Abstract
Efficient and accurate replication of the eukaryotic nuclear genome requires DNA polymerases (Pols) α, δ and ε. In all current replication fork models, polymerase α initiates replication. However, several models have been proposed for the roles of Pol δ and Pol ε in subsequent chain elongation and the division of labor between these two polymerases is still unclear. Here, we revisit this issue, considering recent studies with diagnostic mutator polymerases that support a model wherein Pol ε is primarily responsible for copying the leading-strand template and Pol δ is primarily responsible for copying the lagging-strand template. We also review earlier studies in light of this model and then consider prospects for future investigations of possible variations on this simple division of labor.
Introduction
Replication of eukaryotic chromosomes is initiated at replication origins spaced ∼30-100 kb apart. Each origin directs the assembly of two divergently migrating replication forks that faithfully replicate their portion of the chromosome. Substantial evidence indicates that the default replication apparatus in eukaryotes uses three DNA polymerases for normal fork propagation, polymerase (Pol) α, Pol δ and Pol ε [1,2] (Table 1). Because Pol α has limited processivity and lacks intrinsic 3′ exonuclease activity for proofreading errors, it is not well suited to efficiently and accurately copy long templates. Indeed, current evidence indicates an essential, but more limited, role for Pol α in replication (i.e. initiating replication at origins and during lagging-strand synthesis of Okazaki fragments). Pol δ and/or Pol ε are better suited than Pol α for efficient, accurate and extensive chain elongation because, when operating with their accessory proteins, they are the most processive of the nuclear DNA polymerases and they have the highest fidelity [3,4], partly owing to their intrinsic 3′ exonucleolytic proofreading activities. Based on these properties and on genetic evidence that Pol δ and Pol ε are both required for efficient replication in yeast (for a review, see Ref. [1]), it is reasonable to place both polymerases at the replication fork. Interestingly, unlike Escherichia coli, which replicates both DNA strands using a dimeric holoenzyme containing two identical DNA polymerase III core complexes, Pol δ and Pol ε are monomeric with regard to their catalytic cores and there is, as yet, no evidence that they form either homodimers or heterodimers. This leads to the main question to be considered here. What is the division of labor between Pol δ and Pol ε in replicating the leading- and lagging-strand templates?
Table 1. Major DNA polymerases at the replication forka.
| Pol α-primase | Pol δ | Pol ε | |
|---|---|---|---|
| Subunit organization | 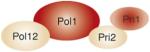 |
 |
|
| Genes and subunit sizes | |||
| S. cerevisiae | Pol1-p167 Pol12-p79 Pri1-p48 Pri2-p62 |
Pol3-p125 Pol31-p55 Pol32-p40 - |
Pol2-p256 Dpb2-p78 Dpb3-p23 Dpb4-p22 |
| S. pombe | Pol1-p159 Pol12-p64 Pri1-p52 Spp2-p53 |
Pol3-p124 Cdc1-p51 Cdc27-p42 Cdm1-p19 |
Pol2-p253 Dpb2-p67 Dpb3-p22 Dpb4-p24 |
| Human | PolA1-p166 PolA2-p68 Prim1-p48 Prim2A-p58 |
PolD1-p124 PolD2-p51 PolD3-p66 PolD4-p12 |
PolE-p261 PolE2-p59 PolE3-p17 PolE4-p12 |
| Activity | Polymerase Primase |
Polymerase 3′-exonuclease |
Polymerase 3′-exonuclease double-strand-DNA binding |
| Fidelity | 10-4-10-5 | 10-6-10-7 | 10-6-10-7 |
| Function | Initiation of replication Initiation of Okazaki fragments |
Elongation and maturation of Okazaki fragments DNA repair Mutagenesis |
Replisome assembly Leading-strand synthesis Replication checkpoint |
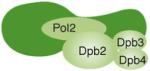
The nomenclature for the cartoon depictions is for S. cerevisiae genes. For Pol δ, a fourth subunit (p12) is shown, which is found in humans but not in S. cerevisiae. Specific subunit interactions are as shown. For a review, see Ref. [1]. The largest subunit of each complex contains the polymerase activity and, for Pol δ and Pol ε, the 3′-exonuclease activity. The Pri1 subunit of Pol α is the catalytic primase subunit. Proposed replication functions and additional functions are as indicated.
Over the past 20 years, several models have been proposed to answer this question: (i) model one proposes that Pol δ and Pol ε replicate the lagging and leading strands, respectively [1,5]; (ii) model two proposes that the opposite is true (i.e. that Pol δ and Pol ε replicate the leading and lagging strands, respectively) [2]; and (iii) model three proposes that Pol δ performs the majority of synthesis on both strands with Pol ε being responsible for only a modest portion of total replication [6,7]. In this review, we revisit these three possibilities by describing an experimental strategy and two recent studies that strongly support the first model. We then consider earlier studies in light of this model and, finally, mention other, non-exclusive possibilities that deserve further investigation in the future.
