during acid treatment and storage of IgG.
EXPERIMENTAL PROCEDURES
Materials—Polyclonal human IgG was purchased from ICN/Cappel Biomedicals, Aurora, OH. Human transferrin, human serum albumin, bovine serum albumin (BSA),4 and Staphylococcus aureus Protein A were purchased from Sigma. Human IgG Fc fragment (IgG-Fc), human IgA, and human IgE were purchased from Athens Research & Technology, Athens, GA. Mouse IgG, mouse IgA, and mouse IgE were purchased from PharMingen, San Diego, CA. Gelatin was purchased from Wako Chemicals, Arcadia, CA. The humanized anti-human interleukin-6 receptor (IL-6R) therapeutic antibody Tocilizumab (MRA) and the humanized anti-HER2 therapeutic antibody Trastuzumab (Herceptin) were kindly provided by Chugai Pharmaceutical Corporation Ltd., Tokyo, Japan. Recombinant human IL-6R and the CTLA4/Fc fusion protein were purchased fromR&D Systems, Minneapolis, MN. Recombinant human Fcγ receptor (FcγR) Ia, IIb, and IIIa were purchased from Abnova Corporation, Taipei, Taiwan.
Random Peptide T7 Phage Libraries and Biopanning against Human IgG-Fc—The T7 phage libraries displaying CX7-10C or X3CX7-10CX3 random peptides used here (X is a mixture of twenty amino acid residues) were constructed as described previously (12).
Microplate wells (Nunc Maxisorp) were coated with human IgG-Fc (1 μg/300 μl/well) and blocked with 0.5% BSA in PBS. The T7 phage libraries (1.5 × 1011 pfu) of CX7-10C or X3CX7-10CX3 were added to the wells and incubated for 1 h at room temperature. After washing the wells five times with PBS containing 0.1% Tween 20 (0.1% PBST) (second round, 0.1% PBST, 10 s incubation × five times; third round, 0.3% PBST, 10 s incubation × 10 times; fourth and fifth rounds, 0.5% PBST, 2 min incubation × 10 times), BLT5615 Escherichia coli cells were added, infected by phages, and cultured for phage propagation. The phages recovered from the culture supernatants were precipitated with polyethylene glycol, dissolved in PBS, filtered through a 0.22-μm nitrocellulose filter, and used for the next round of panning, according to the manufacturer's recommendation (Novagen). After five rounds of panning, the phages were monocloned on the culture plates forming phage plaques.
Preparation of Synthetic Peptides—Biotinylated ethylene glycol spacerarmed peptides IMG1 (biotin-PEO4-GCGYWRSEWGLCG), IMG-1E6Q (biotin-PEO4-GCGYWR-SQWGLCG), IMG4 (biotin-PEO4-GCTGYWPKAWGLCG), IMG4-K6R (biotin-PEO4-GCTGYW-PRAWGLCG), CS (biotin-PEO4-GSGYWRSEWGLSG), and Fc-III (biotin-PEO4-GDCAWHLGELV-WCT, C-terminal amidate blocked) were chemically synthesized by Fmoc protection chemistry and subsequent biotinylation with NHS-PEO4-Biotin (Sigma). After the protecting groups were removed, all peptides were purified on C-18 reversed-phase columns. Peptide cyclization via disulfide bond formation was performed in dimethyl sulfoxide by air oxidation. The intramolecular disulfide formation was confirmed by liquid chromatographic mass spectrometry on the Acquity SQD ultra-performance liquid chromatography system (Waters).
Detection of Human IgG Interaction with Phages or Synthetic Peptides by ELISA—Each microplate well (Nunc Maxisorp) was coated with human IgG, other control proteins (100 ng/50 μl/well), or 100-fold diluted human serum in PBS, and blocked with 0.5% BSA in PBS. Phages were added to each well and incubated. The bound phages were detected with biotinylated anti-T7 phage mouse antibody (Novagen) and horseradish peroxidase-conjugated streptavidin (SA) (Vector Laboratories, Peterborough, UK). To estimate the content of bound phages, tetramethylbenzidine colorimetry solution (Wako Chemicals), a horseradish peroxidase-conjugated peroxidase substrate, was added to each well, and the absorbance at 450 nm was measured using a microplate reader.
For peptide ELISA, biotinylated IMG4K6R, Fc-III, or CS peptide (500 nm) were pre-incubated with alkaline phosphatase-conjugated (AP-conjugated) SA (120 nm, Vector) to form an SA-peptide complex, and the mixture was added to the wells. After incubation for 1 h, the wells were washed three times with 0.1% PBST. To estimate the content of the bound peptide complex, p-nitrophenyl phosphate (Wako Chemicals), an AP substrate, was added to each well, and the absorbance at 405 nm was measured.
