Abstract
Phosphatidylinositol 4-kinases play essential roles in cell signaling and membrane trafficking. They are divided into type II and III families, which have distinct structural and enzymatic properties and are essentially unrelated in sequence. Mammalian cells express two type II isoforms, phosphatidylinositol 4-kinase IIα (PI4KIIα) and IIβ (PI4KIIβ). Nearly all of PI4KIIα, and about half of PI4KIIβ, associates integrally with membranes, requiring detergent for solubilization. This tight membrane association is because of palmitoylation of a cysteine-rich motif, CCPCC, located within the catalytic domains of both type II isoforms. Deletion of this motif from PI4KIIα converts the kinase from an integral to a tightly bound peripheral membrane protein and abrogates its catalytic activity (Barylko, B., Gerber, S. H., Binns, D. D., Grichine, N., Khvotchev, M., Sudhof, T. C., and Albanesi, J. P. (2001) J. Biol. Chem. 276 ,7705 -7708). Here we identify the first two cysteines in the CCPCC motif as the principal sites of palmitoylation under basal conditions, and we demonstrate the importance of the central proline for enzymatic activity, although not for membrane binding. We further show that palmitoylation is critical for targeting PI4KIIα to the trans-Golgi network and for enhancement of its association with low buoyant density membrane fractions, commonly termed lipid rafts. Replacement of the four cysteines in CCPCC with a hydrophobic residue, phenylalanine, substantially restores catalytic activity of PI4KIIα in vitro and in cells without restoring integral membrane binding. Although this FFPFF mutant displays a perinuclear distribution, it does not strongly co-localize with wild-type PI4KIIα and associates more weakly with lipid rafts.
Phosphatidylinositol 4-phosphate (PI4P)3 is a regulator of membrane trafficking as well as a substrate in the biosynthesis of other phosphoinositides. It is especially abundant in the Golgi apparatus where it partners with the Arf1 GTPase to recruit adaptor proteins essential in vesicular trafficking from the trans-Golgi network (TGN) (1, 2). The generation of PI4P is catalyzed by two distinct classes of phosphatidylinositol 4-kinase, type II and type III. Mammals have an α and β form of each type (3-5). PI4KIIα and PI4KIIIβ contribute primarily to Golgi-localized PI4P, and they are responsible for about 50% of the PI4P in the Golgi. The pools of Golgi PI4P produced by each enzyme are distinct but interconnected by cross-talk (6).
PI4KIIα resides primarily in the TGN and in endosomes (7-9). The critical roles of PI4KIIα in Golgi and endosome functions are evident by the dramatic effects of PI4KIIα depletion by RNA interference. These include impaired recruitment of clathrin adaptor protein AP-1 complexes and GGA to the TGN (10, 11), inhibition of constitutive secretion from the TGN (10), inhibition of phagocytosis (12), and decreased trafficking of epidermal growth factor receptors through the late endosomal pathway (13). In neuronal cells, PI4KIIα is also found on secretory granules and synaptic vesicles (14, 15), and it has been implicated in exocytosis and endocytosis of regulated secretory vesicles (15, 16).
Despite the importance of PI4KIIα, very little is known about its enzymatic regulation or the basis of its localization in cells. It behaves like an integral membrane protein, requiring detergent for solubilization, despite its lack of a bona fide transmembrane domain (14, 17). We have previously reported that this behavior is probably because of palmitoylation in a cysteine-rich motif, residues 173CCPCC177 in rat PI4KIIα, as en bloc deletion of these five residues results in loss of palmitoylation and in conversion of the kinase from an integral to a peripheral membrane protein. In addition, the deletion mutant was catalytically inactive (14). However, we did not definitively prove that palmitoylation, rather than the deletion itself, was responsible for these altered properties. The loss of catalytic activity is especially problematic because the CCPCC motif resides within the catalytic domain, in close proximity to two residues that we had shown to be essential for kinase activity (18). Moreover, the proline residue within the motif is also necessary for expression of activity (see below). Here we use multiple alternative approaches to show conclusively that palmitoylation is indeed required for critical properties of the kinase, including enzymatic activity, intracellular localization, integral membrane association, and partitioning into “rafts.” We identify two cysteine residues within the CCPCC motif that are preferentially palmitoylated under basal conditions. In addition, replacing the four cysteines with highly hydrophobic phenylalanine residues can partially restore endomembrane targeting and catalytic activity, although the phenylalanine mutant behaves as a peripheral protein.
EXPERIMENTAL PROCEDURES
Materials—l-α-Phosphatidylinositol was from Avanti Polar Lipids, Inc. (Alabaster, AL). [γ-32P]ATP and [9,10-3H]palmitic acid were from PerkinElmer Life Sciences. Dimethyl pimelimidate was from Pierce. Primers were obtained from Sigma Genosys and IDT (Coralville, IA). Cloning and mutagenesis reagents were from Stratagene (La Jolla, CA). Lipofectamine 2000 and cDNA purification kits were from Invitrogen. Nickel nitrilotriacetic acid (Ni2+-NTA)-agarose was from Qiagen (Valencia, CA). Triton X-100 and reagents for electrophoresis and immunoblotting were from Bio-Rad. Other reagents, including 2-bromopalmitate, ATP, Brij 98, buffers, and protease inhibitors, were from Sigma. Polyclonal anti-PI4KIIα antibodies were prepared as described in Ref. 18. Monoclonal anti-Myc antibody 9E10 was obtained from the National Cell Culture Center (Minneapolis, MN); recombinant protein G-Sepharose 4B conjugate was from Zymed Laboratories Inc., and sheep anti-TGN46 was from Serotec (Raleigh, NC). 125I-Labeled secondary antibodies were from Amersham Biosciences, and fluorescently conjugated secondary antibodies were from Molecular Probes (Eugene, OR). TLC plates were from Whatman.
Purification of Recombinant PI4KIIα—Recombinant PI4KIIα containing a His6 tag at its amino terminus was expressed in Sf9 cells and in Escherichia coli. The procedure for Sf9 expression is described in Ref. 18. For expression in E. coli, PI4KIIα was subcloned from the pCMV5myc1 vector into a pQE-80L vector (Qiagen) using the flanking BamHI (5′) and PstI (3′) restriction sites. Cells were resuspended in buffer A containing 1% Triton X-100, 20 mm Hepes (pH 8.0), 100 mm NaCl, 1 mm β-mercaptoethanol, a protease inhibitor mixture consisting of 10 μg/ml each of Nα-p-tosyl-l-lysine chloromethyl ester, Nα-p-tosyl-l-arginine methyl ester, Nα-p-tosyl-l-lysine chloromethyl ketone, leupeptin, pepstatin A, and 0.2 mm phenylmethylsulfonyl fluoride (PMSF). In the case of E. coli, lysozyme (0.05 mg/ml) was added to extract the kinase. Cell lysates were centrifuged at 100,000 × g for 30 min at 4 °C, and the supernatants were mixed with Ni2+-NTA resin for 1 h at 4 °C. The resin was washed with buffer A supplemented with 30 mm imidazole (pH 8.0) and 300 mm NaCl but containing 0.1% Triton X-100. The kinase was then eluted with 20 mm Hepes (pH 8.0), 150 mm imidazole (pH 8.0), 100 mm NaCl, 0.1% Triton X-100, 1 mm β-mercaptoethanol, and 0.2 mm PMSF. When prepared from E. coli, the kinase eluted from Ni2+-NTA resin was further purified on an SP-Sepharose column using a 0.05-0.3 m NaCl linear gradient without detergent. Aliquots of the protein were frozen in liquid N2 and could be stored at -70 °C for several months without significant loss of activity.