Strategy to infer which polymerase copies which DNA strand during replication in vivo
Just as seminal studies of Pol δ and Pol ε were emerging in the 1990s, so too were structure-function studies of DNA polymerases that provided insights into DNA replication fidelity. Several amino acid substitutions at or near the binding pocket for the nascent base pair of several different DNA polymerases were found to reduce the fidelity of DNA synthesis (for a review, see Ref. [8]). Among these was a mutant derivative of the large Klenow fragment of E. coli DNA polymerase I containing an alanine substituted for Glu710, a residue that interacts with the ribose of the incoming deoxynucleoside triphosphate (dNTP). The alanine mutant not only had reduced fidelity [9], but it was selectively error-prone for A-deoxycytidine triphosphate (A-dCTP) mismatches compared with T-dGTP mismatches [10]. Interestingly, these are the two mismatches that could give rise to an A-T to G-C mutation in vivo, depending on which of the two template strands was being copied while the error was generated. A replicative polymerase with this property could, in principle, be used to infer which strand it copies in vivo. This prompted the search for analogous ‘asymmetric’ mutator alleles of the three major replicative polymerases in Saccharomyces cerevisiae. The goal was to identify polymerases with six properties: (i) the polymerases should retain robust catalytic activity to enable normal replication and cell growth; (ii) those polymerases that have an intrinsic 3′ exounuclease activity (Pols δ and ε) should retain this activity for functions other than proofreading, for example, processing the 5′ ends of Okazaki fragments; (iii) the polymerases should have reduced replication fidelity despite retaining 3′ exonuclease activity; (iv) the polymerases should have asymmetric error rates in vitro that can be used for strand assignment in vivo; (v) they should generate a mutator phenotype in yeast, but not so strong as to cause error catastrophe; and finally (vi) the mutator polymerases should generate a unique error signature in vivo that can be used to infer which strand(s) they replicate. Using a previously developed strategy [11] to study mutagenesis during leading- versus lagging-strand replication, the intent was to use yeast strains harboring these mutator alleles to study spontaneous mutational specificity in the URA3 reporter gene placed adjacent to ARS306, an origin of replication on chromosome III that fires in early S phase in >90% of yeast cells in a population. URA3 was positioned much closer to ARS306 than to its nearest neighbors, ARS305 and ARS307. It was placed in each of the two orientations and either to the right or left of ARS306. Because replication forks emerging from all three of these early firing origins move at similar rates, the fork emerging from ARS306 copies the URA3 reporter long before the forks emerging from ARS305 and ARS307 could travel to URA3. Thus, the location and orientation of URA3 clearly identifies the leading- or lagging-strand replication machinery that replicates each strand of URA3.
Evidence that S. cerevisiae Pol ε participates in leading-strand-DNA replication
The strategy described was applied to the catalytic Pol2 subunit of Pol ε containing glycine substituted for Met644 [12]. This methionine was targeted for two reasons. First, the X ray crystal structure of RB69 DNA polymerase [13], the replicative DNA polymerase for bacteriophage RB69 and a homolog of Pol α, Pol δ and Pol ε, indicated that Met644 in Pol ε is adjacent to an invariant residue tyrosine in this family of DNA polymerases that has the same function as Glu710 in the large Klenow fragment of E. coli DNA polymerase I; the residue that, when changed, yielded asymmetric error rates [10]. Second, earlier genetic and biochemical studies of several B family polymerases[14-21] indicated that the residue at this position was important for replication fidelity and could be changed without substantial loss of catalytic efficiency. Fortunately, the pol2-M644G allele and the Pol ε protein it encodes were found to fulfill each of the criteria mentioned in the preceding section. This includes the fact that M644G Pol ε has an error rate for T-dTTP mismatches in vitro that is at least 39-fold that of A-dATP mismatches (Figure 1a). Consistent with this property, a preponderance of spontaneous ura3 mutants observed in a pol2-M644G mutator strain contained A-T to T-A substitutions. Most importantly, these were generated at much higher rates when T-dTTP mismatches would be leading-strand errors rather than lagging-strand errors (Figure 1b,c; see additional examples in Figure 2 of Ref. [12]). The patterns of mutagenesis varied by URA3 location and orientation, in a manner consistent with much greater participation of Pol ε in leading-strand replication than in lagging-strand replication.
Figure 1.
