Abstract
Histone covalent modifications and 26S proteasome-mediated proteolysis modulate many regulatory events in eukaryotes. In Saccharomyces cerevisiae, heterochromatin mediates transcriptional silencing at telomeres, HM loci and rDNA array. Here, we show that proteasome-associated Sem1p and its interacting partner, Ubp6p (a deubiquitinating enzyme), are essential to maintain telomeric silencing. Simultaneous deletion of SEM1 and UBP6 induces dramatic silencing defect accompanied by significantly increased level of ubiquitinated-histone H2B and markedly reduced levels of acetylated-lysine 14 and 23 on histone H3 at the telomeres. Further, the loss of Sem1p and Ubp6p triggers relocation of silencing factors (e.g. Sir proteins) from telomere to HM loci and rDNA array. Such relocation of silencing factors enhances gene silencing at HM loci and rDNA array, but diminishes telomeric silencing. Interestingly, both Sem1p and Ubp6p participate in the proteolytic function of the proteasome. However, we find that the telomeric silencing is not influenced by proteolysis. Taken together, our data demonstrate that Sem1p and Ubp6p maintain telomeric heterochromatin structure (and hence silencing) through modulation of histone covalent modifications and association of silencing factors independently of the proteolytic function of the proteasome, thus offering a new regulatory mechanism of telomeric silencing.
INTRODUCTION
In Saccharomyces cerevisiae, genes in three distinct genomic regions: telomeres, mating-type loci (HMR and HML) and rDNA array, are transcriptionally silenced in a nonspecific fashion. Establishment of silencing requires formation of silent chromatin and the proper function of the silent information regulatory (SIR) complex composed of Sir2p, Sir3p and Sir4p (1,2). This locus-specific silencing is known to be sensitive to the status changes in methylation, acetylation and ubiquitination of the core histones (3–5). These changes alter the binding of silencing proteins to chromatin. Many histone modification enzymes, such as Set1p, Dot1p, Ubp10p and Rad6p, have been shown to be involved in silencing. Rad6p, an E2 ubiquitin-conjugating enzyme, functions together with E3 ubiquitin ligase Bre1p to attach ubiquitin to lysine 123 (K123) of histone H2B (4–7). H2B ubiquitination is required for functions of Set1p and Dot1p (8–10). Set1p methylates K4 in histone H3 N-terminal tail (4–6,11), while Dot1p methylates K79 in the core domain of histone H3 (4–6,12). Simultaneous deletion of DOT1 and SET1 significantly reduces the binding of Sir proteins to telomeres, indicating that these two modifications function together to mediate silencing. Recently, a deubiquitinating enzyme Ubp10p was found to be involved in silencing (13,14). Either ubp10Δ or mutation in the catalytic domain of Ubp10p results in reduced silencing, especially at telomeres. Ubp10p has been implicated to participate in H2B deubiquitination which influences H3K4 and H3K79 methylation in silent chromatin regions (13,14). Thus, a delicate equilibrium between H2B ubiquitination and deubiquitination is critical for establishing methylation pattern of H3K4 and H3K79 in silent chromatin domains.
Several studies implicate acetylation of lysine residues on histone N-terminal tails to transcriptional activation while deacetylation is more frequently associated with silent chromatin. The status of histone acetylation is controlled by a dynamic equilibrium between histone acetyltransferases (HATs) and histone deacetylases (HDACs). Many enzymes modulating the status of histone acetylation, such as Esa1p, Sas2p, Sir2p and Hat1p, contribute to silencing in budding yeast (15–18). Among the four acetylable lysines in the N-terminal tail of histone H4, only mutation of H4K16 significantly affects telomeric silencing (19). Among the five acetylable lysine residues in the N-terminal tail of histone H3, K14 and K23 (H3K14/K23) are more important than K9 or K18 in telomeric silencing (17). Recently, Taverna et al. (20) have shown that histone H3 K14 acetylation is correlated with histone H2B ubiquitination via H3 K4 methylation. Thus, the enzymes involved in histone H2B deubiquitination can potentially regulate telomeric silencing.