Generation of Point Mutants—Rat pCMV5-Myc-PI4KIIα cDNAs was used as a template for generation of constructs containing mutations in the 173CCPCC177 palmitoylation motif. Site-directed mutagenesis was performed using the Stratagene QuickChange site-directed mutagenesis kit according to the manufacturer's protocol.
Preparation of Cytosol and Membrane Fractions—Twenty hours after transfection, COS-7 cells expressing Myc-PI4KIIα were washed with ice-cold phosphate-buffered saline (PBS) and scraped from Petri dishes in a solution containing 0.25 m sucrose, 20 mm Tris-HCl (pH 7.5), 0.1 m NaCl, 1 mm EDTA, protease inhibitors as described above and phosphatase inhibitors (50 mm sodium fluoride, 50 mm glycerophosphate, 1 mm sodium orthovanadate, 10 μm microcystin). Cells were lysed by two freeze-thaw cycles and passage through a 27-gauge needle. The lysates were centrifuged at 1,000 × g for 5 min to remove cell debris and nuclei. The supernatant (post-nuclear supernatant) was then centrifuged at 200,000 × g for 15 min to separate cytosol from membranes. The resulting membrane pellets were homogenized in a solution containing 20 mm Tris-HCl (pH 7.5), 1 m NaCl, and 1 mm EDTA and then centrifuged at 200,000 × g for 15 min to obtain weakly binding peripheral membrane proteins in the supernatant. The resulting pellets were then homogenized in 0.1 m sodium carbonate (pH 11) and centrifuged as above to obtain tightly bound peripheral membrane proteins in the supernatant. Finally, the pellets of this centrifugation were homogenized in 1% Triton X-100 and centrifuged to obtain integral membrane proteins in the supernatant. All solutions contained the protease and phosphatase inhibitors described above.
Subcellular Fractionation of Cells—Post-nuclear supernatants of COS-7 cells prepared as described above were fractionated on a discontinuous sucrose gradient according to Waugh et al. (8). Briefly, samples were layered on a gradient consisting of successive layers of 10, 15, 20, 25, 30, and 40% sucrose and centrifuged for 20 h at 180,000 × g in an SW40 rotor. One-ml fractions were collected from the bottom of the tubes, and equal aliquots were subjected to SDS-gel electrophoresis and immunoblotting.
Preparation of Low Buoyant Density Membrane Fractions—Low buoyant density “membrane raft” fractions were prepared using Brij 98 as described by Claas et al. (19). Cells were homogenized in a solution containing 1% Brij 98, 150 mm NaCl, 5 mm MgCl2, 25 mm MES (pH 6.5), 10 mm sodium pyrophosphate, 2 mm sodium fluoride, 0.1 mm sodium orthovanadate, and protease inhibitors. The lysate was adjusted to 40% sucrose (1.5 ml of lysate mixed with 1.5 ml of 80% w/v sucrose) and overlaid with 5.5 ml of 35% sucrose and 3.5 ml of 5% sucrose in the buffer described above but without Brij 98. Following centrifugation at 210,000 × g for 16 h at 4 °C in an SW40 rotor, 1-ml fractions were collected from the bottom of the tubes, and equal aliquots were subjected to SDS-gel electrophoresis and immunoblotting.
Analysis of [3H]Palmitate Incorporation—Sixteen hours after transfection with plasmids encoding wild-type (WT) or mutant PI4KIIα, COS-7 cells were labeled with [3H]palmitate (0.3 mCi/ml) for 3 h as described by Linder et al. (20). Cells were washed with ice-cold PBS and solubilized in RIPA buffer consisting of 0.15 m NaCl, 50 mm Tris-HCl (pH 8.0), 1% Nonidet P40, 0.5% deoxycholate, 0.05% SDS, 2 mm EDTA, 0.2 mm PMSF, and the protease and phosphatase inhibitors described above. The kinases were immunoprecipitated with anti-Myc antibody and, after five washes with RIPA buffer, each immunoprecipitate was divided into 2 aliquots and electrophoresed. One gel was stained with Coomassie Blue, and [3H]palmitate incorporation was detected by autoradiography. The radioactive band (containing PI4KIIα) was cut from the gel, solubilized with 30% H2O2 for 16 h at 60 °C, and subjected to liquid scintillation counting. The other aliquot was used for immunoblotting with anti-Myc antibodies and 125I-labeled secondary antibodies to estimate relative amounts of PI4KIIα. Blots were scanned by phosphorimaging (Fujifilm BAS 1500). Levels of [3H]palmitate incorporation were normalized to the amount of kinase in each sample.
Immunoprecipitation—Total cell lysates or solubilized membranes were cleared of insoluble material by centrifugation at 15,000 × g for 15 min. Resulting supernatants were then precleared by incubation with recombinant protein G-Sepharose 4B conjugate for 30 min. Myc-tagged proteins were immunoprecipitated following 4 h of incubation with anti-Myc antibodies that were chemically cross-linked with dimethyl pimelimidate to protein G-Sepharose. Cross-linking prevents the elution from protein G-Sepharose of the IgG heavy chain, which has a similar molecular weight as PI4KIIα. The immunoprecipitates were washed extensively before analysis.
Inhibition of Palmitoylation with 2-Bromopalmitate (2-BP)—COS-7 cells expressing WT Myc-PI4KIIα were treated with 2-BP according to a procedure described by Webb et al. (21). Briefly, 12 h after transfection, COS-7 cells were incubated for an additional 12 h in DMEM containing 2.5% dialyzed fetal bovine serum (FBS), 0.25% defatted BSA, with or without 100 μm 2-BP (from a 0.1 m stock solution dissolved in ethanol). To measure [3H]palmitate incorporation into PI4KIIα,[3H]palmitate was added for the last 4 h of incubation with 2-BP or vehicle.