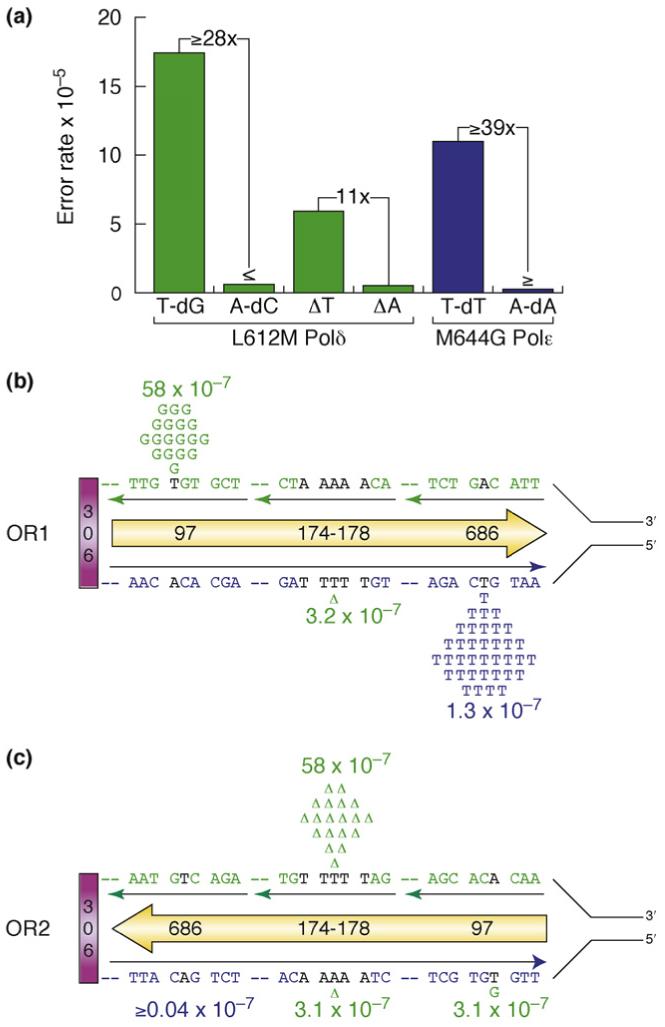
Assigning yeast Pol δ and Pol ε to specific strands. (a) During DNA synthesis in vitro, L612 M Pol δ generates T-dGMP errors at a rate that is at least 28-fold that of A-dCMP errors (green). (b) In a pol3-L612 M msh2- yeast strain, the T-A to C-G mutation rate (depicted earlier as the T to C substitution and depicted here as the inferred T-G mismatch [23]) is high (58 × 10-7) at base pair 97 in URA3 when present in orientation 1. Given the biased error rates in panel (a), these mutations are inferred to result from T-dGMP errors during lagging-strand synthesis by Pol δ. (c) In orientation 2, the mutation rate for the same base pair (base pair 97) in the same neighboring sequence context is much lower (3.10-7), implying that Pol δ has little role in leading-strand synthesis. Using this same logic, because Pol δ deletes template T in homopolymeric runs at a rate 11-fold that of what it deletes in template A [see (a)], L612 M Pol δ is inferred to delete a T-A base pair from a run of five T-A base pairs (174-178) during lagging-strand replication of template Ts [see (c)], but not during leading-strand replication of template As [see (b)]. Finally, note that M644G Pol ε generates T-dTMP errors at a rate ≥39-fold that of A-dAMP errors [blue in part (a)]. In a pol2-M644G strain, the T-A to A-T mutation rate is higher at base pair 686 in URA3 orientation 1. Given the biased error rates in panel (a), these mutations are inferred to result from T-dTMP errors during leading-strand replication by M644G Pol ε. In orientation 2 [see (c)], the mutation rate for the same base pair (base pair 686) in the same neighboring sequence context is much lower, implying that Pol ε has little role in lagging-strand synthesis. Additional examples of mutational specificity that are consistent with these interpretations can be found in Refs [12,23]. Part (a) adapted from Refs. [12,22]. Parts (b,c) adapted from Refs [12,23].
Figure 2.