Acid Treatment of Human IgG—Acid treatment at low pH was performed using 0.1 m glycine-HCl buffer (pH 3.5-2.2) for specified incubation times at room temperature and subsequently neutralized by adding 1 m Tris-HCl buffer (pH 9.1). After treatment, IgG was stored at 4 °C until use.
Immunoprecipitation of Human IgG by Magnetic Beads—Immobilization of biotinylated IMG1, IMG1E6Q, IMG4, IMG4K6R, and Fc-III peptides (200 pmol) on Dynabeads M-280 SA magnetic beads (0.2 mg) (Dynal Biotech, Oslo, Norway) was performed according to the manufacturer's protocols. After blocking with 0.5% BSA in PBS, the beads were reacted with 100 μl of commercially available polyclonal human IgG (200 μg/ml) in PBS containing 0.5% BSA or human serum diluted with PBS (100-fold) on a rotary incubator for 1 h. After washing three times with 0.1% PBST, beads were added to SDS sample buffer containing 2% 2-mercaptoethanol, boiled, and then applied to a 12.5% SDS-PAGE gel. After electrophoresis, the proteins were electroblotted onto a polyvinylidene difluoride membrane (Millipore Corp. Billercia, MA). The membrane was blocked with 0.5% BSA in PBS and subjected to Western blot analysis using biotin-conjugated anti-human IgG goat antibody (Vector) and horseradish peroxidase-conjugated SA (Vector Laboratories). The blotted protein bands were detected by chemiluminescence using Chemi-Lumi One (Nacalai Tesque, Inc., Kyoto, Japan).
Purification of Acid Conformer of Human IgG—Biotinylated IMG4K6R peptide was immobilized on a SA HiTrap column (1 ml; GE Healthcare, Chalfont St. Giles, UK), according to the manufacturer's instructions. After blocking the column with 0.5% BSA in PBS and washing with 20 ml of PBS, 1 ml MRA solution treated with acidic conditions (pH 2.7 × 60 min) was applied to the IMG4K6R peptide column. PBS (1 ml) was injected into the column, and the eluate was collected as the excluded fraction. The column was then washed with 20 ml PBS, and the bound proteins were eluted with 1 ml of 0.1 m glycine-HCl (pH 2.7), neutralized immediately by adding 100 μl of 1 m Tris-HCl buffer (pH 9.1), and stored at 4 °C until use. After use, the column was regenerated with 20 ml of PBS and stored at 4 °C.
Surface Plasmon Resonance (SPR) Analysis—SPR analysis was performed on a BIAcore 2000 (GE Healthcare) at 25 °C. All reagents and sensor chips were purchased from GE Healthcare. Immobilization of the biotinylated IMG4K6R peptide on the SA sensor chip was done by injecting 100 μm peptide solution at pH 7.0. The amount of the immobilized peptide was adjusted to 200-400 response units (RU). To regenerate the sensor chip, 0.1 m glycine-HCl buffer (pH 2.7) was used. The association reaction was monitored by injecting several concentrations of IgG dissolved in HBS buffer (10 mm HEPES (pH 7.4), containing 0.15 m NaCl, 3 mm EDTA, and 0.005% Tween 20) into the sensor chip at a flow rate of 10 or 100 μl/min. The dissociation phase was monitored by HBS buffer flow. The binding kinetic parameters were calculated by BIAevaluation 3.2 software (BIAcore, Uppsala, Sweden).
To assess the inhibitory activity of Protein A toward the interaction between IMG4K6R peptide and the acid conformer, the acid-treated MRA (100 ng/μl, 660 nm) was preincubated with Protein A (1-100 μg/μl, 0.24-24 μm) for 1 h and injected into an IMG4K6R peptide-immobilized flow cell.
To estimate the binding activity of the acid conformer to the antigen or FcγRs, the acid-treated MRA (100 ng/μl, 660 nm) was injected into the IMG4K6R peptide-immobilized flow cell. Subsequently, human IL-6R (antigen) or human FcγR (FcRIa, IIb, or IIIa; 5-20 ng/μl) was injected to monitor the association and dissociation reactions.
RESULTS
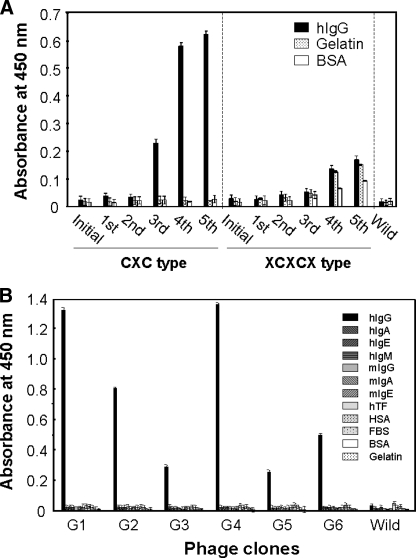
Isolation of Human IgG-specific Phage Clones from Random Peptide Phage Libraries—Five rounds of biopanning against human IgG-Fc were performed using T7 phage libraries displaying CX7-10C or X3CX7-10CX3. Human IgG-Fc binding phages were enriched in CX7-10C libraries at the fourth and fifth rounds, but not in the X3CX7-10CX3 libraries (Fig. 1A). The binding assays for the phage clones isolated after the fourth and fifth panning rounds were done by ELISA, and the DNA sequences of the positive clones were determined. Six individual phage clones (G1-6) were identified (Table 1) and showed specific binding to human IgG but not to other immunoglobulins or control proteins (Fig. 1B).
