Abstract
Patients treated with anticancer chemotherapy exhibit variation, both in terms of tumor response and the incidence and severity of adverse effects. The etiology of this variation is multifactorial with genetic factors likely contributing to a significant extent. Pharmacogenetic and genomic studies can be used to identify the genetic variants that contribute to interindividual variation in susceptibility to chemotherapy-induced cytotoxicity. This review will describe candidate and whole-genome approaches, describe the advantages and disadvantages of each, and illustrate how they can be used to obtain clinically relevant information. Specific emphasis is given to recent advances emerging from the International HapMap Project and to the development of genetic signatures, as opposed to expression signatures, to explain drug sensitivity and resistance.
Keywords: chemotherapy, cytotoxicity, lymphoblastoid cell lines, pharmacogenomics, whole-genome
The goal of pharmacogenetic and genomic research is to individualize therapy in an effort to maximize efficacy and minimize toxicity for each patient. These goals are extremely relevant to the field of oncology, where most drugs have a very narrow therapeutic index. In addition, patients treated with the same agent display wide variation in both response and in the incidence and severity of side effects. Response to therapy is likely to be partly attributed to genetic factors [1] and identifying the genetic variants that contribute to susceptibility to the cytotoxic effects of chemotherapy will provide physicians a means by which to tailor therapy and decrease the chance of adverse events. The genetic heterogeneity of tumors, even among tumors of the same histological subtype, is another factor that contributes to variability in response to therapy. Identifying the genetic features specific to a particular tumor will also contribute to the ability of physicians to tailor therapy to each individual patient. By choosing drugs most likely to be effective in the context of the genetic aberrations within a specific tumor, the chances for maximizing efficacy will increase. Somatic mutations within the tumor and germline polymorphisms may contribute to tumor response, while germline polymorphisms contribute to host toxicity. Although the process of identifying these genetic variants is still evolving, there are a number of different approaches that can be employed.
The traditional approach of pharmacogenomic studies has been the candidate gene approach, in which a gene or pathway is identified as potentially important based on literature evidence and then subjected to further study. However, the sequencing of the human genome and the genetic resource provided by the International HapMap Project [2] have allowed researchers to greatly expand the focus of pharmacogenomic studies and perform genome-wide studies. Although the genome-wide approach provides an enormous amount of information, along with that information comes false positive findings as a result of multiple testing. A major advantage of the genome-wide approach is that it opens up the possibility of identifying previously unknown genetic variants that contribute to chemotherapy-induced cytotoxicity.
This review will present a brief summary of candidate and whole-genome approaches and will provide examples from the literature to illustrate both the utility and the challenges that accompany each approach. The focus will be on the use of these applications for the identification of genetic variants that contribute to chemotherapy-induced cytotoxicity, with the ultimate goal of individualizing chemotherapy for each patient.
Candidate gene approach
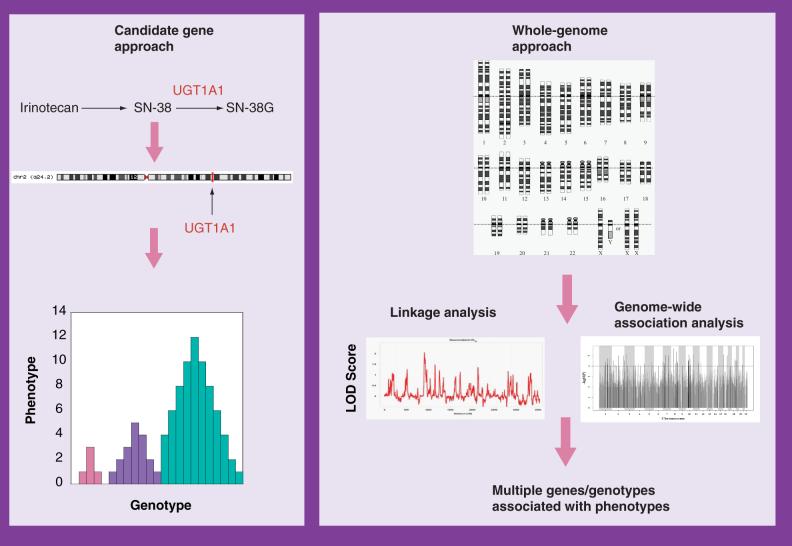
Candidate gene approaches focus on one or a small number of genes known to be important in the pharmacokinetics or pharmacodynamics of a particular drug (Figure 1). This approach has yielded important and clinically relevant information within the field of oncology, mainly with the study of genes involved in drug metabolism, transport and DNA repair (Table 1) [3,4].
Figure 1. An overview of the candidate gene and whole-genome approaches.
For the candidate gene approach, the pharmacokinetic and pharmacodynamic pathways of the drug are used to identify a gene (or a small group of genes) that is potentially important. The single gene is then studied in order to determine if variants in the gene contribute to variation in susceptibility to the cytotoxic effects of the drug. By contrast, with the whole-genome approach, the entire genome is studied, either by association analyses or linkage analyses or both, in order to identify candidate genes.
Table 1.