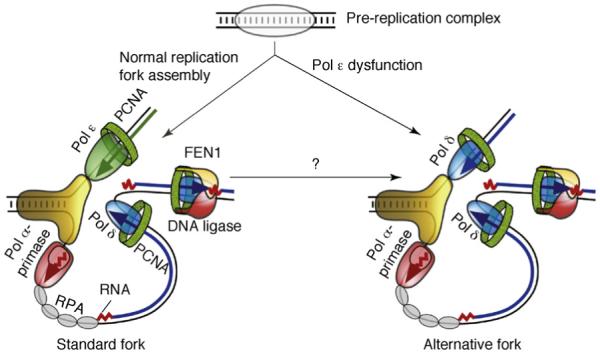
Models for eukaryotic DNA replication forks. On the left is a model illustrating primary roles for Pol ε and Pol δ in leading- and lagging-strand replication, respectively. Other proteins shown include the Pol α-primase (red), the MCM helicase (yellow), the eukaryotic single-stranded-DNA-binding protein, replication protein A (RPA; gray), the sliding clamp proliferating cell nuclear antigen (PCNA; green) and the FEN1-DNA ligase complex (yellow-red). On the right is a model wherein Pol ε dysfunction causes formation of an alternative fork. Conditions other than Pol ε dysfunction might also cause formation of alternative forks and such forks could be assembled at origins, by remodeling the normal fork or during replication restart after an encounter with a natural replication barrier or a lesion.
Evidence for an equal division of labor between Pol ε and Pol δ
Although this study indicates that Pol ε participates in leading-strand replication, by itself it does not define how the workload is shared between Pol ε and Pol δ with respect to replication of the two template strands. For example, even though Pol ε participates more in leading- than lagging-strand replication, it could be that its overall workload is relatively small and that Pol δ has the major role in replicating both strands (model 3 in the Introduction). To distinguish this from a more equal division of labor, the same experimental strategy was applied to the catalytic Pol3 subunit of Pol δ containing a methionine substituted for Leu612 [22,23]. In this case, the pol3-L612M allele and the Pol δ protein it encodes also fulfill each of the six criteria mentioned earlier. As one example, L612M Pol δ has an error rate for T-dGTP mismatches in vitro that is at least 28-fold that of A-dCTP mismatches [22] (Figure 1a). Consistent with a high rate of T-G mismatches, a large number of spontaneous ura3 mutants observed in a pol3-L612M mutator mutant strain contained T-A to C-G substitutions in URA3 (depicted here as the inferred T-G mismatched intermediate), especially at base pair 97. Importantly, the mutation rate at this hotspot was much higher in orientation 1 (Figure 1b), in which T-G mismatches would be lagging-strand errors, than in orientation 2, in which they would be leading-strand errors (Figure 1c). Similar orientation-dependent biases consistent with lagging-strand errors were also observed for deletion of a T-A base pair (Figure 1) and at four other mutational hotspots (Figure 1 in Ref. [23]), and for mutations scattered throughout the URA3 gene (Figure 2 in Ref. [23]). These data imply greater participation of Pol δ in lagging-strand replication than in leading-strand replication. Combined with the inference that M644G Pol ε participates more in leading- than lagging-strand replication [12], the results strongly support a model for normal DNA replication wherein Pol ε is the major leading-strand polymerase and Pol δ is the major lagging-strand polymerase (Figure 2).
Consistency with earlier studies
The model for a nearly equal division of replication labor between Pol ε and Pol δ (Figure 2) is consistent with several earlier studies that have already provided substantial evidence that Pol δ, but perhaps not Pol ε, participates in lagging-strand replication. For example, Pol α, which initiates Okazaki fragments on the lagging strand, interacts with the Pol 32 subunit of Pol δ [24,25]. This is consistent with Pol δ operating on the lagging strand. Moreover, the flap endonuclease FEN1 degrades the initiator RNA during Okazaki-fragment maturation. In this precisely regulated process, strand displacement synthesis by Pol δ is tightly coupled to 5′-flap cutting by FEN1 and Pol ε will not substitute in this process [26]. Excessive strand-displacement synthesis by the polymerase could be deleterious to the cell; for example, duplication mutations can arise if the displaced strand is not removed. Mutational studies have shown strong genetic interactions between mutations in the exonuclease domain of Pol δ and mutations in RAD27, the gene for FEN1 [27]. Most pol3-exo- rad27 double mutants confer lethality. Rare viable double mutants with mild mutations in both genes accumulate small duplications, which is consistent with a defect in Okazaki-fragment maturation. These data indicate that Pol δ functions in the maturation of Okazaki fragments (i.e. it operates on the lagging strand).
Additional evidence that Pol δ operates during lagging-strand replication comes from DNA-replication studies in extracts of Xenopus, which is a eukaryotic replication system amenable to robust biochemical analysis. Xenopus extracts depleted for Pol δ show a marked decrease in DNA synthesis that can be attributed to defects in elongation [28]. Single-stranded gaps accumulate in the absence of Pol δ, which is consistent with a defect in lagging-strand-DNA synthesis. Studies in Xenopus extracts also indicate that Pol ε is required for efficient chromosomal DNA replication [29], although these studies did not address whether Pol ε was replicating the leading or lagging strand.