Lipid Kinase Assays—PI4K activity was determined by measuring phosphorylation of phosphatidylinositol (PI) using [γ-32P]ATP (10 mCi/ml) as radioactive phosphate donor. Typically, assays were performed on 1% Triton X-100 extracts of membranes from cells expressing WT or mutant Myc-PI4KIIα. These extracts were incubated with PI/Triton X-100 micelles at a 1:10 molar ratio of lipid to detergent, and assays were initiated by addition of MgCl2 and ATP. Final concentrations of PI, ATP, and MgCl2 were 1, 0.5, and 15 mm, respectively. The PI/Triton X-100 micelles were prepared by sonicating PI dissolved (after evaporating chloroform under N2) in a solution containing 50 mm Tris-HCl (pH 7.5), 6 mm Triton X-100, and 0.5 mg/ml BSA. Following incubation, lipids were extracted according to Bligh and Dyer (22) and separated by TLC in a solvent system consisting of n-propyl alcohol/H2O/NH4OH (65:20:15). Radioactive PI4P spots were detected by autoradiography, scraped, and subjected to scintillation counting to quantify radioactivity. The relative amounts of kinase in samples were estimated by quantitative immunoblotting using 125I-labeled secondary antibodies. When assaying purified kinases (as in Fig. 3B), PI4P production was measured directly by scintillation counting without resolving the lipids on TLC. In some experiments (Fig. 7, C and D), PI4K activity was measured in intact membranes in the absence of detergent, with or without PC/PI liposomes. To prepare liposomes, PC and PI (in chloroform) were mixed at the molar ratio of 9:1, dried under N2, and resuspended in 0.3 m sucrose solution to obtain final concentration of 18 and 2 mm PC and PI, respectively. After overnight incubation at 4 °C on a rotator, the suspensions were passed 20 times through a 0.1 μm filter using a Mini-Extruder (Avanti Polar Lipids). Activities of the expressed kinase were estimated by subtracting the activity of membrane extracts from mock-transfected cells. Expression levels were similar as determined by immunoblotting.
FIGURE 3.
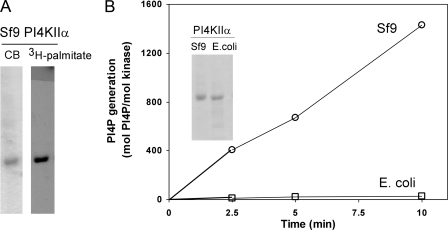
Kinase activities of rat PI4KIIα purified from Sf9 cells or E. coli. A, palmitoylation of recombinant PI4KIIα expressed in Sf9 cells. Sf9 cells overexpressing His6-PI4KIIα were labeled for 4 h with [3H]palmitate, and the kinase was isolated by affinity chromatography using Ni3+-resin. The eluted samples were subjected to electrophoresis and autoradiography. Left lane, CB, Coomassie Blue staining; right lane, autoradiogram. B, activities of PI4KIIα purified on Ni3+-resin after expression in Sf9 cells or E. coli. Assays were carried out using PI/Triton X-100 micelles as substrate. Values represent activities from one Sf9 preparations and three E. coli preparations, each assay done in triplicate. Inset shows Coomassie Blue-stained gel of typical preparations.
FIGURE 7.
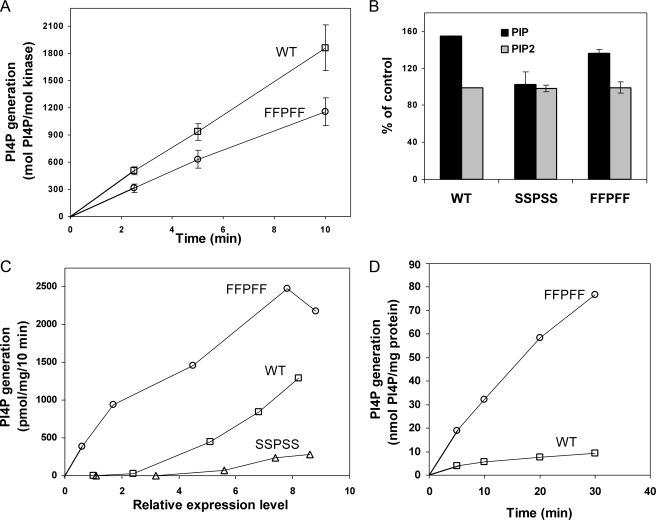
Activity of WT and FFPFF mutant of PI4KIIα. Kinase activities of WT PI4KIIα and the FFPFF mutant are shown. A, activities assayed using PI/Triton X-100 micelles as substrate. Membranes prepared from transfected COS-7 cells were extracted with Triton X-100, and the extracts were assayed as described in Fig. 2B. Data are from triplicate measurements of three experiments. B, phosphorylation of PI and PIP in COS-7 cells transfected with WT PI4KIIα or the SSPSS or FFPFF mutants. Cells were labeled with 32Pi for 4 h, and lipids were extracted and separated by TLC. Levels of radioactive PIP and PIP2 are expressed as relative to mock-transfected cells (control). Results represent the mean ± S.E. of single measurements from six experiments. Expression levels of WT and mutant kinases were nearly identical within each experiment, as determined by immunoblotting. C, phosphorylation of endogenous PI in membranes isolated from transfects of COS-7 cells. Membranes were incubated for 10 min with 0.5 mm [γ-32P] ATP, and then lipids were extracted and subjected to TLC to estimate incorporation of radioactive phosphate into PI4P. Activities of membranes from mock-transfected cells (∼2-3 pmol/mg/min) were subtracted. Relative expression levels were estimated by immunoblotting with 125I-labeled secondary antibodies. Results are the average of duplicate measurements from a representative experiment. D, activities assayed using PI/PC liposomes as substrate. Intact membranes from transfected COS-7 cells were preincubated with 0.5 mm [γ-32P] ATP for 30 min to maximally phosphorylate endogenous PI and then further incubated for the indicated time with PC/PI liposomes (18 mm PC, 2 mm PI). The basal activity in the absence of exogenous PI was subtracted. Results are the average of duplicate measurements of one experiment, which is representative of three experiments.
Quantification of Phosphoinositides in Cells—Sixteen hours after transfection with plasmids encoding WT or mutant PI4KIIα, COS-7 cells were labeled for 4 h with 25 μCi/ml [32P]orthophosphate in phosphate-free DMEM, then scraped into methanol/concentrated HCl (10:1), and transferred into tubes, and lipids were extracted with chloroform. The organic phase was collected and washed twice with an equal volume of methanol and 1 n HCl (1:1). Aliquots were counted in a scintillation counter, and equal amounts of radioactivity were spotted onto TLC plates and developed as described above.
Immunofluorescence Microscopy—COS-7 and HeLa cells grown on coverslips were transfected with WT or mutant MycPI4KIIα. Sixteen hours after transfection, cells were fixed with 3.7% formaldehyde in PBS for 10 min at room temperature, permeabilized with 0.1% Triton X-100 in PBS for 5 min on ice, blocked with 1% BSA and 3% donkey serum in PBS, and stained with anti-Myc antibody followed by rhodamine-conjugated secondary antibody. Fluorescence was visualized using a Zeiss LSM510 confocal microscope.
Other Procedures—COS-7 and HeLa cells were grown in DMEM containing 10% fetal bovine serum (FBS) and antibiotics. Cells were transiently transfected using Lipofectamine 2000 according to the manufacturer's instructions and were used 16-24 h after transfection. Protein concentrations were determined using a modified Lowry method (23, 24) with BSA as a standard. SDS-PAGE was carried out according to the method of Laemmli (25). Immunoblot analysis was carried out by the method of Towbin et al. (26) as described previously (27).