FIGURE 1.
The isolation of human IgG-specific phage clones from random peptide libraries constructed on the T7 phage display system. A, biopannings were performed against human IgG-Fc fixed on a plastic plate using two types of disulfide-constrained peptide libraries (CXC type: CX7-10C or XCXCX type: X3CX7-10CX3). The enrichment of human IgG-specific phages was confirmed only from the CXC library by phage ELISA. B, phages obtained after the fourth and fifth pannings from the CXC library were cloned and used for the binding assay and DNA sequence analysis. Six individual clones specific to human IgG were obtained and the binding specificities of each phage clone were examined by monoclonal phage ELISA. The abbreviations used in the panel are as follows: hIg, human immunoglobulin; mIg, mouse immunoglobulin; hTF, human transferrin; HSA, human serum albumin; FBS, fetal bovine serum; and BSA, bovine serum albumin.
TABLE 1.
Comparison of the amino acid sequences of human IgG-binding peptides type II (G1-6) and type I (Fc-III)
Asterisks indicate highly (more than 5/6) conserved amino acid positions among the type II motifs. The Fc-III sequence is aligned with the type II peptide sequences, with fixed positions of Cys residues.
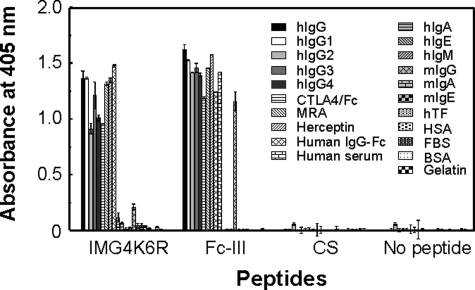
Binding Specificity of Type II Synthetic Peptide to Human IgG—The peptide sequences displayed on the phage clones shared a high consensus [C (T/-) (G) YW (P/A) (R) (E) WGLC; ≥80% of the preserved amino acids are indicated outside parentheses, while those preserved at ≥30% are within] and were named the type II motif. This was clearly different from the type I motif (Fc-III, DCAWHLGELVWCT) reported by DeLano et al. (9) (Table). The binding specificities of the type I (Fc-III) and type II (IMG4K6R: CTGYWPRAWGLC) synthetic peptides were compared by ELISA using various antibodies. Both peptides bound to human IgGs including therapeutic and IgG1-4 isotype human antibodies (Fig. 2). Moreover, IMG4K6R exhibited binding activity toward the CTLA4/human Fc fusion protein, indicating that the binding site of the type II peptide was specific to the Fc region. While IMG4K6R was highly specific to human IgG-Fc and did not bind the mouse IgG, Fc-III recognized rabbit and goat IgG as well as mouse and human IgG, as shown in Fig. 2, suggesting that IMG4K6R and Fc-III possess different binding properties.
FIGURE 2.
The binding specificities of type I (Fc-III) and type II (IMG4K6R) peptides in ELISA. IMG4K6R peptide is a derivative in which the 6th Lys in the G4 peptide sequence was replaced with Arg and was used as a representative of the type II peptide. CS indicates an IMG1 peptide derivative in which two Cys residues were substituted with Ser. The abbreviations used in the panel and not listed in Fig. 1 are as follows: CTLA4/Fc, a fusion protein of the extracellular domain of human CTLA4 (CD152) and human Fc (IgG1); MRA, anti-human IL-6R human antibody Tocilizumab (IgG1); Herceptin, anti-HER2 (Erb2) human antibody Trastuzumab (IgG1).