Examples of pharmacogenomic candidate genes and the frequency of variants of these genes within Caucasian and African-American populations.
| Gene | Variant | Frequency Caucasian (%) | Frequency African-American (%) | Ref. |
|---|---|---|---|---|
| ABCB1 (MDR1) | MDR1*2 | 27 | 6.5 | [51] |
| ABCG2 | ABCG2 -421C>A | 12 | 1-5 | [51] |
| DPD | IVS14+1G>A | 0.91 | [52] | |
| ERCC1 | ERCC1-496C>T | 58 | 0 | [51] |
| ERCC2 | ERCC2 -2251A>C | 44 | 16 | [51] |
| GSTP1 | GSTP1 -105 | 33 | 42 | [53] |
| MTHFR | MTHFR -677C>T | 36 | 13 | [54] |
| TPMT | TPMT*2 | 0.17-0.5 | 0.4 | [55] |
| TPMT*3A | 3.2-5.7 | 0.8 | ||
| TPMT*3C | 0.17-4.8 | 2.4 | ||
| TS | TSER*2 | 40 | 46 | [56,57] |
| UGT1A1 | UGT1A1*28 | 45 | 39 | [58] |
| XRCC1 | XRCC1-1301 | 35 | 36 | [51] |
Perhaps the best known example of successful application of the candidate gene approach is thiopurine methyltransferase (TPMT). TPMT catalyzes the S-methylation of 6-mercaptopurine (6-MP) and other thiopurines to relatively inactive metabolites, and thereby decreases the formation of the active thioguanine nucleotides [5]. TPMT is inherited as an autosomal codominant genetic trait in which a number of allelic variants with decreased enzymatic activity have been identified [6]. Patients who are heterozygous or homozygous for a variant allele have decreased methylation activity, increased formation of thioguanine nucleotides and an increased risk of toxicity when exposed to thiopurines [7]. For example, patients with acute lymphoblastic leukemia who were treated with 6-MP and who were homozygous or heterozygous for a variant allele exhibited increased toxicity and an increased need for 6-MP dose reductions compared with those who were homozygous wild-type [8]. By genotyping the three variant alleles that account for more than 90% of cases with low or intermediate TPMT enzyme activity [5] prior to therapy, most patients who are at greatest risk for severe thiopurine-induced cytotoxicity can be identified and thus the chemotherapeutic regimen can be tailored. TPMT genotyping to identify patients at risk for toxicity, albeit important to patient care, is performed at a limited number of academic centers.
Another example of successful application of the candidate gene approach involves the study of UDP-glucuronosyltransferase 1A1 (UGT1A1) polymorphisms. UGT1A1 is responsible for the glucuronidation of the active metabolite of irinotecan, SN-38, to an inactive metabolite [9]. UGT1A1 exhibits variation in expression related to the number of (TA) repeats in the promoter region of the gene. The substitution of six repeats (TA6) by additional repeats (TA7) is associated with a decrease in UGT1A1 expression and consequently decreased glucuronidation of its targets [10]. A prospective study of adult cancer patients who received irinotecan mono-therapy demonstrated that patients who carried two (TA7) alleles had a 50% incidence of grade 4 neutropenia, while those who were heterozygous for (TA7) or carried no (TA7) alleles had a 12.5 and 0% incidence, respectively. This study clearly demonstrated that patients who are homozygous for (TA7) have an increased risk of severe neutropenia [11].
TPMT and UGT1A1 are two well-studied examples in which the candidate gene approach has successfully provided clinically relevant information. Based on the strong pharmacogenetic evidence, the FDA has included genotyping information in the drug labels of both 6-MP and irinotecan [12,13]. By performing simple genotyping tests, oncologists are better informed as to whether the patient is at increased risk for developing chemotherapy-associated toxicities, and can therefore tailor the dose to the individual patient in order to avoid undue adverse effects. However, there are a multitude of drugs for which the candidate gene approach has not been equally successful. For example, cytarabine arabinoside (ara-C) is an antimetabolite chemotherapeutic agent used for the treatment of hematologic malignancies. There are a number of genes within the pharmacokinetic pathway of ara-C that have been studied with the goal of identifying genetic variants that contribute to the cytotoxic effect of the drug. These include deoxycytidine kinase, cytidine deaminase, 5′-nucleotidases and the human equilibrative nucleotide transporters [14,15]. The expression of the 5′-nucleotidase cN-II was measured in leukemic blasts from 96 patients with acute myeloid leukemia and correlated with clinical outcome [16]. High levels of cN-II mRNA significantly correlated with both worse disease-free and overall survival, as well as with ara-C IC50. There was not, however, a correlation between cN-II mRNA levels and response to induction chemotherapy. In another study by the same group, cN-II mRNA was measured in 77 patients and in this study there was a correlation between cN-II mRNA and response to ara-C, with levels being significantly lower in responders compared with nonresponders [17]. It has been postulated that cN-II may be a marker of disease aggressiveness rather than a specific predictor of ara-C effectiveness [18]. Another group measured the levels of mRNA of a number of genes related to ara-C metabolism in myeloid blasts from patients with leukemia [19]. In this study only the hENT1 expression correlated with in vitro sensitivity to ara-C. While the studies are beginning to point to some potential pharmacogenomic markers of sensitivity to ara-C, they have not, as yet, yielded conclusive findings that have translated to the patient and allowed for the a priori identification of patients at increased or decreased risk of ara-C-induced cytotoxicity.
Chemotherapy-induced cytotoxicity is most likely a multigenic trait. Therefore, the candidate gene approach that focuses on a single gene may not provide the most optimal results for the majority of chemotherapeutic agents. The approach is most successful if the gene involves a key step in the metabolic pathway of the drug, has a variant allele that significantly alters the metabolism of the drug, and is present in a significant percentage of the population. Since this is not the case for most chemotherapeutic agents, a broader approach is needed.