Ubp6p is one of the two deubiquitinating enzymes associated with the ‘lid’ subcomplex of the 26S proteasome (1,21–25). Association of Ubp6p with the proteasome is critical for the deubiquitinating activity of Ubp6p (26) and for the half-life of ubiquitin (27). Although the exact roles of Ubp6p remain to be discovered, it is widely believed that Ubp6p is involved in proteasome-mediated protein degradation (22,28). Notably, affinity capture-MS has identified the physical interaction between Ubp6p and Sem1p, a subunit of the 26S proteasome lid subcomplex (21). Thus, Ubp6p and Sem1p form a structural module with the lid subcomplex of the proteasome. Like Ubp6p, Sem1p is involved in proteasome-dependent proteolysis (29). Further, Sem1p has been shown to be required for DNA double-strand break repair (29).
Several lines of evidence indicate that H2B deubiquitination is important in the maintenance of heterochromatin structure at telomeres, and hence telomeric silencing. Therefore, H2B deubiquitinating enzymes are potential regulators of telomeric silencing. Recent studies (13,14) have implicated a H2B ubiquitin protease Ubp10p, but not SAGA-associated Ubp8p, in controlling H2B ubiquitination at the telomere. However, the role of the proteasome-associated Ubp6p in regulation of H2B ubiquitination and gene expression at telomere has not yet been analyzed, even though a large number of studies (30) have implicated proteasome in transcriptional regulation. Here, we have analyzed whether Ubp6p is involved in H2B deubiquitination and telomeric silencing. Our data demonstrate that Ubp6p in conjunction with Sem1p participates in telomeric silencing by promoting histone H2B deubiquitination, H3 acetylation and association of silencing factors. Further, we show that Sem1p and Ubp6p maintain telomeric silencing independently of the proteolytic function of the proteasome. Thus, these two proteins perform two distinct functions (i.e. heterochromatin maintenance and protein degradation) in separate pathways.
MATERIALS AND METHODS
Yeast strains
Genotypes of yeast strains used in this study are described in Table 1. Yeast genetic manipulation was performed following standard methods. Deletion mutant strains were generated via PCR-mediated gene disruption method as previously described (31), and were confirmed by PCR analysis. Multiple myc-epitope tags were added at the C-terminals of Sem1p, Ubp6p and Sir2p as described previously (32,33), and were confirmed by PCR and western blot analyses.
Table 1.
Relevant yeast strains
| Strain | Genotype | Reference or source |
|---|---|---|
| BY4705α | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 | (31) |
| SQY1002 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 sem1::HIS3 | This study |
| SQY1079 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 sem1::HIS ubp6::URA3 | This study |
| SQY1084 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 UBP6::13myc::TRP1 | This study |
| SQY1085 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 SEM1::13myc::TRP1 | This study |
| SQY1090 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 SIR2::13myc::TRP1 | This study |
| SQY1091 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 sem1::HIS3 ubp6::URA3 SIR2::13myc::TRP1 | This study |
| UCC606 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL | Gottschling |
| SQY1012 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL sem1::TRP1 | This study |
| SQY1039 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL ubp6::HIS3 | This study |
| SQY1040 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL sem1::TRP1 ubp6::HIS3 | This study |
| SQY1046 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL sir2::HIS3 | This study |
| SQY1048 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL ubp10::HIS3 | This study |
| SQY1050 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL ubp6::HIS3 ubp10::TRP1 | This study |
| SQY1054 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL rad6::HIS3 | This study |
| UCC6394 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hta1-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pRG422 | (14) |
| SQY1067 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hta1-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pRG422 ubp6::TRP1 | This study |