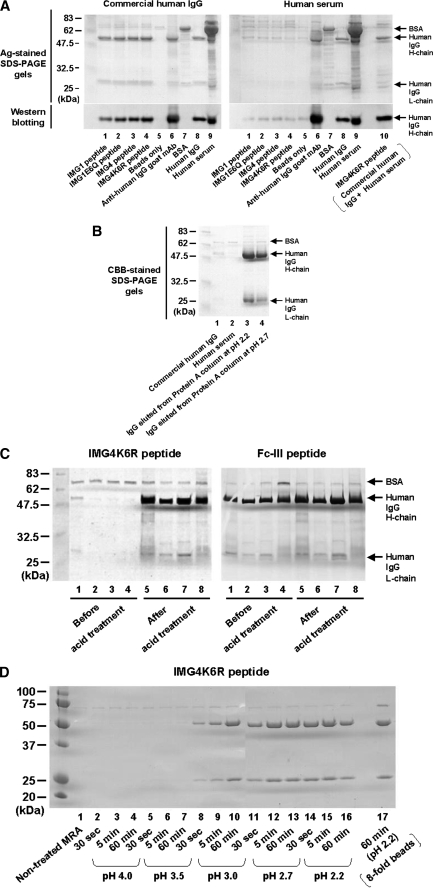
Recognition of the Acid Conformer of Human IgG Using the Type II Peptide—Despite sufficient binding of Fc-III to the human serum containing IgG, IMG4K6R did not show clear binding in the ELISA (Fig. 2). To confirm this observation, binding experiments were performed by immunoprecipitation with type II peptide-immobilized magnetic beads using human serum or commercial polyclonal human IgG (Fig. 3A). IgG was recovered from the polyclonal human IgG solution (Fig. 3A, left panel, lanes 1-4) with several type II peptides, but almost no IgG was pulled down from human serum (Fig. 3A, right panel, lanes 1-4). To explain this observation, we suggest that type II peptides recognize abnormal (irregular) structures that are contained in commercial human IgG but not in serum.
FIGURE 3.
Detection of the acid conformer recognized by the type II peptide. A, precipitation of the antibody from commercial polyclonal human IgG (200 μg/ml) or human serum (100× dilution) with type II peptides (IMG1, IMG1E6Q, IMG4, IMG4K6R) or anti-human IgG goat antibody immobilized magnetic beads. The beads were subjected to SDS-PAGE under reduced conditions, and the captured human IgG was detected by silver staining or Western blotting. B, enhanced reactivity of the type II peptide to human IgG in serum after Protein A column purification. Human serum was applied to the Protein A column (Protein A HiTrap column, GE Healthcare), followed by washing, elution with 0.1 m glycine-HCl (pH 2.2 or 2.7) and neutralization. Reactivity of the purified antibody to the type II peptide IMG4K6R was determined by bead precipitation and subsequent SDS-PAGE in the same manner as A. C, augmentation of the reactivity of human IgG to the type II peptide by low pH treatment. Using polyclonal human IgG (lanes 1 and 5), MRA (lanes 2 and 6), Herceptin (lanes 3 and 7), and human serum (lanes 4 and 8), the binding reactivity to type II (IMG4K6R) or type I (Fc-III) peptide was examined before and after acid treatment (pH 2.7). D, effect of pH and treatment time on the generation of the acid conformer. MRA was incubated at pH 2.2-4.0 for 30 s, 5 min, or 1 h at room temperature, immediately neutralized, and stored at 4 °C until use.
Because commercial polyclonal human IgG is generally purified using Protein A columns, we speculated that the irregular structures might have arisen from the purification process. Therefore, we purified IgG from human serum using a Protein A column with acidic pH elution (pH 2.2 and 2.7), followed by immediate neutralization. Although no IgG was recovered from human serum with type II peptide beads before Protein A purification (Fig. 3B, lane 2), a considerable amount of IgG was precipitated from the human IgG solution after purification (Fig. 3B, lanes 3 and 4). One plausible explanation for this observation is that the low pH used for the elution generates structures other than normal human IgG. To investigate this finding, we examined the binding of IgG to type II peptide beads before and after acid treatment (pH 2.7). The binding of IgG to the IMG4K6R beads was very weak before acid treatment, but increased dramatically after treatment (Fig. 3C, left panel). This suggests that exposure of human IgG to acidic conditions induces a conformational change to an alternative structure, i.e. the acid conformer, which is specifically recognized by type II peptides. In contrast, the apparent binding between human IgG and Fc-III did not change before and after treatment (Fig. 3C, right panel). Generation of the acid conformer, as detected by the type II peptide, was observed at a pH lower than 3.0 at room temperature within a short period of time (30 s), as shown in Fig. 3D.