Whole-genome approach
In contrast to the candidate gene approach, the entire genome is taken into consideration without bias towards a particular gene or pathway in the whole-genome approach (Figure 1). Advances in genomics technology including the manufacturing of SNP chip and gene-expression platforms, and the development of software to analyze large data sets generated by such platforms have provided pharmacogenomic researchers with new tools in the quest to find important genetic or expression variation to explain pharmacologic variation. These tools have mainly been applied to finding genetic variants associated with disease, or expression changes associated with disease progression and response to therapeutics [20-23]. However, these tools can also be applied to pharmacologic questions [24-26], such as sensitivity to chemotherapy. While there are numerous studies identifying gene expression-based signatures associated with tumor sensitivity to drugs, only recently has there been attention directed at identifying genetic variation associated with sensitivity to drugs.
The International HapMap Project as a tool for pharmacogenomic research
An important resource for understanding genetic variation is the International HapMap Project. This work was initiated in 2002 in an effort to characterize common variations in DNA sequence among four different populations and to construct haplotype maps [2]. Cell lines derived from individuals from four different populations were extensively genotyped [27]:
90 Utah residents with ancestry from northern and western Europe (CEU)
90 Yoruba in Ibadan, Nigeria (YRI)
45 Japanese in Tokyo, Japan (JPT)
45 Han Chinese in Beijing, China (CHB)
Phase I of the project included genotypes of approximately 1.1 million SNPs in each of the four populations. In 2005, Phase II was completed, providing the scientific community with genotypic information on more than 6 million SNPs for each cell line. Phase III will include cell lines derived from additional populations, including African-American and Mexican-American. This genotype data represents an incredibly useful resource for pharmacogenomic studies. By using cell lines that are part of the HapMap Project as the source for in vitro experiments, researchers have genotype data readily available which provides a more extensive interrogation of the genome than is currently possible with any other resource. In addition, because four different populations were evaluated, the effect of genetic differences among these populations on cellular phenotypes can be explored.
As a result of the ability to easily obtain extensive genotype data in the public domain, or from the use of commercially available genotyping platforms, a new approach has evolved that can be applied to pharmacogenomic-related questions [28,29]. HapMap samples have been used to study the expression level of a gene as the quantitative trait of interest and analysis of SNP genotypes is used to identify the genes that are associated with expression quantitative trait loci (eQTL) [30]. Studies applying this methodology to lymphoblastoid cell lines have illustrated that whole-genome genotyping can be used to perform a genome-wide interrogation of the regulatory mechanisms that underlie gene-expression differences in humans [31].
Monks et al. used lymphoblastoid cell lines from 15 Centre d'Etude du Polymorphisme Humain (CEPH) families to study the heritability of gene expression and to identify eQTLs [32]. Significant heritability was identified for 31% of the 2340 genes that were differentially expressed and eQTLs were detected for 33 genes. Stranger et al. tested 60 unrelated CEU cell lines for eQTLs for 374 genes and then defined each eQTL as either cis (located within 1 Mb from the midpoint of the genomic regions of the corresponding gene) or trans [33]. The majority of eQTLs identified were cis in this study.
Morley et al. measured gene expression levels (quantitative traits) using the Affymetrix Human Genome Focus Array for 94 unrelated CEU cell lines obtained from CEPH pedigrees [34]. Genotype data for the same cell lines, obtained from the SNP Consortium, was utilized to perform a genome-wide linkage analysis to identify regions of the genome containing genetic determinants that contribute to the expression phenotypes. Of the 3554 gene expressions analyzed, approximately 1000 were found to be linked to a particular genetic determinant that influenced its expression. The genetic determinants were located within or close to the target gene (i.e., cis-acting) and distant from the target gene (i.e., trans-acting). Of the target genes, 19% had only cis-acting genetic determinants, 77.5% had only trans-acting, and 3.5% had both a cis-acting and a trans-acting associated SNP. In a separate study of 57 CEU cell lines, the same group performed association analyses for those gene-expression phenotypes identified in the previous analysis as having a cis-regulating SNP [35]. For 374 phenotypes, the association analysis was performed with a dense set of HapMap SNPs located only in regions with strong evidence of linkage. This analysis confirmed the linkage results and further narrowed down the regulatory region. For 27 gene expression phenotypes a genome-wide association analysis was performed using more than 700,000 SNPs. A total of 14 of the 27 expression phenotypes were significantly associated with a SNP, demonstrating the ability of dense genome-wide association studies to identify eQTLs. A better understanding of genetic elements associated with variation in expression of pharmacogenetic genes can contribute to our understanding of variation in pharmacologic effects.