| SQY1068 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hta1-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pRG422 ubp10::TRP1 | This study |
| SQY1080 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hta1-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pRG422 rad6::TRP1 | This study |
| SQY1081 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hta1-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pRG422 ubp6::TRP1 ubp10::LYS2 | This study |
| SQY1086 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hta1-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pRG422 sem1::TRP1 | This study |
| SQY1088 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hta1-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pRG422 sir2::TRP1 | This study |
| SQY1089 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 hta1-htb1::MET15 hta2-htb2::LEU2 ADE2-TEL-VR URA3-TEL-VIIL pRG422 ubp6::TRP1 Δsem1::LYS2 | This study |
| CCFY101 | W303–1A MATα ade2–1 ura3–1 trp1–289 leu2–3,112 his3–11,15 can1–100 hmrΔE::TRP1 rDNA::ADE2-CAN1 URA3-TEL-VR | (37) (38) |
| SQY1092 | W303–1A MATα ade2–1 ura3–1 trp1–289 leu2–3,112 his3–11,15 can1–100 hmrΔE::TRP1 rDNA::ADE2-CAN1 URA3-TEL-VR ubp6::HIS3 | This study |
| SQY1093 | W303–1A MATα ade2–1 ura3–1 trp1–289 leu2–3,112 his3–11,15 can1–100 hmrΔE::TRP1 rDNA::ADE2-CAN1 URA3-TEL-VR sem1::HIS3 | This study |
| SQY1094 | W303–1A MATα ade2–1 ura3–1 trp1–289 leu2–3,112 his3–11,15 can1–100 hmrΔE::TRP1 rDNA::ADE2-CAN1 URA3-TEL-VR sir2::HIS3 | This study |
| SQY1096 | W303–1A MATα ade2–1 ura3–1 trp1–289 leu2–3,112 his3–11,15 hmrΔE::TRP1 rDNA::ADE2-CAN1 URA3-TEL-VR sem1::HIS3 ubp6::LEU2 | This study |
| SQY1103 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL pdr5::LEU2 | This study |
| SQY1104 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR URA3-TEL-VIIL sem1::TRP1 ubp6::HIS3 pdr5::LEU2 | This study |
| YAF182 | MATa hta1-htb1::LEU2 hta2-htb2, ade2-1, ura3–1 trp1–289 leu2–3,112 his3–11, can1–100 GAPDH-HA-UBI4::URA3<PZS145 HTA1-FLAG-HTB1, CEN, HIS3> | Osley |
| YAF183 | MATa hta1-htb1::LEU2 hta2-htb2, ade2-1, ura3–1 trp1–289 leu2–3,112 his3–11, can1–100 GAPDH-HA-UBI4::URA3<PZS145 HTA1-FLAG-htb1-K123R, CEN, HIS3> | Osley |
| SQY1107 | MATa hta1-htb1::LEU2 hta2-htb2, ade2-1, ura3–1 trp1–289 leu2–3,112 his3–11, can1–100 GAPDH-HA-UBI4::URA3<PZS145 HTA1-FLAG-htb1-K123R, CEN, HIS3> sem1::ADE2 ubp6::TRP1 | This study |
Spot test assay
Spot tests were done essentially the same as previously described with minor changes (17,25). Five-milliliter yeast cultures were grown overnight at 30°C. For each strain 5 × 107 cells were resuspended in 200 µl of distilled water. Ten-fold serial dilutions of the cell suspensions were made, and 10 µl of each dilution was spotted onto synthetic complete (SC) plates, synthetic complete plates containing 0.1% 5-fluoroorotic acid (SC+FOA). These plates were incubated at 30°C for 3 to 7 days, and photographs were then taken with a digital camera.
Western blotting
Whole-cell extracts were prepared as previously described with minor modifications (34). Briefly, 50-ml cultures were grown overnight at 30°C to mid-log phase. Cells were harvested by centrifugation and then lyzed in 200 μl of SUME buffer (1% sodium dodecyl sulfate, 8 M urea, 10 mM morpholino-propane-sulfonic acid, pH 6.8, 10 mM EDTA) by vortexing with acid-washed glass beads. For each sample 25 μg or 50 μg of total protein was resolved by SDS–PAGE. Resolved SDS–PAGE gels were transferred onto nitrocellulose membranes, and proteins were analyzed with appropriate primary and secondary antibodies. For quantitation, films were scanned, and protein bands were quantitated with Alpha Imager 2000 software.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
RT-PCR was performed essentially as previously described with minor modifications (24). The Pdr5▵ strain that is capable of uptaking MG132 were generated, and used for preparing mRNA by standard TriZol-based RNA extraction protocol. Total mRNA (200 ng) and 10 pmol of oligo (dT) primer were used to generate complementary DNA (cDNA) by RT (Invitrogen). Ten percent of the cDNA products were then used for 20 to 25 cycles of PCR. PCR products were resolved on 2% agarose gels, which were then photographed. DNA bands were quantitated with Alpha Imager 2000 software. Primer sequences are available upon request.