Support from studies of proofreading
Support for a simple division of labor at the replication fork (Figure 2) also comes from studies that have investigated the role of proofreading by Pol δ and Pol ε during DNA replication.
Studies of proofreading-deficient yeast strains
Proofreading-deficient forms of Pol δ (pol3-exo-) and Pol ε (pol2-exo-) can be tolerated in yeast and such mutants show an increase in mutation rates [30,31]. The pol2-exo- mutator phenotype is multiplicative with that of a mismatch repair mutant, indicating that the insertion errors made by Pol ε, which remain uncorrected because of its proofreading defect, are subsequently corrected by mismatch repair. This clearly indicates that some replication is carried out by Pol ε. The pol3-exo- mutator phenotype is also multiplicative with that of a mismatch repair mutant, again consistent with the participation of Pol δ in DNA replication. Mutational spectra in URA3 placed in both orientations near an origin led the authors to suggest that the 3′ exonucleases of Pol δ and Pol ε function on opposite DNA strands. This same interpretation was derived from another elegant genetic study [32], in which pol2-exo- or pol3-exo- mutants were used to characterize mutagenesis induced by the base analog, 6-N-hydroxylaminopurine (HAP). HAP base pairs ambiguously with both T and C, leading to G-C to A-T and A-T to G-C transitions, depending on whether HAP is the incoming nucleotide or in the template. HAP mutagenesis is unaffected by mismatch repair, recombination or post-replication repair. Therefore, mutations induced by HAP are a direct consequence of mis-insertion by the polymerase and of its ability to proofread these mis-insertions, and the lack thereof, in the exonuclease mutants. In a pol2-exo- mutant, the frequencies of HAP-induced reversion of several mis-sense mutations in URA3 changed dramatically in magnitude when the orientation of the URA3 gene was switched in relation to the nearby ARS306 replication origin. In the pol3-exo- mutant, a similar large change in reversion frequencies was observed upon target reversal. Importantly, for each of the four mis-sense mutations investigated, the direction of the change was opposite for the pol2-exo-- mutant compared with the pol3-exo- mutant. Kunz and coworkers found analogous strand biases with regard to the direction of replication when they examined spontaneous mutation rates in specific positions in the SUP4 gene in pol2-exo- and pol3-exo-- mutants [33]. Although these data do not reveal which polymerase operates on which strand, they are consistent with the idea that Pol ε and Pol δ largely replicate opposite strands [32]. With evidence that Pol δ participates in lagging-strand replication, this implies that Pol ε performs leading-strand replication (Figure 2).
Surprisingly, pol2-exo- mutants have a weaker mutator phenotype than pol3-exo- mutants and, although haploid pol2-exo- mutants are viable when combined with a mismatch repair defect, haploid pol3-exo- mutants combined with a mismatch repair defect are inviable owing to error catastrophe [30,31]. At face value, these results are consistent with model 3, wherein Pol ε is less involved than Pol δ in bulk DNA replication. However, Pol ε has been reported to have higher fidelity than Pol δ for base substitutions [34], for single base deletions [3,4] and for large deletions between direct repeats [35]. Thus, the weaker mutator phenotype of a pol2-exo- strain does not necessarily imply that Pol ε does less work, but could rather reflect the possibility that Pol ε simply generates fewer replication errors in vivo than does Pol δ.
Extrinsic proofreading
Another relevant study involving proofreading used a Pol α L868M mutant that, like the homologous Pol δ L612M mutant, confers a mutator phenotype on yeast [17]. When the pol1-L868M mutation was combined with a proofreading exonuclease-deficiency in Pol δ, the mutation rate in the double mutant was much higher than the sum of mutation rates of the single mutants [20]. These data are consistent with ‘extrinsic’ proofreading, wherein the exonuclease activity of Pol δ proofreads errors made by Pol α, which itself is naturally exonuclease deficient and cannot proofread its own replication errors. These data imply that Pol α and Pol δ both operate on the same strand at a replication fork (i.e. the lagging strand). By contrast, no hypermutability was observed when L868M Pol α was combined with an exonuclease deficiency in Pol ε, thereby indicating that Pol ε does not proofread errors made by Pol α, which is the expected result if Pol ε primarily replicates the leading strand (Figure 2).