A similar approach was employed to identify genetic variants that explain gene-expression differences among populations [36]. In this study, 142 cell lines representing three different populations of the HapMap Project (CEU, CHB and JPT) were phenotyped for expression with the Affymetrix Human Genome Focus Array. SNP genotype data was obtained from the International HapMap Project and a genome-wide analysis was performed between the genotypes of approximately 2 million SNPs and the levels of gene expression. The expression of 1097 genes differed between the CEU and the CHB + JPT populations. A subset of these genes was identified, for which expression was significantly associated with a SNP, and for which differences in SNP allele frequency among the populations could explain the differences in gene expression that was observed. Storey et al. also looked at gene-expression differences among populations by comparing levels of expression from the Affymetrix Human Genome Focus Arrays of eight CEU cell lines to that of eight YRI cell lines [37]. A total of 50 genes were differentially expressed at a false discovery rate of less than or equal to 20%. The authors further studied SH2B3, one of the most differentially expressed genes, and used allele-specific PCR to demonstrate that there was a difference in expression between the two alleles of the gene, suggesting a cis-regulatory effect. Other studies, including a study in mice, have also demonstrated the presence of both cis-acting and/or trans-acting genetic regulatory elements for gene expression [38,39].
These studies illustrate that whole-genome approaches can be applied to the study of complex traits (eg., gene expression) in cell lines and can further be used to identify the genetic basis for differences in quantitative traits among populations. As susceptibility to drug-induced cytotoxicity is a multigenic quantitative trait, applying similar techniques will allow the identification of genetic determinants that play an important role for this complex trait.
Whole-genome genetics of chemotherapy-induced cytotoxicity
An early study used lymphoblastoid cell lines derived from patients with a history of Wilm's tumor, their first-degree relatives and normal controls to study sensitivity to the DNA damaging agent mitomycin C [40]. After exposure to mitomycin C, cells from both Wilm's tumor patients and their relatives were more sensitive to the drug compared with normal controls, as measured by both cell viability and the frequency of chromosome aberrations, suggesting a genetic basis for susceptibility to the drug. Poot et al. studied sensitivity to camptothecin-induced apoptosis in patients with Werner syndrome and their unaffected siblings [41]. Cell lines from affected individuals exhibited decreased survival, increased cell death and an increase in the number of apoptotic cells after exposure to camptothecin. These earlier studies demonstrate the utility of using lymphoblastoid cell lines to study the genetic basis for susceptibility to chemotherapy-induced cytotoxicity and provide evidence to support the genetic nature of susceptibility.
Cloos et al. studied human susceptibility to carcinogenesis by incubating cultured peripheral blood lymphocytes with bleomycin and counting the number of chromatid breaks as a measure of mutagen sensitivity [42]. This analysis was performed on cells from a total of 135 healthy volunteers from 53 different pedigrees and a heritability estimate of 75% was calculated, providing strong evidence for the genetic basis for susceptibility to bleomycin-induced chromatid breaks. This was further explored by microarray analysis of seven lymphoblastoid cell lines that were highly sensitive (n = 7) and relatively insensitive (n = 7) to the mutagenic effects of bleomycin, as defined by the number of chromatid breaks after exposure to bleomycin [43]. After exposure to bleomycin, microarray analysis of gene expression was performed. In all 101 genes identified as differentially expressed between the sensitive and insensitive cell lines, there were genes involved in signal transduction and cell growth and/or maintenance. The susceptibility to carcinogenesis as described in these experiments may also be related to one's susceptibility to adverse effects from cancer chemotherapy, including the development of therapy-related cancers.
Dolan et al. used CEPH cell lines (147 cell lines from ten CEPH pedigrees) to study sensitivity to cisplatin-induced cytotoxicity [25]. The cells exhibited wide variation in percent survival after exposure to cisplatin. Heritability was calculated based on the variance in percent survival and found to be 47% for cisplatin. Watters et al. [44] utilized a similar approach with CEPH cell lines (427 cell lines from 38 pedigrees) to study the contribution of genetics to human variation in 5-fluorouracil (5-FU) and docetaxel-induced cytotoxicity. Heritability estimates ranged from 0.26-0.65 for 5-FU, and from 0.21-0.70 for docetaxel, depending upon the dose. Genome-wide linkage analysis using 983 microsatellite markers available from the CEPH database identified an area on chromosome 9 with significant linkage to susceptibility to 5-FU and regions on chromosomes 5 and 9 with significant linkage for docetaxel. These results demonstrate that the genetic contribution to susceptibility to chemotherapy-induced cytotoxicity can vary depending on the concentration of the drug, and may also imply that different genetic variants are important for low versus high concentrations of drug.
In a study of daunorubicin-induced cytotoxicity, Duan et al. performed similar heritability for 324 CEPH cell lines in 24 pedigrees, but took the analysis further by performing linkage-directed association studies [45]. This approach decreased multiple testing problems inherent with whole-genome association by focusing association only within suggestive linkage regions. Therefore, instead of testing 3.2 million SNPs throughout the genome, 31,312 SNPs representing 1281 genes within linkage regions (lod ≥1.5) were tested, with SNPs within 30 genes showing significant association with cellular susceptibility to daunorubicin. Their findings suggest that a proportion of susceptibility to chemotherapeutic-induced cytotoxicity may be controlled by genetic determinants and further showed heritability and linkage differences at different concentrations of drug suggesting unique genetic variants contributing to cytotoxicity for low versus high drug dosages.