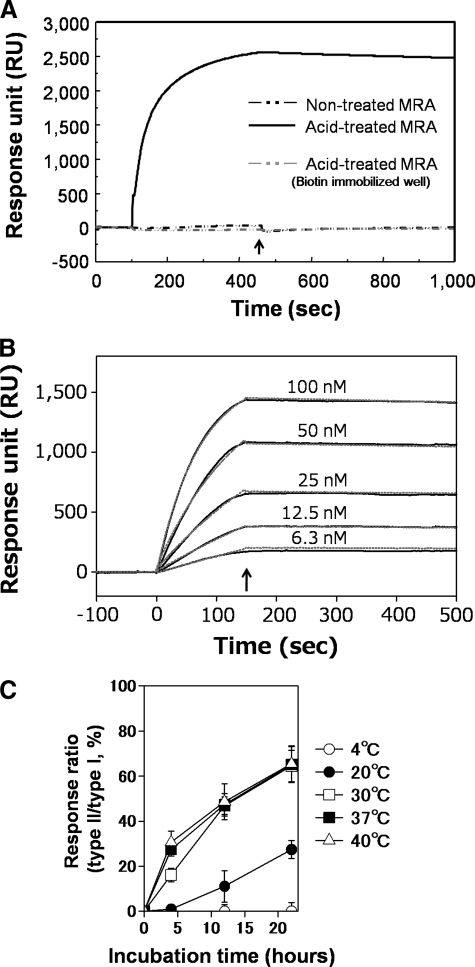
Detection of the Acid Conformer by SPR Analysis—The generation of the acid conformer was examined by SPR analysis using a type II peptide-immobilized sensor chip. Although there were no binding responses on the sensorgram before acid treatment at pH 2.7, a remarkable increase in the signal (∼2500 RU at 350 s after injection) was observed after acid treatment (Fig. 4A). This finding indicates that SPR analysis using the type II peptide-immobilized sensor chip is a sensitive and convenient method for monitoring generation of the acid conformer. The binding between the immobilized type II peptide and the purified acid conformer was very strong with an apparent dissociation constant (Kd) of 0.5 nm (Fig. 4B).
FIGURE 4.
SPR analysis of the acid conformer using the type II peptide. A, detection of the acid conformer generated by acid treatment. MRA (660 nm) before and after acid treatment was analyzed on a type II peptide (IMG4K6R)-immobilized sensor chip using the BIAcore 2000. At the time indicated by the arrow (450 s), the injection phase was stopped and the flow cells were washed with HBS buffer. B, estimation of the kinetic parameters in binding of the acid conformer to the type II peptide IMG4K6R on SPR analysis. The acid conformer (6.3-100 nm) purified by the type II peptide affinity column was injected into the type II peptide-immobilized flow cell on the SA sensor chip at a flow rate of 100 μl/min, and the association/dissociation reactions were monitored. The amount of the immobilized type II peptide was 220 RU. At the time indicated by the arrow (150 s), the dissociation phase was started by eluting with HBS buffer. The sensorgrams were subtracted with that of the control cell, which was immobilized with 0.1 mm biotin and including in the fitting calculation. The kinetic parameters were evaluated assuming a 1:1 binding model and using the global fitting calculation, as follows: (1) Kd of 0.5 nm; (2) ka of 1.4 × 105 m-1s-1; and (3) kd of 7.0 × 10-5 s-1. The solid and broken lines indicate the experimental and fitting data (Chi2 = 201), respectively. C, effects of acid-treatment temperature on the generation of the acid conformer. Human antibody (Herceptin) incubated at pH 2.7 and at each temperature for 0-22 h was immediately neutralized and injected into the flow cells immobilized with type I (Fc-III) or type II (IMG4K6R) peptides. The response units (RU) of the type I and type II peptides were measured at 300 s after injection, and the % ratio (type II/type I) was plotted against the incubation time at acidic pH.
The effect of temperature on the generation of acid conformer was examined using the SPR system (Fig. 4C). The rate of acid conformer generation at pH 2.7 increased as temperature increased from 4-40 °C. This dependence of the generation rate on temperature may reflect the finding that the mobility or fluctuation of IgG induced by the increased temperature is favorable for conformational changes to the acid conformer.
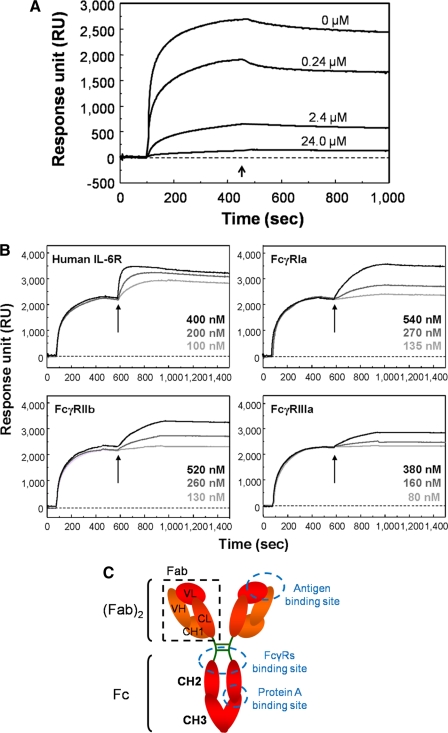
Characterization of the Acid Conformer—To characterize the binding sites of the type II peptide on the acid conformer, competitive inhibition assays of type II peptide with Protein A were performed using SPR analysis (Fig. 5A). The SPR signal arising from the binding of the acid conformer to the immobilized type II peptide was reduced by the addition of Protein A (0.24-24 μm) in a concentration-dependent manner, indicating that the binding site of the type II peptide overlapped with or was adjacent to the region recognized by Protein A.