RESULTS
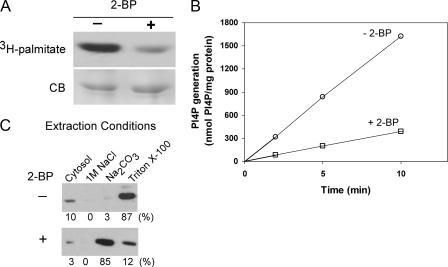
Effect of Palmitoylation on the Enzymatic Activity of PI4KIIα—This study was motivated by our earlier observation that deletion of a cysteine-rich motif, 173CCPCC177, from rat PI4KIIα abolishes both its ability to incorporate [3H]palmitate in cells and its catalytic activity in vitro (14). To ascertain that the loss of activity is because of the failure of the mutant to undergo palmitoylation, and not to the deletion itself, we inhibited palmitoylation using the nonmetabolizable palmitate analog 2-BP (21). Treatment of cells with 100 μm 2-BP for 12 h reduced [3H]palmitate incorporation into Myc-tagged PI4KIIα by 75% (Fig. 1A) and its kinase activity by 80% (Fig. 1B). Despite this reduction in palmitoylation, the kinase remained tightly associated with membranes, even in the presence of 1 m NaCl (Fig. 1C). However, unlike PI4KIIα from untreated cells, which exists almost entirely as an integral membrane protein, PI4KIIα from 2-BP-treated cells behaved predominantly as a peripheral membrane protein that was extractible by 0.1 m Na2CO3 (pH 11) in the absence of detergent.
FIGURE 1.
Effect of 2-BP on activity and membrane association of PI4KIIα. A, inhibition of PI4KIIα palmitoylation by 2-BP. COS-7 cells expressing Myc-PI4KIIα were treated with 100 μm 2-BP (or vehicle) for 12 h and then labeled (in the continued presence of 2-BP or vehicle) for 4 h with [3H]palmitate. Kinase was immunoprecipitated using anti-Myc antibodies and electrophoresed. Gels were stained with Coomassie Blue (CB) and then subjected to autoradiography to detect [3H]palmitate incorporation. B, kinase activity of Myc-PI4KIIα from untreated or 2-BP treated cells. Post-nuclear supernatants from transfected COS-7 cells were centrifuged at 200,000 × g for 15 min to separate cytosol from membranes. The membranes were then extracted using 1% Triton X-100, and the extracts were assayed in the presence of PI/Triton X-100 micelles and [γ-32P]ATP as described under “Experimental Procedures.” Data represent the averages of triplicate measurements from a single experiment. C, effect of 2-BP on the association of PI4KIIα with membranes. Membranes from transfected COS-7 cells were extracted sequentially with 1 m NaCl (to release loosely bound peripheral membrane proteins), 0.1 m Na2CO3 (pH 11) (to release tightly bound peripheral membrane proteins), and 1% Triton X-100 (to release integral membrane proteins). Equivalent amounts of each fraction were analyzed by immunoblotting with anti-Myc antibodies followed by 125I-labeled secondary antibody for quantification.
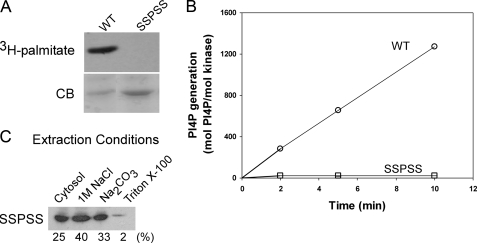
Similar results were obtained when PI4KIIα palmitoylation was prevented by mutating all four cysteines in the 173CCPCC177 motif to serines. However, in this case no palmitoylation was detectable (Fig. 2A), and the mutant expressed much lower catalytic activity (Fig. 2B). As expected, this SSPSS mutant did not bind integrally to membranes (Fig. 2C). Moreover, its association with membranes was considerably weaker than that of the wild-type kinase from 2-BP treated cells, with 25% of the mutant being cytosolic (extractible in 0.1 m NaCl) and another 40% solubilized by 1 m NaCl. This weaker membrane binding may be due to the greater intrinsic hydrophobicity of cysteine than serine (28).
FIGURE 2.
Membrane association and catalytic activity of the unpalmitoylated SSPSS mutant of PI4KIIα. COS-7 cells were transfected with cDNA encoding a Myc-tagged PI4KIIα mutant in which all four cysteines of the palmitoylation motif (CCPCC) were replaced by serines. The catalytic activities and membrane binding properties of the mutant were analyzed as described in Fig. 1. A, failure of the SSPSS mutant to incorporate [3H]palmitate. B, kinase activities of Triton X-100 extracts of membranes expressing WT or SSPSS forms of PI4KIIα, using PI/Triton X-100 micelles as substrate. Kinase levels were estimated by quantitative immunoblotting. Data are from triplicate measurements of two experiments. C, distribution of the SSPSS mutant among cytosolic, peripheral membrane, and integral membrane pools. CB, Coomassie Blue.
To further demonstrate the importance of palmitoylation for catalysis, we compared the activities of recombinant PI4KIIα purified from Sf9 cells or E. coli. The bacterially expressed kinase is unpalmitoylated, as E. coli cells do not express protein acyltransferases, whereas PI4KIIα incorporates [3H]palmitate when expressed in Sf9 cells (Fig. 3A). The Sf9-derived kinase had an activity of ∼150 min-1, whereas the activity of bacterially expressed PI4KIIα never exceeded 10 min-1 (Fig. 3B).
These results confirm that palmitoylation strongly enhances PI4KIIα activity. Moreover, because the unpalmitoylated enzymes are largely membrane-associated, albeit not integrally, the role of this modification in catalysis is not merely to tether PI4KIIα to substrate-containing membranes or micelles.
Mutational Analysis of the Palmitoylation Motif of PI4KIIα—Site-directed mutagenesis was used to identify the acylated residue(s) within the 173CCPCC177 motif. Individual and multiple cysteines in the motif were mutated to serine, a structurally similar amino acid. Individual mutations of Cys-173, Cys-174, or Cys-176 resulted in 35-54% decrease in [3H]palmitate incorporation, which corresponds closely to the reduction in integral membrane binding of the mutants (Fig. 4A). In contrast, the C177S mutant was palmitoylated normally, although it showed a 20% reduction in integral association with membranes. Perhaps this cysteine contributes to membrane binding by virtue of its intrinsic hydrophobicity (28) rather than by undergoing acylation. Kinase activity was more strongly inhibited by mutation of Cys-173 or Cys-174 than of Cys-176 or Cys-177, suggesting that palmitoylation of either of the first two residue of the CCPCC motif contributes slightly more to activity than does palmitoylation of Cys-176 or the intrinsic hydrophobicity of Cys-177.
FIGURE 4.