Huang et al. used lymphoblastoid cell lines from the HapMap Project that represented two different populations (CEU and YRI) to study population and gender differences in susceptibility to chemotherapy-induced cytotoxicity [26]. Cell lines from YRI and CEU demonstrated large interindividual variation in sensitivity to carboplatin, cisplatin, daunorubicin and etoposide-induced cytotoxicity with YRI significantly less sensitive than CEU for carboplatin and daunorubicin. The analysis of gender revealed that within the YRI population the cell lines from females were less sensitive to both carboplatin and cisplatin compared with cell lines from males; while within the CEU population, the cell lines from females were more sensitive to etoposide compared with those from males. A three-way model was built to identify the specific genetic variants that contribute through gene expression to sensitivity to cisplatin [46] and etoposide [47]. In the first step, genome-wide association analyses were performed between the genotypes of over 300,000 of the most informative SNPs from the HapMap and the IC50 for each drug. In the second step, the SNPs coming out of the first step (significant association with IC50) were evaluated for association with gene expression as measured on the Affymetrix Gene-Chip® Human Exon 1.0ST Array. Finally, those genes whose expression were found to be significantly associated with a SNP were evaluated for linear correlation between the level of gene expression and IC50. The final set of SNPs can be considered a genetic signature that acts through baseline gene expression and are associated with susceptibility to chemotherapeutic-induced cytotoxicity. Both the host genes for these SNPs, as well as the target genes these SNPs control, may be interesting candidates for further study.
These studies demonstrate that in vitro cell line models can be used to study the contribution of genetic variants to chemotherapy-induced cytotoxity. They also illustrate that whole-genome genotype and expression information can be combined to identify specific SNPs and genes that contribute to the variation in response to a particular drug. These methods identify a set of novel candidate genes that can then be studied further with more traditional techniques.
It is important to note that the relevance of SNPs identified with these methods will, in some instances, depend on the ethnicity of the population being studied. This point was nicely illustrated in a study by Giovannetti et al. [48] with respect to polymorphisms of thymidylate synthase (TSER) and methylenetetrahydrofolate reductase (MTHFR C677T), two genes involved in the metabolism of methotrexate and therefore relevant in the context of the treatment of childhood leukemia. In this study, the frequency of polymorphisms in Indonesian children was compared with that of Caucasian Dutch individuals. The frequency of the TT and CT genotypes of MTHFR were found to be twofold higher in Caucasians while homozygosity for the TSER 3R/3R polymorphism was threefold higher in the Indonesian children. Since the TSER 3R/3R polymorphism has been shown to be associated with poorer outcome in children with acute lymphoblastic leukemia [49] it may be a useful pharmacogenomic marker but it will be more prevalent in the Indonesian population compared with the Caucasian.
Expert commentary
Without a doubt, enormous strides have been made in the fields of genomics and pharmacogenomics in recent years. Researchers have many new tools at their disposal, some that are still being developed to their fullest potential. While the whole-genome approaches described in this review bring exciting new methods that can be employed to identify genetic variation that explains human variation in expression and chemotherapy-induced cytotoxicity, there are challenges with this approach as well. The genetic signatures and associated genes arising from these new approaches will need to be validated before they can be utilized to influence the care of patients with cancer.
Both the candidate gene and whole-genome approaches have unique advantages and disadvantages. The techniques are well established for the candidate gene approach, the genetic analyses are straightforward and the cost is considerably less. Furthermore, the candidate gene approach leads to a more thorough understanding of the genetic variation within a gene or pathway and its functional consequences. The approach provides rationale for moving functionally relevant SNPs to a clinical setting. One significant drawback of the method is that only one or a small group of genes are evaluated, thereby consequences of other changes within the cell are not considered. Since chemotherapeutic-induced toxicity is likely to be multigenic, the candidate gene approach will, in some cases, have limited applicability.
By contrast, the whole-genome approach evaluates multiple genes and therefore, allows for the identification of known and previously unknown genetic variants contributing to the phenotype without bias. The multiple genes identified can be used to evaluate pathways in addition to individual genes. Subtle changes of several genes within a pathway may be more important than a dramatic change in a single gene. Disadvantages of the whole-genome approach are the high false-discovery rate and likelihood of missing rare genetic variants that could be important. Bioinformatic and biostatistical approaches for data analysis are continually being developed for these large datasets to reduce the false-discovery rate. Experiments need to be well thought out owing to the expense associated with whole-genome studies. Once genes are identified, prioritizing genes for functional validation is an important next step. There are challenges to performing siRNA for gene knockdown on more than a single gene.
There are different experimental models used to study how genetic variation influences gene expression and/or toxicity associated with chemotherapy. The most relevant is the human system because the whole system encompasses pharmacokinetic and pharmacodynamic effects of drugs. However, using chemotherapy for family-based studies in nonaffected individuals is not possible, owing to toxicities associated with this class of drugs. The use of cell lines from related individuals is a reasonable alternative, particularly HapMap cell lines that have publicly available genotype data. Additional benefits of cell-based models are that they are well controlled, easily manipulated, and avoid confounding factors such as drug-drug interactions or comorbidities [50]. There are a number of disadvantages including that only one particular tissue type is being studied and therefore, the data may not be applicable to other tissues exhibiting known toxicities to chemotherapy. In addition, the full effects of Epstein-Barr virus-transformation on gene expression have yet to be specifically elucidated. The lymphoblastoid cell lines can be seen as representing germline genetic variation, which may differ in important ways from the genetic makeup of a tumor. Thus, the findings from these cell lines are probably most applicable to the identification of genetic factors that contribute to the host toxicity of a particular chemotherapeutic agent. However, some findings may also be relevant to tumor sensitivity to the drug since some germline genetic variants present in tumor cells may contribute to drug sensitivity. Finally, how findings from cell lines will translate to the patient, where pharmacokinetics, comorbid conditions and drug-drug interactions play a role, is yet to be determined.