Observations consistent with alternative replication fork models
SV40 viral DNA replication
The functions of Pol α-primase and Pol δ at the fork were initially established through elegant biochemical studies of simian virus 40 (SV40) viral DNA replication [36]. Using its large T antigen as both initiator and DNA helicase, the SV40 virus appropriates cellular enzymes for all other functions required to replicate duplex DNA. Interestingly, Pol ε is not required for SV40 origin-dependent replication in vitro and, although crosslinking studies confirm the participation of Pol α and Pol δ in SV40 DNA replication in vivo, Pol ε was not observed to crosslink to replicating SV40 chromatin, but did crosslink to replicating cellular chromatin [37]. Although it is possible that replication of extrachromosomal SV40 viral DNA might not completely recapitulate chromosomal replication enzymology, it is also possible that Pol ε is dispensable for some types of chromosomal replication.
Yeast strains defective in Pol ε catalytic activity
S. cerevisiae and Schizosaccharomyces pombe strains with in-frame deletions of the N-terminal region of the pol2 gene that inactivate Pol ε catalytic activity can grow and divide [7,38,39]. Thus, at least under this unusual circumstance, some replication can proceed without Pol ε catalytic activity, perhaps catalyzed by Pol δ. This is just one example of the remarkable ability of the cell to adapt to the absence of a DNA polymerase. Other examples of functional redundancy are also known among the several polymerases thought to participate in base excision repair, nucleotide excision repair, non-homologous end joining of DNA double-strand breaks and translesion DNA synthesis (see Figure 4 in Ref. [40]). Nonetheless, yeast strains with in-frame deletions of the N-terminal region of the pol2 gene are far from healthy; they show severe phenotypic defects in the progression of DNA replication [41]. In addition, point mutations in the polymerase active site that inactivate Pol ε do confer lethality [38]. Thus, the presence of a catalytically active polymerase domain is essential when Pol ε is actually incorporated into the replisome. That Pol ε actually is incorporated into the replisome, and travels with it during the elongation phase of DNA replication, is supported by chromatin immunoprecipitation studies in yeast [42].
Concluding remarks and future perspectives
How general is the model for a simple division of labor? In the initial study of replicational mutagenesis using URA3 in opposite orientations [11], mutations due to template 8-oxo-G-A mispairs on one strand and template C-HAP mispairs on the other strand both occurred with distinctive strand biases and these biases were maintained throughout an entire 34-kilobase replicon between ARS306 and ARS307 on chromosome III (Figure 3). This indicates that fidelity determinants that are assembled at two independent replication forks in early S phase are maintained until the forks merge and complete their tasks. In addition, the orientation-dependent biases in mutagenesis in the pol2-M644G strain that were seen at ARS306 (Figure 1), which fires in early S phase, were also observed at ARS501 [12], an origin on chromosome V that fires later in S phase. These results support the possibility that the model on the left side of Figure 3 could be broadly applicable. Nonetheless, results so far are with only a few origins that fire frequently and are located in euchromatic DNA. This is like ‘looking under the lamp post’ because eukaryotic genomes are large and there are many origins and these origins vary in frequency of use and time of firing in S phase. The genome varies in transcriptional activity and in sequence content and some sequences (e.g. palindromes, repetitive sequences, fragile sites) can be more problematic than others for replication fork progression. The genome is highly organized by chromatin content (heterochromatin, centromeres, telomeres and subtelomeric regions). Possible influences of chromatin structure are interesting because one subunit of the four-subunit Pol ε holoenzyme has a role in chromatin remodeling and transcriptional silencing (for a review, see Ref. [43]). In fact, there is some evidence to indicate that Pol ε could be especially important for replication of heterochromatic DNA late in S phase [44]. Given these many variables, future studies will be required to determine whether or not the simple model shown in Figure 2 applies throughout the genome. It is remarkable that, 40 years after the key contributions to the field by Okazaki and coworkers that gave rise to the concept of a leading and a lagging strand, we are still struggling to identify and place the factors that replicate each strand [45]. It seems possible that the protein architecture at the fork is more plastic than originally thought. Under certain circumstances, the complex eukaryotic fork with three DNA polymerases might collapse to a simpler fork (Figure 2) that was initially discovered through biochemical studies of SV40 viral DNA replication. That a cell can actually limp along with Pol δ perhaps replicating both strands is astonishing in itself. However, it could also reflect a specialized form of the replication fork that can assemble in a wild-type strain under certain circumstances. Assembly of such an alternative fork could occur at origins, through remodeling of pre-existing normal forks or perhaps upon replication restart after a normal fork stalls. These considerations might be particularly important during replication of the large and complex nuclear genomes of mammalian cells, in which the division of labor during replication has not yet been investigated.
Figure 3.