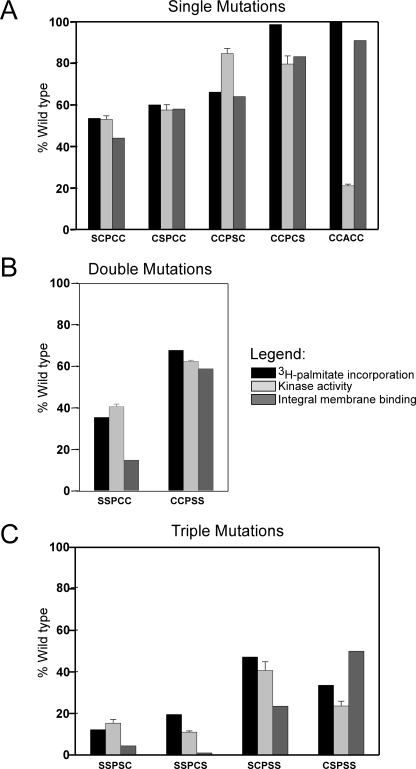
Effect of mutations in the palmitoylation motif of PI4KIIα. Single (A), double (B), and triple (C) mutations were introduced into the palmitoylated motif, 173CCPCC177, of Myc-PI4KIIα, and the proteins were expressed in COS-7 cells. Incorporation of [3H]palmitate was determined as described under “Experimental Procedures.” Values are the averages of duplicate measurements from two experiments for the SSPCC mutant, of three experiments for the CCPCS mutant, or of single experiment for all other constructs. Kinase activities were measured in detergent extracts of membranes from unlabeled cells, subtracting the background activity from untransfected cells membranes, and using PI/Triton X-100 micelles as substrate. Data represent the mean ± S.E. of triplicate kinase activity measurements from two separate preparations of each construct. Distributions of kinase among cytosolic, peripheral, and integral membrane pools were estimated as described in Figs. 1 and 2, with results representing the average of duplicate measurements from single preparations.
The greater importance of the first two cysteines was confirmed by analysis of the double and triple mutants. The SSPCC mutant showed far greater reduction in [3H]palmitate incorporation, integral membrane binding, and catalytic activity than did the CCPSS mutant (Fig. 4B). When only Cys-176 or Cys-177 was available for acylation, as in the SSPCS and SSPSC mutants, maximal levels of palmitoylation were only 20 and 12% of the wild-type, respectively (Fig. 4C). In contrast, palmitoylation was ∼50% of wild type if only Cys-174 was available (mutant SCPSS), and 35% if only Cys-173 was available (CSPSS mutant). Because the SCPSS mutant incorporates half the amount of [3H]palmitate as the wild-type kinase, despite having only one palmitoylatable residue, we conclude that the average stoichiometry of PI4KIIα palmitoylation is no greater than 2 mol/mol.
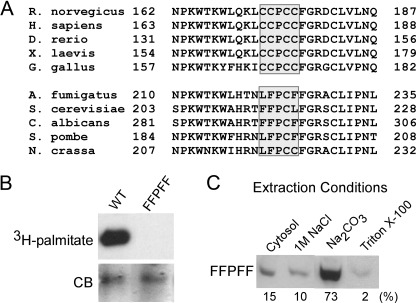
Because the central proline of the CCPCC motif is conserved among all known PI4KII orthologs from human to yeast (Fig. 6A), we mutated this residue to assess its functional significance. The P175A mutant was palmitoylated normally and associated integrally with membranes (see Fig. 6A), indicating that the proline is not required for the interaction of PI4KIIα with its acyltransferase or for membrane binding. However, this mutant expressed only ∼20% of the activity of the wild-type kinase. Perhaps the central proline helps to maintain the correct orientation of the flanking cysteines with respect to the bilayer, thereby allowing a more productive interaction between the kinase active site and its lipid substrate.
FIGURE 6.
Effect of substituting the four palmitoylated cysteines in PI4KIIα with phenylalanines. A, sequence alignment of the putative membrane anchoring loops of selected type II PI 4-kinases from vertebrates (top) and fungi (bottom). B and C, COS-7 cells were transfected with a Myc-PI4KIIα mutant (FFPFF) in which the four cysteines of palmitoylation motif were converted to phenylalanines. Palmitate incorporation (B) and distribution among cytosolic, peripheral membrane, and integral membrane fractions (C) were measured as described in Fig. 1. CB, Coomassie Blue.
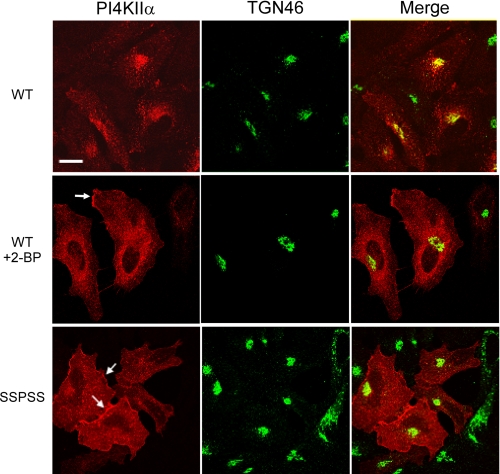
Role of Palmitoylation in the Subcellular Localization of PI4KIIα—We previously reported that endogenous and recombinant PI4KIIα are concentrated in the perinuclear region of cells, where it co-localizes with the TGN marker, TGN46 (10). It is also present in endosomal structures (7, 13, 29-31) but is nearly undetectable at the plasma membrane. Because palmitoylation controls the subcellular distribution of several proteins (32-34), we determined the effect of reducing or eliminating palmitoylation on PI4KIIα localization. Treatment of cells with 2-BP induced the dispersal of Myc-tagged PI4KIIα from the perinuclear region to punctate structures throughout the cytoplasm (Fig. 5, middle panels). This mislocalization of the kinase was not because of disruption of the TGN per se, as TGN46 staining remained perinuclear, albeit slightly expanded compared with control cells. Strikingly, the presence of PI4KIIα on the plasma membrane became evident in 2-BP-treated cells (Fig. 5, middle panel, arrow), in contrast to its almost complete absence from the plasma membrane in untreated cells.
FIGURE 5.
Role of palmitoylation in the subcellular targeting of PI4KIIα. HeLa cells were transfected with either Myc-WT or Myc-SSPSS mutant PI4KIIα. Sixteen hours after transfection, cells were fixed, permeabilized, and stained with anti-Myc antibodies followed by rhodamine-labeled secondary antibodies to detect kinases, and with anti-TGN46 followed by fluorescein-labeled secondary antibodies to identify the trans-Golgi network. Cells transfected with WT PI4KIIα were treated with vehicle (top panels) or 100 μm 2-BP (middle panels) for 12 h prior to fixation and staining. Scale bar, 20 μm.
The importance of palmitoylation for PI4KIIα targeting was further demonstrated by examination of the SSPSS mutant. Like the wild-type kinase in 2-BP-treated cells, this mutant was dispersed throughout the cytoplasm, and it showed more pronounced localization at the plasma membrane (Fig. 5, bottom panel, arrows). These results demonstrate that unpalmitoylated PI4KIIα has access to a much larger variety of membranes than does the palmitoylated kinase, although it is unable to phosphorylate PI within those membranes.