While there are still some questions to be answered, pharmacogenomic researchers now have improved tools to take cancer treatment to the next level. By applying the type of whole-genome technology discussed in this review, genetic determinants important in susceptibility to chemotherapy-induced cytotoxicity will likely be identified. This will give oncologists a new means by which to make treatment decisions for their patients and allow them to maximize benefit and minimize toxicity for each patient based on the genetic makeup of the individual.
Future Perspective
The International HapMap project began 5 years ago. In just that short amount of time, the field of pharmacogenetics and genomics has been revolutionized. Never before have researchers had such a wealth of genetic information readily available. As researchers develop more sophisticated techniques for the analysis of whole-genome data and methods for the validation of targets identified by these studies, eventually the results of these studies will directly affect how patients are treated. It is reasonable to believe that within 10 years' time we will have at our disposal genetic signatures for susceptibility to the toxic effects of multiple chemotherapeutic agents. By genotyping a patient for these variants to identify those at risk for either toxicity or nonresponse, this information can be utilized to personalize chemotherapy in order to maximize the therapeutic benefit and minimize the adverse side effects for each individual patient.
Executive summary.
Susceptibility to chemotherapy-induced cytotoxicity is likely a multigenic trait.
Methods to identify the genetic variants that contribute to chemotherapy-induced cytotoxicity include the candidate gene approach and the whole-genome approach.
TPMT and UGT1A1 are examples of the successful application of the candidate gene approach.
During to the multigenic basis for susceptibility to most drugs, the whole-genome approach will, in some cases, provide a better means to identify novel genetic variants that contribute to chemotherapy-induced cytotoxicity.
Advances in pharmacogenomic research such as gene expression and genotyping platforms and the International HapMap Project have provided the means by which whole-genome studies can be performed.
Whole-genome techniques have been successfully applied to the identification of genetic variants that contribute to the variation in quantitative traits such as gene expression, and are now beginning to be applied to the identification of genetic variants that contribute to chemotherapy-induced cytotoxicity.
Lymphoblastoid cell lines that are part of the International HapMap Project are extremely useful for identifying genetic variants that contribute to chemotherapy-induced cytotoxicity. However, how the findings from these cell lines will translate into patients is yet to be determined.
Further development of data-analysis software to reduce false discovery and prioritize candidate genes for whole-genome studies is needed.
Eventually, combinations of whole-genome approaches and candidate gene approaches should allow for the identification of novel genetic variants that contribute to chemotherapy-induced cytotoxicity and result in greater individualization of cancer therapy to maximize efficacy and minimize toxicity for each patient.
Acknowledgements
We are grateful to Shiwei Duan, Rong Stephanie Huang and Sunita Shukla for helpful suggestions.
Financial disclosure Some of the work in this article was supported by the Pharmacogenetics of Anticancer Agents Research (PAAR) Group (www.pharmacogenetics.org), NIH/NIGMS grants GM61393 and GM61374 (www.pharmGKB.org). ME Dolan is on the Scientific Advisory Board for NIGMS Human Genetic Cell Repository.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Spitz MR, Wu X, Mills G. Integrative epidemiology: from risk assessment to outcome prediction. J. Clin. Oncol. 2005;23(2):267–275. doi: 10.1200/JCO.2005.05.122. [DOI] [PubMed] [Google Scholar]; • Provides examples of single genes that are involved in exposure to carcinogens, cancer predisposition and response to therapy.
- 2.The International HapMap Project Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 3.Bosch TM, Meijerman I, Beijnen JH, Schellens JH. Genetic polymorphisms of drug-metabolising enzymes and drug transporters in the chemotherapeutic treatment of cancer. Clin. Pharmacokinet. 2006;45(3):253–285. doi: 10.2165/00003088-200645030-00003. [DOI] [PubMed] [Google Scholar]
- 4.Efferth T, Volm M. Pharmacogenetics for individualized cancer chemotherapy. Pharmacol. Ther. 2005;107(2):155–176. doi: 10.1016/j.pharmthera.2005.02.005. [DOI] [PubMed] [Google Scholar]; • Reviews the application of pharmacogenetics to the treatment of cancer and provides multiple examples of candidate genes.