FIGURE 5.
The binding properties of the acid conformer to Protein A, antigen and Fcγ receptors. A, inhibition of the binding between the type II peptide and the acid conformer by Protein A. Acid-treated MRA (660 nm) was injected to the type II peptide (IMG4K6R)-immobilized sensor chip in the presence of several concentrations of Protein A (0-24 μm). At the time indicated by the arrow (450 s), the injection phase was stopped and the cells were washed with HBS buffer. B, antigen- and receptor-binding properties of the acid conformer trapped on the type II peptide-immobilized sensor chip. Acid-treated MRA (660 nm) was trapped on the sensor chip at 2200 RU. The antigen (human IL-6R) or FcγR (human FcγRIa, FcγRIIb, or FcγRIIIa) was subsequently injected at each concentration (times indicated by arrows). C, schematic diagram of human IgG. Acid treatment induces conformational changes on the Fc region but not on the Fabs or antigen binding region.
To examine the functionality of the acid conformer, we evaluated the antigenand FcγR -binding capacities of the acid conformer. The acid conformer was trapped on the type II peptide-immobilized sensor chip on BIAcore and an antigen (IL-6R) or FcγR were subsequently injected (Fig. 5B). The acid conformer maintained binding activities to both the antigen and the FcγR, suggesting that conformational changes of the acid conformer are probably limited to the Fc regions (Fig. 5C). In addition, a circular dichroism (CD) spectrum of the purified acid conformer indicated a high content of beta-sheet structures despite a slight increase in random structures (supplemental Fig. 1). Furthermore, gel permeation chromatography of the purified acid conformer revealed that ≥90% of the molecules were present as monomers (supplemental Fig. 2). Combining these data, we tentatively concluded that the conformational changes of the acid conformer is limited to the CH2 and CH3 regions, and does not have features typical of denatured structures.
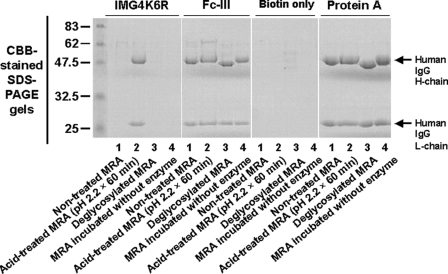
Conformational Changes Were Not Correlated with Removal of the Carbohydrate in IgG—Conformational changes of the acid conformer seemed to occur predominantly in the Fc region (Fig. 5). Recently, it was reported that the FcγR-binding activity of IgG is modulated by the removal of a carbohydrate structure attached to the CH2 domain of IgG through dynamic or static structural changes around the hinge region, although the mechanism is not fully understood (17, 18). We assessed the binding ability of the carbohydrate-free IgG to the type II peptide using a precipitation-binding assay with type II peptide-immobilized beads (Fig. 6). The carbohydrate-free IgG was prepared by endoglycosidase digestion and characterized by the decrease in the apparent molecular size of the heavy chain (48 kDa) on SDS-PAGE. This molecule did not bind to the type II peptide, indicating that there is no correlation between the acid conformer and the structural changes induced by removing the carbohydrate from IgG. In contrast, the binding of type I peptide (Fc-III) and Protein A to human IgG was not affected by removing the carbohydrate from IgG.
FIGURE 6.
Reactivity of the type II peptide to carbohydrate-free human IgG. The N-glycosidic sugar chain of MRA was removed by incubation with endoglycosidase PNGase F (New England BioLabs Inc., MA) in 50 mm acetate buffer (pH 5.5) at 37 °C for 24 h. After deglycosylation, the human IgG was subjected to the immunoprecipitation assay using magnetic beads.
DISCUSSION
In this study, we identified a new and unique peptide motif that exhibits specific binding to the non-native conformation of human IgG, which was induced by acid treatment. Although several IgG-binding peptide sequences have already been reported (5-9), such peptides cannot recognize such conformationally changed IgG species. Several explanations are possible for the reason why we could isolate a type II peptide motif, even though the acid conformer is a minor component of the human IgG samples. One reason is the difference between the peptide libraries expressed by filamentous M13 and lytic T7 phage display systems. T7 phage display library show less bias in the displayed amino acids, which allows greater coverage of a variety of peptide sequences in the library (12). The display of a part of the peptide sequences in the library may be restricted in the M13 display system. In fact, the display of the Trp cage motif for peptide library construction was successful in the T7 phage but not in the M13 phage (19), probably because of the incompatibility of the Trp cage motif and the biological constraints of the M13 system, which needs the displayed peptides to penetrate the cell membrane (20). The other reason is the slow dissociation rate of the complex between the acid conformer and the type II peptide (kd = 7.0 × 10-5 s-1, Fig. 4B). Because we used a strict washing procedure in biopanning selections, the T7 phages that are specific to normal human IgG might be washed out early. In fact, we also succeeded in isolating peptide motifs that were very similar to the type I peptide (data not shown). However, this succeeded only when the type II peptide binding sites of the human IgG-Fc on the plastic plate were masked in advance by adding the IMG4K6R synthetic peptide to the biopanning solution and the washing procedure was gentle.