Retention of Activity by Replacement of Cysteines in the CCPCC Motif with Phenylalanines—The results presented thus far demonstrate the importance of acylated cysteines for integral membrane association, intracellular targeting, and catalytic activity of PI4KIIα. Therefore, it is interesting that in some lower eukaryotes two or even three of the cysteines in the corresponding position are replaced by hydrophobic residues. For example, the type II kinase from Saccharomyces cerevisiae, Lsb6p, contains the sequence FFPCL (Fig. 6A). Moreover, the single cysteine in that motif is not palmitoylated, but instead Lsb6p is palmitoylated in a carboxyl-terminal cysteine (35).4
To determine whether hydrophobic residues can mimic the anchoring function of cysteine acylation, we generated a PI4KIIα FFPFF mutant. As expected, this mutant neither incorporated [3H]palmitate nor associated integrally with membranes (Fig. 6, B and C). However, it bound much more strongly to membranes than the SSPSS mutant (compare Fig. 6C with Fig. 2C).
The most striking difference between the FFPFF and SSPSS mutants was the retention of near wild-type catalytic activity of the FFPFF mutant. When assayed using PI/Triton X-100 micelles as substrate, the FFPFF mutant consistently expressed at least 50% of the kinase activity of WT PI4KIIα (Fig. 7A). The FFPFF mutant was also able to increase 32P incorporation into PI4P in cells (Fig. 7B). Overexpression of the WT kinase nearly doubled the incorporation of radioactive phosphate into PIP, with almost no effect on incorporation into PIP2. Overexpression of FFPFF to similar levels increased radioactive PIP by 1.5-fold, with again no change in incorporation into PIP2. Overexpression of the SSPSS mutant failed to increase incorporation of phosphate into either lipid. Interestingly, the increase in incorporation of 32P into cellular PIP (1.5-2.0-fold) was not commensurate with the extent of overexpression of PI4KIIαWT or FFPFF (greater than 10-fold over endogenous PI4KIIα levels), presumably reflecting homeostatic processes that prevent unphysiologically high phosphoinositide levels.
Unexpectedly, the FFPFF mutant was even more active than WT PI4KIIα in phosphorylating endogenous PI in membrane fragments (Fig. 7C). WT PI4KIIα and the SSPSS and FFPFF mutants were expressed to various levels in COS-7 cells, and membranes were then prepared from these cells. At relatively low expression levels, the FFPFF mutant was able to phosphorylate more than 20 times the amount of endogenous PI phosphorylatable in these membranes by the WT kinase. At higher levels of expression, there is still an increase in activity, although the extent of increase is decreased, probably because of substrate depletion. Unlike the FFPFF mutant, the SSPSS mutant showed little activity even at high levels of expression.
We propose that the FFPFF mutant can phosphorylate a larger pool of endogenous PI because, unlike WT PI4KIIα, it may shuttle from one membrane fragment to another. To test this possibility we compared the effect of adding exogenous PI liposomes in trans to membranes containing overexpressed WT or FFPFF PI4KIIα. Initially, membranes containing comparable amounts of overexpressed WT and mutant PI4KIIα were incubated with ATP to allow maximal phosphorylation of endogenous PI. We found that after a 30-min incubation, the extent of PI4P generation reached a plateau, suggesting that the endogenous PI was maximally converted to PI4P (data not shown). Following this 30-min incubation, PI/PC liposomes (consisting of 2 mm PI and 18 mm PC) were introduced, and phosphorylation of PI in liposomes was measured over a 30-min time course. Under this condition, the rate of PI4P generation was 2.5-fold higher for the FFPFF mutant compared with WT. Therefore, these results support the hypothesis that the FFPFF mutant is able to access the liposomal PI far more efficiently than WT PI4KIIα.
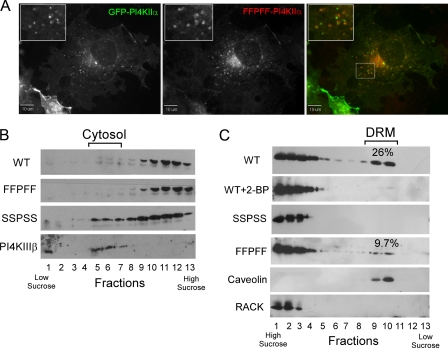
Subcellular Distribution of the FFPFF Mutant—The above results suggested that the FFPFF mutant can move more freely among membranes than the WT kinase, which is anchored by its covalently attached palmitoyl chains. However, immunofluorescence microscopy (Fig. 8A) and subcellular fractionation (Fig. 8B) indicate that the FFPFF mutant has a more restricted distribution than the SSPSS mutant. Like WT PI4KIIα, the FFPFF mutant is enriched in the perinuclear region but is nearly absent from the plasma membrane. Both proteins were also found on cytoplasmic punctae, consistent with the reported distribution of PI4KIIα to organelles in the endosomal pathway (7, 13, 29). Interestingly, many of the punctae contained either the FFPFF mutant or WT PI4KIIα (expressed as an enhanced green fluorescent fusion protein) but not both.
FIGURE 8.
Comparison of the subcellular distribution of WT and FFPFF forms of PI4KIIα. A, COS-7 cells were co-transfected with GFP-PI4KIIα and the Myc-tagged FFPFF mutant. Left panel shows localization of the WT kinase; middle panel shows localization of the mutant detected by anti-Myc antibodies and rhodamine-labeled secondary antibodies; and the right panel shows the merged images. B, fractionation of post-nuclear supernatants of COS-7 cells expressing Myc-tagged WT and mutant PI4KIIαs. Post-nuclear supernatants were subjected to centrifugation on discontinuous sucrose gradients (10-40%) as described under “Experimental Procedures.” Proteins were detected by immunoblotting with anti-Myc antibodies. PI4KIIIβ distribution is shown as a cytosolic marker. C, association with low buoyant density DRM. COS-7 cells were homogenized in 1% Brij 98 and subjected to centrifugation in a discontinuous sucrose step gradient as described under “Experimental Procedures.” Protein distribution in the collected fractions was monitored by immunoblotting with anti-Myc (PI4KIIα), anti-caveolin to identify DRMs, and anti-RACK (Receptor for Activated C Kinase) to identify non-DRMs. The numbers show the amount of kinase in DRM as a percentage of total from three experiments.
WT PI4KIIα and the FFPFF mutant distributed similarly, in high density fractions, upon sucrose gradient centrifugation of post-nuclear supernatants (Fig. 8B). In contrast, the SSPSS mutant distributed among all fractions, including those containing the cytosol as determined by the presence of PI4KIIIβ, which is predominantly cytosolic.