Chromatin immunoprecipitation (ChIP) and sequential chromatin immunoprecipitation (SeqChIP) assays
Except for minor protocol modifications, ChIP and SeqChIP assays were performed essentially as previously described (33). For ChIP, 50-ml yeast cultures were incubated overnight at 30°C in YPD (yeast extract, peptone plus dextrose), and grown upto an OD600 of ∼0.8. Cells were fixed by adding formaldehyde (1% final concentration) and collected by centrifugation. Fixed cells were washed, lyzed and sonicated to generate DNA fragments in the range of 500–800 bp. Anti-myc antibody was used to immunoprecipitate myc-tagged Sem1p, Ubp6p and Sir2p. For SeqChIP, 500-ml yeast cultures were grown overnight in YPD upto an OD600 of ∼0.8. Cells were then fixed, lyzed and sonicated to generate DNA fragments in the range of 500–800 bp as described above. Chromatin was then isolated and incubated with an anti-Flag antibody (Sigma, M5) and protein A-Sepharose. Precipitated protein complexes were washed, eluted and subjected to a second immunoprecipitation with an anti-HA antibody (Convance). ChIP and SeqChIP assays were replicated three times. PCR was used to quantitate the amounts of immunoprecipitated DNA fragments and input samples. Each PCR was performed with cycling conditions as follows: 95°C for 5 min, followed by 22 to 25 cycles consisting of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. Optimal amplification in the linear range was empirically determined. DNA bands were quantitated with Alpha Imager 2000 software. The primer sequences are available upon request.
RESULTS AND DISCUSSION
Sem1p and Ubp6p participate in telomeric silencing
Telomeric silencing (also known as telomere position effect, TPE) refers to epigenetically repressed transcription of a gene placed near the chromosome ends due to the formation of heterochromatin-like structures at the telomeres (35). Telomeric silencing can be sensitively analyzed with a reporter gene, i.e. URA3 or ADE2 integrated into telomeric ends (36). Gene product of an URA3 gene at an internal chromosomal site can convert 5-FOA into a toxin, 5′ fluorouridine monophosphate, which severely inhibits cellular growth. When URA3 is placed near a telomere, its expression is repressed in 30–50%of cells which will be able to grow in the presence of 5-FOA. With the disruption of telomeric silencing, increased expression of the telomeric URA3 will cause repressed cellular growth in medium containing 5-FOA (35). Similarly, Ade2p catalyzes a step in the de novo purine nucleotide biosynthetic pathway. Yeast cells with an internal ADE2 form white colonies and deletion of ADE2 will cause accumulation of P-ribosylamino imidazole which results in formation of red color. Due to epigenetic repression, cells with a telomeric ADE2 form red-sectored colonies.
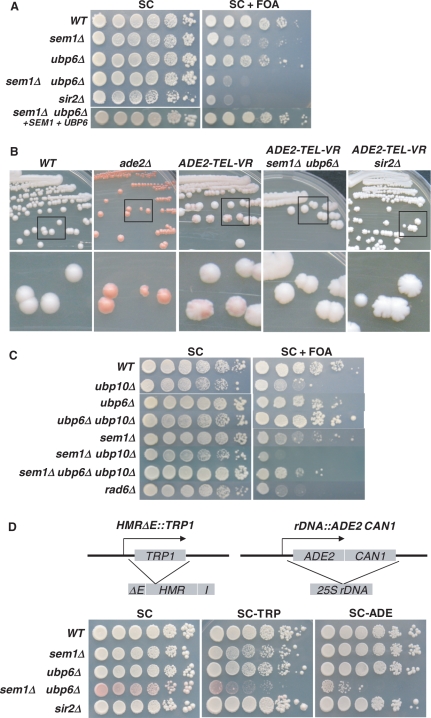
To investigate the roles of Sem1p and Ubp6p in telomeric silencing, we deleted SEM1 or UBP6 in UCC606, which contains an URA3 gene in the right end of chromosome VII and an ADE2 gene in the right end of chromosome V, and studied the corresponding effect on the expression of telomeric URA3 and ADE2. As shown in Figure 1A, sem1Δ or ubp6Δ alone had slight repressive effect on cellular growth in the presence of 5-FOA. However, deletion of SEM1 together with and UBP6 caused significant sensitivity to 5-FOA. This suggests that telomeric silencing is disrupted by simultaneous loss of Sem1p and Ubp6p. Moreover, the 5-FOA sensitivity of the sem1Δ ubp6Δ mutant was indistinguishable from that induced by deleting SIR2 (Figure 1A). A screening study of double mutants containing either sem1Δ or ubp6Δ and deletion of a nonessential proteasomal gene failed to identify any significant 5-FOA sensitivity (data not shown). This indicated that the observed effects of Sem1p and Ubp6p in telomeric silencing are quite specific.