- 5.Petros WP, Evans WE. Pharmacogenomics in cancer therapy: is host genome variability important? Trends Pharmacol. Sci. 2004;25(9):457–464. doi: 10.1016/j.tips.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Krynetski EY, Evans WE. Genetic polymorphism of thiopurine S-methyltransferase: molecular mechanisms and clinical importance. Pharmacology. 2000;61(3):136–146. doi: 10.1159/000028394. [DOI] [PubMed] [Google Scholar]
- 7.McLeod HL, Krynetski EY, Relling MV, Evans WE. Genetic polymorphism of thiopurine methyltransferase and its clinical relevance for childhood acute lymphoblastic leukemia. Leukemia. 2000;14(4):567–572. doi: 10.1038/sj.leu.2401723. [DOI] [PubMed] [Google Scholar]
- 8.Relling MV, Hancock ML, Rivera GK, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J. Natl Cancer Inst. 1999;91(23):2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 9.Iyer L, King CD, Whitington PF, et al. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J. Clin. Invest. 1998;101(4):847–854. doi: 10.1172/JCI915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosma PJ, Chowdhury JR, Bakker C, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N. Engl. J. Med. 1995;333(18):1171–1175. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 11.Innocenti F, Undevia SD, Iyer L, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J. Clin. Oncol. 2004;22(8):1382–1388. doi: 10.1200/JCO.2004.07.173. [DOI] [PubMed] [Google Scholar]
- 12.Ratain MJ. From bedside to bench to bedside to clinical practice: an odyssey with irinotecan. Clin. Cancer Res. 2006;12(6):1658–1660. doi: 10.1158/1078-0432.CCR-06-0159. [DOI] [PubMed] [Google Scholar]
- 13.Maitland ML, Vasisht K, Ratain MJ. TPMT, UGT1A1 and DPYD: genotyping to ensure safer cancer therapy? Trends Pharmacol. Sci. 2006;27(8):432–437. doi: 10.1016/j.tips.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Galmarini CM, Mackey JR, Dumontet C. Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia. 2001;15(6):875–890. doi: 10.1038/sj.leu.2402114. [DOI] [PubMed] [Google Scholar]
- 15.Maring JG, Groen HJ, Wachters FM, Uges DR, de Vries EG. Genetic factors influencing pyrimidine-antagonist chemotherapy. Pharmacogenomics J. 2005;5(4):226–243. doi: 10.1038/sj.tpj.6500320. [DOI] [PubMed] [Google Scholar]
- 16.Galmarini CM, Cros E, Thomas X, Jordheim L, Dumontet C. The prognostic value of cN-II and cN-III enzymes in adult acute myeloid leukemia. Haematologica. 2005;90(12):1699–1701. [PubMed] [Google Scholar]
- 17.Galmarini CM, Thomas X, Calvo F, et al. Potential mechanisms of resistance to cytarabine in AML patients. Leuk. Res. 2002;26(7):621–629. doi: 10.1016/s0145-2126(01)00184-9. [DOI] [PubMed] [Google Scholar]
- 18.Galmarini CM. What does over-expression of cN-II enzyme signify in haematological malignancies? Leuk. Res. 2007 doi: 10.1016/j.leukres.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Hubeek I, Stam RW, Peters GJ, et al. The human equilibrative nucleoside transporter 1 mediates in vitro cytarabine sensitivity in childhood acute myeloid leukaemia. Br. J. Cancer. 2005;93(12):1388–1394. doi: 10.1038/sj.bjc.6602881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J. Clin. Oncol. 2005;23(26):6306–6315. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 21.Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 22.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 23.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]; •• Uses genome-wide SNP analysis to identify molecular lesions in acute lymphoblastic leukemia.
- 24.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat. Med. 2006;12(11):1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]; •• Describes the development of gene-expression signatures that predict sensitivity to individual chemotherapeutic drugs.
- 25.Dolan ME, Newbold KG, Nagasubramanian R, et al. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64(12):4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 26.Huang RS, Kistner EO, Bleibel WK, Shukla SJ, Dolan ME. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Mol. Cancer. Ther. 2007;6(1):31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Demonstrates the use of lymphoblastoid cell lines to identify population differences in response to chemotherapy.
- 27.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Overview of the International HapMap Project as a resource for genetic association analyses.
- 28.Jansen RC, Nap JP. Genetical Genomics: the added value from segregation. Trends Genet. 2001;17(7):388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]; • Introduces the concept of genetical genomics and the study of gene expression as a quantitative trait.
- 29.Li J, Burmeister M. Genetical genomics: combining genetics with gene expression analysis. Hum. Mol. Genet. 2005;14(Spec No 2):R163–R169. doi: 10.1093/hmg/ddi267. [DOI] [PubMed] [Google Scholar]
- 30.Cheung VG, Spielman RS. The genetics of variation in gene expression. Nat. Genet. 2002;32(Suppl):522–525. doi: 10.1038/ng1036. [DOI] [PubMed] [Google Scholar]; •• Discusses the analysis of gene expression as a complex trait, provides examples from experimental organisms in which the genetic analysis of gene expression has been successful, and how similar studies might be performed in humans.
- 31.McVean G, Spencer CC, Chaix R. Perspectives on human genetic variation from the HapMap Project. PLoS Genet. 2005;1(4):E54. doi: 10.1371/journal.pgen.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monks SA, Leonardson A, Zhu H, et al. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 2004;75(6):1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stranger BE, Forrest MS, Clark AG, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1(6):E78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morley M, Molony CM, Weber TM, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430(7001):743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung VG, Spielman RS, Ewens KG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437(7063):1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spielman RS, Bastone LA, Burdick JT, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nat. Genet. 2007;39(2):226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Characterized variation in gene expression in HapMap lymphoblastoid cell lines from European and Asian ancestry, compared this quantitative trait among the populations, and identified cis-linked and trans-linked SNPs associated with expression.