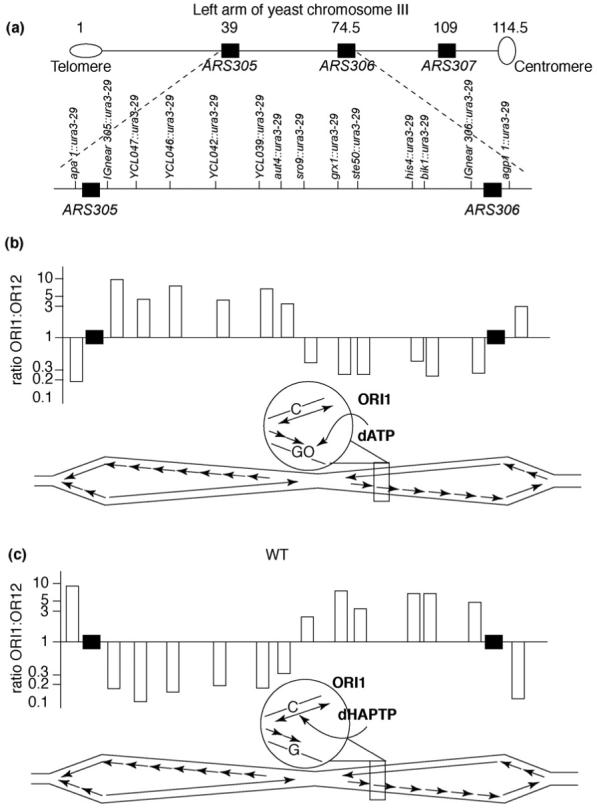
Fork integrity maintained over a 34 kb replicon. (a) Map of the left arm of yeast chromosome III. Distances between elements are in thousands of base pairs and are shown above the chromosome. The region examined is expanded below the chromosome with locations and names of insertion alleles. Replication origins are shown as black rectangles. (b) Ratio of reversion rate of the ura3-29 allele in orientation 1 versus orientation 2 at different locations in chromosome III in an ogg1 strain that is defective in OGG1, the DNA glycosylase that removes the pre-mutagenic 8-oxoG lesion from DNA. Black rectangles indicate two functional origins. Each bar represents the reversion rate ratio at the location corresponding to the position in panel (a). The scheme below each of the bar graphs depicts the region of chromosome III undergoing bidirectional replication initiated at ARS305 and the ARS306. Continuous arrows are for leading-strand replication and multiple arrows represent lagging-strand replication. Replication forks move to the left and to the right from each origin and meet at a site that is equidistant from both origins. The encircled region indicates the reporter allele in orientation 1 showing dATP incorporation opposite 8-oxo-G during lagging-strand replication. (c) As in panel (b), but showing the ratio of HAP-induced reversion frequencies. The encircled region depicts dHAPTP incorporation opposite template C during leading-strand replication. Figure reproduced, with permission, from Ref. [11].
Acknowledgements
The authors thank Stephanie A. Nick McElhinny for help in preparing Figure 1 and Stephanie A. Nick McElhinny and Zachary Pursell for thoughtful comments on the manuscript. The research conducted by the authors was supported, in part, by National Institutes of Health Grant GM032431 to P.M.B. and, in part, by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to T.A.K.
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues. Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited. In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier’s archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright
References
- 1.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 2.Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 3.Fortune JM, et al. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 4.Shcherbakova PV, et al. Unique error signature of the four-subunit yeast DNA polymerase ε. J. Biol. Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 5.Morrison A, et al. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- 6.Burgers PM. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma. 1998;107:218–227. doi: 10.1007/s004120050300. [DOI] [PubMed] [Google Scholar]
- 7.Kesti T, et al. DNA polymerase ε catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 9.Minnick DT, et al. Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I (Klenow fragment) J. Biol. Chem. 1999;274:3067–3075. doi: 10.1074/jbc.274.5.3067. [DOI] [PubMed] [Google Scholar]
- 10.Minnick DT, et al. Discrimination against purine-pyrimidine mispairs in the polymerase active site of DNA polymerase I: a structural explanation. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1194–1199. doi: 10.1073/pnas.032457899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavlov YI, et al. Yeast origins establish a strand bias for replicational mutagenesis. Mol. Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- 12.Pursell ZF, et al. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin MC, et al. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 14.Reha-Krantz LJ, Nonay RL. Motif A of bacteriophage T4 DNA polymerase: role in primer extension and DNA replication fidelity. Isolation of new antimutator and mutator DNA polymerases. J. Biol. Chem. 1994;269:5635–5643. [PubMed] [Google Scholar]
- 15.Beechem JM, et al. Exonuclease-polymerase active site partitioning of primer-template DNA strands and equilibrium Mg2+ binding properties of bacteriophage T4 DNA polymerase. Biochemistry. 1998;37:10144–10155. doi: 10.1021/bi980074b. [DOI] [PubMed] [Google Scholar]
- 16.Fidalgo da Silva E, et al. Using 2-aminopurine fluorescence to measure incorporation of incorrect nucleotides by wild type and mutant bacteriophage T4 DNA polymerases. J. Biol. Chem. 2002;277:40640–40649. doi: 10.1074/jbc.M203315200. [DOI] [PubMed] [Google Scholar]