A prominent difference between FFPFF and WT PI4KIIα is the greater enrichment of the palmitoylated WT kinase in detergent-resistant membrane (DRM) fractions, commonly termed lipid rafts. We and others had previously shown that type II PI 4-kinases are enriched in these low buoyant density fractions (18, 36), and the implication of this localization for phosphoinositide signaling was discussed (37). Here we found that ∼25% of WT PI4KIIα migrates at low buoyant densities, apparently because of resistance to complete solubilization by the detergent Brij 98 (Fig. 8C). Treatment of cells with 2-BP almost eliminated this low buoyant density pool of the WT kinase, as did mutation of the palmitoylated cysteines to serines. Interestingly, mutation of those cysteines to phenylalanines greatly reduced but did not eliminate raft distribution, as ∼10% of FFPFF was found in low buoyant density fractions in three separate experiments. Thus, palmitoylation of PI4KIIα greatly enhances its association with lipid rafts, but it is not an absolute requirement.
DISCUSSION
Palmitoylation is a potentially reversible post-translational modification that can regulate the membrane association, subcellular targeting, and enzymatic activity of proteins (recently reviewed in Refs. 33, 34, 38). The results presented here demonstrate that in the case of PI4KIIα, all three properties are controlled by palmitoylation of multiple cysteines within the catalytic core of the kinase. These cysteines are clustered in a motif, CCPCC, that is conserved among all vertebrate forms of PI4KII. The CCPCC motif resides within a 26-amino acid segment that is situated between conserved kinase sequence motifs 2 and 3 as defined by Hanks and Hunter (39). This segment, including residues 162-187 of rat PI4KIIα, is rich in hydrophobic and positively charged residues, and the portion amino-terminal to the CCPCC motif is predicted to fold into an amphipathic helix. It is flanked by two residues conserved among nearly all kinases as follows: a lysine (Lys-151 in rat PI4KIIα) that binds the α and β phosphate of ATP, and an aspartic acid (Glu-192 in rat PI4KIIIα) that forms a salt bridge with that lysine (40). We previously reported that mutation of either Lys-151 or Glu-192 strongly reduces kinase activity (18). Our mutational analysis indicates that all four cysteine residues in the CCPCC motif are capable of being acylated in cells, but that the first two are the preferred sites of modification under the conditions used here. We did not attempt to estimate the average stoichiometry of palmitoylation. However, because the SCPSS mutant incorporates half the amount of [3H]palmitate as wild-type PI4KIIα, but contains only one palmitoylatable residue, the stoichiometry of palmitoylation on average apparently does not exceed 2 mol/mol.
Although palmitoylation is required for integral association of PI4KIIα with membranes, the unpalmitoylated kinase from 2-BP-treated cells remains tightly bound to membranes, albeit peripherally. A substantial portion of the SSPSS mutant is also membrane-bound. However, neither unpalmitoylated wild-type PI4KIIα nor SSPSS mutant expresses more than 10% of the catalytic activity of palmitoylated PI4KIIα. In contrast, replacement of the palmitoylated cysteines with phenylalanines allows retention of a substantial portion of wild-type activity. Therefore, we suggest that partial penetration of the hydrophobic core of the bilayer is necessary to properly position the PI4KIIα active site with respect to its substrate, PI, at the membrane interface. Interestingly, the FFPFF mutant is better able than WT PI4KIIα to phosphorylate PI in isolated cell membranes than the wild-type PI4KIIα, perhaps because, unlike the palmitoylated kinase, it can shuttle from one membrane fragment to another and thereby access a larger substrate pool.
Reduction of PI4KIIα palmitoylation, either by treatment of cells with 2-BP or by mutation of the palmitoylated cysteines to serines, causes the kinase to disperse from the perinuclear TGN and endosomes. The mechanism whereby palmitoylation contributes to the proper targeting of the kinase to endomembranes is unknown at this time, but we propose that at least the following three steps are involved. 1) After synthesis on cytoplasmic ribosomes, unpalmitoylated PI4KIIα is rapidly recruited to the Golgi by an as yet unidentified endomembrane targeting motif in the carboxyl-terminal segment (41), which binds a putative Golgi docking protein. 2) The kinase penetrates the Golgi membrane bilayer by virtue of its amphipathic membrane anchoring loop. 3) This positions the CCPCC motif against the membrane and brings it within reach of a Golgilocalized protein acyltransferase. One (or more) cysteine in the motif is palmitoylated, and the covalently attached palmitate group provides a strong secondary anchor to kinetically trap PI4KIIα in the Golgi and in Golgi-derived membranes. Subcellular fractionation and immunofluorescence localization studies showing that PI4KIIα is also found on secretory vesicles and endosomes (13) support this view. Its absence from the plasma membrane has not been explained, but may indicate the operation of a rapid internalization process.
In summary, our results demonstrate the importance of palmitoylation in regulating the cellular functions of PI4KIIα and reveal its multifaceted effects. They are consistent with the growing evidence that palmitoylation is an important post-translational modification that regulates protein targeting to membrane subdomains, membrane trafficking, and enzyme activities (reviewed in Refs. 32, 33, 42, 43). At this time, palmitoylation is the only known mechanism of regulation of PI4IIα activity. Therefore, future studies are directed at identifying the specific acyltransferase(s) responsible for PI4KIIα palmitoylation and determining if its acylation or deacylation is subject to regulation.
Acknowledgments
We thank Dr. Joel Goodman for very valuable comments.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 GM75401 (to J. P. A.) and R01 GM66110 (to H. L. Y.). This work was also supported by American Heart Association Texas Affiliate Grant 0455005Y (to J. P. A. in recognition of HCA Hospitals, North Richland Hills, TX), National Science Foundation Grant 9982061 (to B. B.), and the Welch Foundation Grant I-1200 (to H. L. Y.).
Footnotes
The abbreviations used are: PI4P, phosphatidylinositol 4-phosphate; 2-BP, 2-bromopalmitate; BSA, bovine serum albumin; DMEM, Dulbecco's modified Eagle's medium; PI, phosphatidylinositol; PIP, phosphatidylinositol phosphate; PIP2, phosphatidylinositol bisphosphate; PI4K, phosphatidylinositol 4-kinase; PMSF, phenylmethylsulfonyl fluoride; TGN, trans-Golgi network; MES, 4-morpholineethanesulfonic acid; PBS, phosphate-buffered saline; PC, phosphatidylcholine; Ni2+-NTA, nickel nitrilotriacetic acid; PI4KII, phosphatidylinositol 4-kinase II; WT, wild type; DRM, detergent-resistant membranes.