Using the type II peptide, we identified an alternative conformer of human IgG and we have described the biophysical characteristics of this unique conformer. The existence of an acid conformer has been suggested in several reports. For example, Buchner et al. reported that the treatment of mouse monoclonal antibody MAK33 (IgG1/κ) at pH 3.0 leads to different structures, which exhibited properties similar to the folding intermediates, and the increased hydrophobic molecular surfaces maintained a high content of secondary structures and showed cooperative unfolding against heat and denaturation (15). Martsev et al. reported that, by exposing the rabbit IgG to pH 2, a partially structured conformer was generated, which was characterized by conformational loss, probably in the CH2 domain, and by maintaining the well-defined tertiary structure in other domain regions. Furthermore, the IgG that was refolded at pH 7 after heat denaturation at pH 2 contained a non-native conformer with different binding capacities to the C1q complement and Protein A (16). Recently, Ejima et al. studied the effects of acid exposure on conformational changes in human IgG4 and showed that the molecular species of the non-denatured structures with limited structural changes are generated by exposure to pH 2.7-3.9 by CD and differential scanning colorimetry (14). These studies suggest that acidity induces alternative structures of IgG. Although we must further investigate whether the acid conformer that we identified using the type II peptide is identical to these previously reported structures, they seem to share common characteristics including a high content of secondary structures (beta-sheet), the majority of molecules present in the monomeric form and increased hydrophobicity.
Type II peptides are thought to bind to the CH2-CH3 domain on the Fc region of human IgG, because binding of the type II peptide is inhibited by Protein A (Fig. 5A). As depicted in Fig. 5C, this binding region on IgG involves the hotspot region, which is also recognized by Protein G, neonatal FcR (FcRn) (21), rheumatoid factors (RFs) (22), and type I peptide (9). Competitive inhibition of type II peptide binding using Protein A indicates that the conformational changes in the acid conformer occurred in the region that contains the hotspot. Although we cannot currently identify the precise binding site of the type II peptide, the previous papers described the suggestive but controversial results that the domain is affected by acid treatment. Martsev et al. proposed that the CH2 domain of rabbit IgG is destroyed by acid treatment (16). In contrast, Thies et al. who used the isolated CH3 domain of murine IgG, demonstrated that the CH3 domain could possess an alternative conformational state induced by acidic pH (23). Taken together, the conformational changes in the acid conformer may extend from the CH2 to the CH3 region, although the degree of the structural changes seems to be limited, considering that the Protein A binding ability is maintained by the acid conformer.
The unfavorable biological aspects, including toxicity or immunogenicity of the acid conformer in human body, are still unknown. Martsev et al. suggested that acid-treated IgG exhibited stronger binding to complement C1q than that of native IgG (16), indicating that such species could cause anaphylactic shock via immune stimulation with complement activation. Although the acid conformer is not contained in human therapeutics such as MRA (Tocilizumab) and Herceptin (Trastuzumab), <5% of the acid conformer is detected in commercially available polyclonal IgG (data not shown). Because the generation of the acid conformer is rapidly accelerated at high temperatures (Fig. 4C), the temperature reached during acid treatment should be monitored.
The purified acid conformer remained in a monomeric form at 4 °C under neutral pH in the low concentrations (<10 μg/ml) used in this study. However, when acid-treated IgG was incubated at high temperatures (25 or 37 °C) under high concentration (300 μg/ml), aggregate formations were visible (supplemental Fig. 3). This suggests that the acid conformer may be the starting material of the aggregates formed during the storage of antibody solutions. Based on these findings, the type II peptide described in this study is valuable for the detection or elimination of acid conformers, which potentially cause the aggregation of IgG. In summary, our report described a key tool to elucidate the mechanisms of antibody aggregation and dysfunction in acidic conditions.
Supplementary Material
Acknowledgments
We thank Sayaka Kakoi for experimental assistance and acknowledge Dr. Scott L. Klakamp for help with the discussion and suggestions for the SPR analysis.
This work was supported, in whole or in part, by the National Institutes of Health NCI Center for Cancer Research (Intramural Research Program). This work was also supported by funds from the Japan Science and Technology Agency.
The on-line version of this article (available at http://www.jbc.org) contains Figs. S1 and S2.
Footnotes
The abbreviations used are: BSA, bovine serum albumin; SPR, surface plasmon resonance; IL-6R, interleukin-6 receptor; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline; SA, streptavidin; RU, response unit.