- 37.Storey JD, Madeoy J, Strout JL, et al. Gene-expression variation within and among human populations. Am. J. Hum. Genet. 2007;80(3):502–509. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deutsch S, Lyle R, Dermitzakis ET, et al. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum. Mol. Genet. 2005;14(23):3741–3749. doi: 10.1093/hmg/ddi404. [DOI] [PubMed] [Google Scholar]
- 39.GuhaThakurta D, Xie T, Anand M, et al. Cis-regulatory variations: a study of SNPs around genes showing cis-linkage in segregating mouse populations. BMC Genomics. 2006;7(235) doi: 10.1186/1471-2164-7-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imray FP, Smith PJ, Relf W, Kidson C. Wilms' tumour: association with cellular sensitivity to mitomycin C in patients and first-degree relatives. Lancet. 1984;1(8387):1148–1151. doi: 10.1016/s0140-6736(84)91394-1. [DOI] [PubMed] [Google Scholar]
- 41.Poot M, Gollahon KA, Rabinovitch PS. Werner syndrome lymphoblastoid cells are sensitive to camptothecin-induced apoptosis in S-phase. Hum. Genet. 1999;104(1):10–14. doi: 10.1007/s004390050903. [DOI] [PubMed] [Google Scholar]
- 42.Cloos J, Nieuwenhuis EJ, Boomsma DI, et al. Inherited susceptibility to bleomycin-induced chromatid breaks in cultured peripheral blood lymphocytes. J. Natl Cancer Inst. 1999;91(13):1125–1130. doi: 10.1093/jnci/91.13.1125. [DOI] [PubMed] [Google Scholar]
- 43.Cloos J, de Boer WP, Snel MH, et al. Microarray analysis of bleomycin-exposed lymphoblastoid cells for identifying cancer susceptibility genes. Mol. Cancer Res. 2006;4(2):71–77. doi: 10.1158/1541-7786.MCR-05-0196. [DOI] [PubMed] [Google Scholar]
- 44.Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc. Natl Acad. Sci. USA. 2004;101(32):11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duan S, Huang RS, Shukla SJ, Wu X, Badner J, Dolan ME. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67(11):5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang RS, Shukla S, Kistner EO, et al. Expression quantitative trait loci contributing to cisplatin-induced cytotoxicity. Am. J. Hum. Genet. 2007 doi: 10.1086/519850. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This genome-wide approach provided a model that successfully integrated genotype, gene expression and sensitivity to drug information to identify genetic variants that are important in drug treatment.
- 47.Huang RS, Bleibel WK, Kistner EO, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc. Natl Acad. Sci. USA. 2007;104(23):9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giovannetti E, Ugrasena DG, Supriyadi E, et al. Methylenetetrahydrofolate reductase (MTHFR) C677T and thymidylate synthase promoter (TSER) polymorphisms in Indonesian children with and without leukemia. Leuk. Res. 2007 doi: 10.1016/j.leukres.2007.02.011. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 49.Krajinovic M, Costea I, Chiasson S. Polymorphism of the thymidylate synthase gene and outcome of acute lymphoblastic leukaemia. Lancet. 2002;359(9311):1033–1034. doi: 10.1016/S0140-6736(02)08065-0. [DOI] [PubMed] [Google Scholar]
- 50.Shukla SJ, Dolan ME. Use of CEPH and non-CEPH lymphoblast cell lines in pharmacogenetic studies. Pharmacogenomics. 2005;6(3):303–310. doi: 10.1517/14622416.6.3.303. [DOI] [PubMed] [Google Scholar]
- 51.Pander J, Gelderblom H, Guchelaar HJ. Insights into the role of heritable genetic variation in the pharmacokinetics and pharmacodynamics of anticancer drugs. Expert Opin. Pharmacother. 2007;8(9):1197–1210. doi: 10.1517/14656566.8.9.1197. [DOI] [PubMed] [Google Scholar]
- 52.van Kuilenburg AB, Muller EW, Haasjes J, et al. Lethal outcome of a patient with a complete dihydropyrimidine dehydrogenase (DPD) deficiency after administration of 5-fluorouracil: frequency of the common IVS14+1G>A mutation causing DPD deficiency. Clin. Cancer Res. 2001;7(5):1149–1153. [PubMed] [Google Scholar]
- 53.Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19(2):275–280. doi: 10.1093/carcin/19.2.275. [DOI] [PubMed] [Google Scholar]
- 54.le Marchand L, Wilkens LR, Kolonel LN, Henderson BE. The MTHFR C677T polymorphism and colorectal cancer: the multiethnic cohort study. Cancer Epidemiol. Biomarkers Prev. 2005;14(5):1198–1203. doi: 10.1158/1055-9965.EPI-04-0840. [DOI] [PubMed] [Google Scholar]
- 55.Deeken JF, Figg WD, Bates SE, Sparreboom A. Toward individualized treatment: prediction of anticancer drug disposition and toxicity with pharmacogenetics. Anticancer Drugs. 2007;18(2):111–126. doi: 10.1097/CAD.0b013e3280109411. [DOI] [PubMed] [Google Scholar]
- 56.Marsh S, Ameyaw MM, Githang'a J, et al. Novel thymidylate synthase enhancer region alleles in African populations. Hum. Mutat. 2000;16(6):528. doi: 10.1002/1098-1004(200012)16:6<528::AID-HUMU11>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 57.Marsh S, Collie-Duguid ES, Li T, Liu X, McLeod HL. Ethnic variation in the thymidylate synthase enhancer region polymorphism among Caucasian and Asian populations. Genomics. 1999;58(3):310–312. doi: 10.1006/geno.1999.5833. [DOI] [PubMed] [Google Scholar]
- 58.Kaniwa N, Kurose K, Jinno H, et al. Racial variability in haplotype frequencies of UGT1A1 and glucuronidation activity of a novel single nucleotide polymorphism 686C> T (P229L) found in an African-American. Drug Metab. Dispos. 2005;33(3):458–465. doi: 10.1124/dmd.104.001800. [DOI] [PubMed] [Google Scholar]