B. Barylko, C. Byers, and J. P. Albanesi, unpublished results.
References
- 1.Di Paolo, G., and De Camilli, P. (2006) Nature 443651 -657 [DOI] [PubMed] [Google Scholar]
- 2.De Matteis, M. A., and D'Angelo, G. (2007) Biochem. Soc. Symp. 74107 -116 [DOI] [PubMed] [Google Scholar]
- 3.Fruman, D. A., Meyers, R. E., and Cantley, L. C. (1998) Annu. Rev. Biochem. 67 481-507 [DOI] [PubMed] [Google Scholar]
- 4.Heilmeyer, L. M., Jr., Vereb, G., Jr., Vereb, G., Kakuk, A., and Szivak, I. (2003) IUBMB Life 55 59-65 [DOI] [PubMed] [Google Scholar]
- 5.Balla, A., and Balla, T. (2006) Trends Cell Biol. 16351 -361 [DOI] [PubMed] [Google Scholar]
- 6.Weixel, K. M., Blumental-Perry, A., Watkins, S. C., Aridor, M., and Weisz, O. A. (2005) J. Biol. Chem. 28010501 -10508 [DOI] [PubMed] [Google Scholar]
- 7.Balla, A., Tuymetova, G., Barshishat, M., Geiszt, M., and Balla, T. (2002) J. Biol. Chem. 27720041 -20050 [DOI] [PubMed] [Google Scholar]
- 8.Waugh, M. G., Minogue, S., Anderson, J. S., Balinger, A., Blumenkrantz, D., Calnan, D. P., Cramer, R., and Hsuan, J. J. (2003) Biochem. J. 373 57-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, F. S., Han, G. S., Carman, G. M., and Blumer, K. J. (2005) J. Cell Biol. 171133 -142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, Y. J., Wang, J., Sun, H. Q., Martinez, M., Sun, Y. X., Macia, E., Kirchhausen, T., Albanesi, J. P., Roth, M. G., and Yin, H. L. (2003) Cell 114299 -310 [DOI] [PubMed] [Google Scholar]
- 11.Wang, J., Sun, H. Q., Macia, E., Kirchhausen, T., Watson, H., Bonifacino, J. S., and Yin, H. L. (2007) Mol. Biol. Cell 182646 -2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pizarro-Cerda, J., Payrastre, B., Wang, Y. J., Veiga, E., Yin, H. L., and Cossart, P. (2007) Cell. Microbiol. 92381 -2390 [DOI] [PubMed] [Google Scholar]
- 13.Minogue, S., Waugh, M. G., De Matteis, M. A., Stephens, D. J., Berditchevski, F., and Hsuan, J. J. (2006) J. Cell Sci. 119571 -581 [DOI] [PubMed] [Google Scholar]
- 14.Barylko, B., Gerber, S. H., Binns, D. D., Grichine, N., Khvotchev, M., Sudhof, T. C., and Albanesi, J. P. (2001) J. Biol. Chem. 2767705 -7708 [DOI] [PubMed] [Google Scholar]
- 15.Salazar, G., Craige, B., Wainer, B. H., Guo, J., De Camilli, P., and Faundez, V. (2005) Mol. Biol. Cell 83692 -3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wenk, M. R., and De Camilli, P. (2004) Proc. Natl. Acad. Sci. U. S. A. 1018262 -8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minogue, S., Anderson, J. S., Waugh, M. G., dos Santos, M., Corless, S., Cramer, R., and Hsuan, J. J. (2001) J. Biol. Chem. 27616635 -16640 [DOI] [PubMed] [Google Scholar]
- 18.Barylko, B., Wlodarski, P., Binns, D. D., Gerber, S. H., Earnest, S., Sudhof, T. C., Grichine, N., and Albanesi, J. P. (2002) J. Biol. Chem. 27744366 -44375 [DOI] [PubMed] [Google Scholar]
- 19.Claas, S., Stipp, C. S., and Hemler, M. E. (2001) J. Biol. Chem. 2767974 -7984 [DOI] [PubMed] [Google Scholar]
- 20.Linder, M. E., Kleuss, C., and Mumby, S. M. (1995) Methods Enzymol. 250314 -330 [DOI] [PubMed] [Google Scholar]
- 21.Webb, Y., Hermida-Matsumoto, L., and Resh, M. D. (2000) J. Biol. Chem. 275261 -270 [DOI] [PubMed] [Google Scholar]
- 22.Bligh, E. G., and Dyer, W. J. (1959) Can. J. Med. Sci. 37911 -917 [DOI] [PubMed] [Google Scholar]
- 23.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193265 -275 [PubMed] [Google Scholar]
- 24.Peterson, G. L. (1979) Anal. Biochem. 100201 -220 [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. (1970) Nature 227680 -685 [DOI] [PubMed] [Google Scholar]
- 26.Towbin, H., Staehelin, T., and Gordon, J. (1979) Proc. Natl. Acad. Sci. U. S. A. 764350 -4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner, M. C., Barylko, B., and Albanesi, J. P. (1992) J. Cell Biol. 119163 -170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagano, N., Ota, M., and Nishikawa, K. (1999) FEBS Lett. 458163 -170 [DOI] [PubMed] [Google Scholar]
- 29.Craige, B., Salazar, G., and Faundez, V. (2008) Mol. Biol. Cell 191415 -1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salazar, G., Craige, B., Styers, M. L., Newell-Litwa, K. A., Doucette, M. M., Wainer, B. H., Falcon-Perez, J. M., Dell'Angelica, E. C., Peden, A. A., Werner, E., and Faundez, V. (2006) Mol. Biol. Cell 174014 -4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salazar, G., Zlatic, S., Craige, B., Peden, A. A., Pohl, J., and Faundez, V. (2009) J. Biol. Chem. 2841790 -1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Resh, M. D. (2006) Sci. STKE 2006, RE14. [DOI] [PubMed]
- 33.Linder, M. E., and Deschenes, R. J. (2007) Nat. Rev. Mol. Cell Biol. 8 74-84 [DOI] [PubMed] [Google Scholar]
- 34.Greaves, J., and Chamberlain, L. H. (2007) J. Cell Biol. 176249 -254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth, A. F., Wan, J., Bailey, A. O., Sun, B., Kuchar, J. A., Green, W. N., Phinney, B. S., Yates, J. R., III, and Davis, N. G. (2006) Cell 1251003 -1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waugh, M. G., Lawson, D., and Hsuan, J. J. (1999) Biochem. J. 337591 -597 [PMC free article] [PubMed] [Google Scholar]
- 37.Waugh, M. G., Minogue, S., Anderson, J. S., dos Santos, M., and Hsuan, J. J. (2001) Biochem. Soc. Trans. 29 509-511 [DOI] [PubMed] [Google Scholar]
- 38.Nadolski, M. J., and Linder, M. E. (2007) FEBS J. 2745202 -5210 [DOI] [PubMed] [Google Scholar]
- 39.Hanks, S. K., and Hunter, T. (1995) FASEB J. 9576 -596 [PubMed] [Google Scholar]
- 40.Taylor, S. S., Knighton, D. R., Zheng, J., Sowadski, J. M., Gibbs, C. S., and Zoller, M. J. (1993) Trends Biochem. Sci. 1884 -89 [DOI] [PubMed] [Google Scholar]
- 41.Jung, G., Wang, J., Wlodarski, P., Barylko, B., Binns, D. D., Shu, H., Yin, H. L., and Albanesi, J. P. (2008) Biochem. J. 409501 -509 [DOI] [PubMed] [Google Scholar]
- 42.Greaves, J., Prescott, G. R., Gorleku, O. A., and Chamberlain, L. H. (2008) Mol. Membr. Biol. 26 67-79 [DOI] [PubMed] [Google Scholar]
- 43.Bijlmakers, M. J. (2009) Mol. Membr. Biol. 2693 -103 [DOI] [PubMed] [Google Scholar]