References
- 1.Ey, P. L., Prowse, S. J., and Jenkin, C. R. (1978) Immunochemistry 15 429-436 [DOI] [PubMed] [Google Scholar]
- 2.Food and Drug Administration (2003) Control of Microbiological Contamination, Code of Federal Regulations 21, 211.113, U. S. Government Printing Office, Washington, D. C.
- 3.Li, R., Dowd, V., Stewart, D. J., Burton, S. J., and Lowe, C. R. (1998) Nat. Biotechnol. 16 190-195 [DOI] [PubMed] [Google Scholar]
- 4.Fassina, G., Verdoliva, A., Palombo, G., Ruvo, M., and Cassani, G. (1998) J. Mol. Recognit. 11 128-133 [DOI] [PubMed] [Google Scholar]
- 5.Ehrlich, G. K., and Bailon, P. (1998) J. Mol. Recognit. 11 121-125 [DOI] [PubMed] [Google Scholar]
- 6.Krook, M., Mosbach, K., and Ramstrom, O. (1998) J. Immunol. Methods 221 151-157 [DOI] [PubMed] [Google Scholar]
- 7.Verdoliva, A., Marasco, D., De Capua, A., Saporito, A., Bellofiore, P., Manfredi, V., Fattorusso, R., Pedone, C., and Ruvo, M. (2005) ChemBioChem. 6 1242-1253 [DOI] [PubMed] [Google Scholar]
- 8.D'Agostino, B., Bellofiore, P., De Martino, T., Punzo, C., Rivieccio, V., and Verdoliva, A. (2008) J. Immunol. Methods 333 126-138 [DOI] [PubMed] [Google Scholar]
- 9.DeLano, W. L., Ultsch, M. H., de Vos, A. M., and Wells, J. A. (2000) Science 287 1279-1283 [DOI] [PubMed] [Google Scholar]
- 10.Dias, R. L., Fasan, R., Moehle, K., Renard, A., Obrecht, D., and Robinson, J. A. (2006) J. Am. Chem. Soc. 128 2726-2732 [DOI] [PubMed] [Google Scholar]
- 11.Boonyarattanakalin, S., Martin, S. E., Sun, Q., and Peterson, B. R. (2006) J. Am. Chem. Soc. 128 11463-11470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krumpe, L. R., Atkinson, A. J., Smythers, G. W., Kandel, A., Schumacher, K. M., McMahon, J. B., Makowski, L., and Mori, T. (2006) Proteomics 6 4210-4222 [DOI] [PubMed] [Google Scholar]
- 13.Brorson, K., Krejci, S., Lee, K., Hamilton, E., Stein, K., and Xu, Y. (2003) Biotechnol. Bioeng. 82 321-329 [DOI] [PubMed] [Google Scholar]
- 14.Ejima, D., Tsumoto, K., Fukada, H., Yumioka, R., Nagase, K., Arakawa, T., and Philo, J. S. (2007) Proteins 66 954-962 [DOI] [PubMed] [Google Scholar]
- 15.Buchner, J., Renner, M., Lilie, H., Hinz, H. J., Jaenicke, R., Kiefhabel, T., and Rudolph, R. (1991) Biochemistry 30 6922-6929 [DOI] [PubMed] [Google Scholar]
- 16.Martsev, S. P., Kravchuk, Z. I., Vlasov, A. P., and Lyakhnovich, G. V. (1995) FEBS Lett. 361 173-175 [DOI] [PubMed] [Google Scholar]
- 17.Sethuraman, N., and Stadheim, T. A. (2006) Curr. Opin. Biotechnol. 17 341-346 [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi, Y., Nishimura, M., Nagano, M., Yagi, H., Sasakawa, H., Uchida, K., Shitara, K., and Kato, K. (2006) Biochim. Biophys. Acta 1760 693-700 [DOI] [PubMed] [Google Scholar]
- 19.Herman, R. E., Badders, D., Fuller, M., Makienko, E. G., Houston, M. E., Jr., Quay, S. C., and Johnson, P. H. (2007) J. Biol. Chem. 282 9813-9824 [DOI] [PubMed] [Google Scholar]
- 20.Castagnoli, L., Zucconi, A., Quondam, M., Rossi, M., Vaccaro, P., Panni, S., Paoluzi, S., Santonico, E., Dente, L., and Cesareni, G. (2001) Comb. Chem. High Throughput Screen. 4 121-133 [DOI] [PubMed] [Google Scholar]
- 21.Ghetie, V., and Ward, E. S. (2000) Annu. Rev. Immunol. 18 739-766 [DOI] [PubMed] [Google Scholar]
- 22.Newkirk, M. M. (2002) Clin. Immunol. (Orlando) 104 1-13 [DOI] [PubMed] [Google Scholar]
- 23.Thies, M. J., Kammermeier, R., Richter, K., and Buchner, J. (2001) J. Mol. Biol. 309 1077-1085 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.