- 17.Niimi A, et al. Palm mutants in DNA polymerases α and η alter DNA replication fidelity and translesion activity. Mol. Cell. Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, et al. Sensitivity to phosphonoacetic acid: a new phenotype to probe DNA polymerase δ in Saccharomyces cerevisiae. Genetics. 2005;170:569–580. doi: 10.1534/genetics.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkatesan RN, et al. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase δ. J. Biol. Chem. 2006;281:4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 20.Pavlov YI, et al. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr. Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Pursell ZF, et al. Regulation of B family DNA polymerase fidelity by a conserved active site residue: characterization of M644W, M644L and M644F mutants of yeast DNA polymeraseε. Nucleic Acids Res. 2007;35:3076–3086. doi: 10.1093/nar/gkm132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nick McElhinny SA, et al. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 2007;282:2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nick McElhinny SA, et al. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang ME, et al. The Saccharomyces cerevisiae protein YJR043C (Pol32) interacts with the catalytic subunit of DNA polymerase α and is required for cell cycle progression in G2/M. Mol. Gen. Genet. 1999;260:541–550. doi: 10.1007/s004380050927. [DOI] [PubMed] [Google Scholar]
- 25.Johansson E, et al. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 26.Garg P, et al. Idling by DNA polymerase δ maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin YH, et al. The multiple biological roles of the 3′→5′ exonuclease of Saccharomyces cerevisiae DNA polymerase δ require switching between the polymerase and exonuclease domains. Mol. Cell. Biol. 2005;25:461–471. doi: 10.1128/MCB.25.1.461-471.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukui T, et al. Distinct roles of DNA polymerases δ and ε at the replication fork in Xenopus egg extracts. Genes Cells. 2004;9:179–191. doi: 10.1111/j.1356-9597.2004.00716.x. [DOI] [PubMed] [Google Scholar]
- 29.Waga S, et al. DNA polymerase ε is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4978–4983. doi: 10.1073/pnas.081088798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison A, et al. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison A, Sugino A. The 3′→5′ exonucleases of both DNA polymerases δ and ε participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 32.Shcherbakova PV, Pavlov YI. 3′→5′ exonucleases of DNA polymerases ε and δ correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics. 1996;142:717–726. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karthikeyan R, et al. Evidence from mutational specificity studies that yeast DNA polymerases δ and ε replicate different DNA strands at an intracellular replication fork. J. Mol. Biol. 2000;299:405–419. doi: 10.1006/jmbi.2000.3744. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto K, et al. Fidelity of DNA polymerase δ holoenzyme from Saccharomyces cerevisiae: the sliding clamp proliferating cell nuclear antigen decreases its fidelity. Biochemistry. 2003;42:14207–14213. doi: 10.1021/bi0348359. [DOI] [PubMed] [Google Scholar]
- 35.Fortune JM, et al. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase δ. Nucleic Acids Res. 2006;34:4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waga S, et al. Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 37.Zlotkin T, et al. DNA polymerase ε may be dispensable for SV40-but not cellular-DNA replication. EMBO J. 1996;15:2298–2305. [PMC free article] [PubMed] [Google Scholar]
- 38.Dua R, et al. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ε and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 39.Feng W, D’Urso G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase ε are viable but require the DNA damage checkpoint control. Mol. Cell. Biol. 2001;21:4495–4504. doi: 10.1128/MCB.21.14.4495-4504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 41.Ohya T, et al. The DNA polymerase domain of pol ε is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:28099–28108. doi: 10.1074/jbc.M111573200. [DOI] [PubMed] [Google Scholar]
- 42.Aparicio OM, et al. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 43.Pursell ZF, Kunkel TA. Functions of DNA polymerase ε, a polymerase of unusual size and complexity. Prog. Nucleic Acid Res. Mol. Biol. 2008;82:101–145. doi: 10.1016/S0079-6603(08)00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuss J, Linn S. Human DNA polymerase ε colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J. Biol. Chem. 2002;277:8658–8666. doi: 10.1074/jbc.M110615200. [DOI] [PubMed] [Google Scholar]
- 45.Okazaki R, et al. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc. Natl. Acad. Sci. U. S. A. 1968;59:598–605. doi: 10.1073/pnas.59.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]