Figure 1.
Sem1p and Ubp6p participate in telomeric silencing. (A, B) Sem1p and Ubp6p are required for telomeric silencing. (C) Loss of Ubp6p rescues silencing defects in the ubp10Δ mutant. (D) Sem1p and Ubp6p do not play a direct role in HM and rDNA silencing. Modified hmrΔE locus and rDNA array in CCFY101α were shown on the top. Sensitivity to 5-FOA was performed by spotting 10-fold serial dilutions of cells onto synthetic complete (SC) plates and synthetic complete plates containing 0.1% 5-FOA (SC+FOA), respectively. The red/white colony color assay was conducted by re-streaking cells on YPD plates. Plates were photographed after 5 to 7 days of growth at 30°C. Genotypes of yeast strains are indicated on the left. Assays were repeated three times. WT, wild-type.
To test the validity of these effects, sem1Δ ubp6Δ mutant was transformed with a plasmid carrying wild-type copy of SEM1 or UBP6 gene. Consequently, the mutant cells containing these two plasmids were shown to once again become resistant to 5-FOA (Figure 1A). This more direct evidence demonstrates that specific expression of Sem1p and Ubp6p in mutant background restores the wild-type level of telomeric silencing. To exclude the possibility that the 5-FOA sensitivity of the mutant cells is caused by the toxic effect of the drug, we spotted cells without telomeric URA3 onto 5-FOA-containing medium. As expected, the sem1Δ ubp6Δ mutant grew normally and did not exhibit any 5-FOA sensitivity as compared to the wild-type (data not shown).
We subsequently tested the yeast colony color change caused by simultaneous loss of Sem1p and Ubp6p. As shown in Figure 1B, a wild-type strain containing an internal ADE2 at a normal chromosomal site formed white colonies on YPD medium. In contrast, UCC606, which contains a telomeric ADE2, formed red-sectored colonies. In addition, loss of Sem1p and Ubp6p in UCC606 caused complete disappearance of the red pigmentation. As a control, SIR2 deletion mutant, which is known to disrupt telomeric silencing also resulted in the disappearance of the red pigmentation in UCC606. The colony color change induced by the loss of Sem1p and Ubp6p is consistent with the 5-FOA sensitivity.
The preceding observation led us to investigate if Sem1p and Ubp6p function together with Ubp10p, another deubiquitinating enzyme which is known to participate in silencing at telomeres and HM loci (13,14). Ubp10p exerts its effect by increasing ubiquitinated-histone H2B (ub-H2B) and disrupting the interaction between silencing proteins and telomeres. Interestingly, ubp6Δ, but not sem1Δ, was able to suppress the silencing defects caused by ubp10Δ (Figure 1C). Besides, sem1Δ ubp6Δ ubp10Δ mutant displayed partial 5-FOA sensitivity, which suggested that ubp10Δ could in part rescue the silencing defects caused by simultaneous deletion of SEM1 and UBP6. These results indicated that interaction among Ubp10p, Ubp6p and Sem1p maintain the silencing status at telomeres, possibly by modulating the histone H2B deubiquitination catalyzed by Ubp6p and Ubp10p.
We then investigated the role of SEM1 and UBP6 in the silencing of HM loci and rDNA arrays. For this purpose, we used CCF101α, which contains a TRP1 in hmrΔE (HMR with E silencer removed) locus and an ADE2-CAN1 cassette in the rDNA array (Figure 1D). Expression of TRP1 and ADE2 can be assessed by monitoring the cellular growth on medium lacking tryptophan (SC-TRP) and adenine (SC-ADE), respectively. Alternatively, color change due to de-repression of ADE2 can be used for evaluating rDNA silencing. Under normal conditions, HM loci are completely silenced. Therefore, the E silencer was removed to induce de-repression status. In the absence of Sem1p and Ubp6p, the cellular growth on SC-TRP and SC-ADE was severely inhibited (Figure 1D), demonstrating the repression of TRP1 and ADE2 at both HMR locus and rDNA array due to enhanced internal silencing. On the other hand, SIR2 deletion induced a better cellular growth (larger colony sizes) than wild-type cells on both SC-TRP and SC-ADE plates. These observations prompted us to speculate that Sem1p and Ubp6p are crucial in the formation of silencing machinery in the telomere, or recruitment of silencing factors to the telomere, but are not required for HM and rDNA silencing. These results support a well-established model (37–39), in which loss of silencing at telomeres results in relocation of silencing factors from telomeres to other genomic regions like HM loci and rDNA array, and therefore causes enhanced silencing at those loci.
Sem1p and Ubp6p associate with telomeres
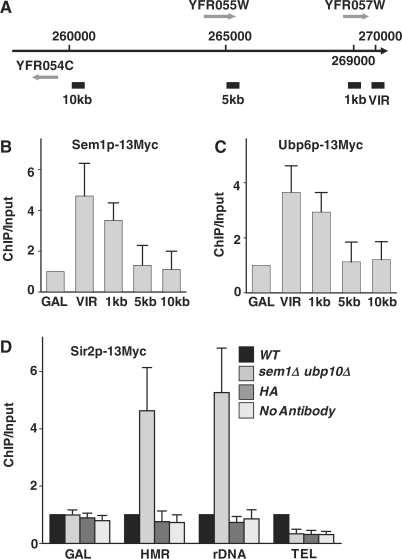
Participation of Sem1p and Ubp6p in the regulation of telomeric silencing indicates that they either directly influence the formation of silent chromatin in telomeres or mediate the interaction between silencing factors and telomeric chromatin. To discern this, we first investigated whether Sem1p and Ubp6p physically associate with telomeres. Thus, SEM1, UBP6 and SIR2 were Myc-tagged via PCR-based tagging method (32). The ChIP assay, using a monoclonal anti-Myc antibody (Sigma), was performed to analyze the interactions between these protein factors and the telomeric DNA sequences. Primers for the ChIP experiments were designed using the DNA sequences on the right end of chromosome VI (Figure 2A) which shares the least homology to the rest of the yeast genome. Our results show that both Sem1p (Figure 2B) and Ubp6p (Figure 2C) were distinctly enriched at telomeres. Moreover, their association with telomere was most significant inside the 1-kb range, but almost disappeared at a 5-kb distance from the telomeric end (Figure 2B and C). These results demonstrate that both Sem1p and Ubp6p are physically associated with telomere.
Figure 2.
Sem1p and Ubp6p associate with telomeres. (A) The ChIP PCR regions in the right arm of chromosome VI are indicated by thick solid bars at VIR, 1 kb, 5 kb and 10 kb. Sem1p (B) and Ubp6p (C) are recruited to telomere. (D) Deletion of SEM1 and UBP6 causes relocation of Sir2p from telomere to HM and rDNA loci. ChIP assays were performed on samples from strains SQY1084 (Ubp6p-Myc), SQY1085 (Sem1p-Myc), SQY1090 (Sir2p-Myc) and SQY1091 (sem1::HIS3 ubp6::URA3 Sir2p-Myc). PCR fragments were generated with primers specific to the GAL1-10 promoter, HMR locus and 25rDNA array, DNA sequences at the right end of chromosome VI, and sequences 1, 5 or 10 kb away from the telomeric end of chromosome VI. Immunoprecipitation was performed using an anti-myc monoclonal antibody (Upstate Biotechnology). An anti-HA monoclonal antibody (Sigma) was used as a nonspecific antibody control. PCR products were quantitated as described in ‘Materials and methods’ section. ChIP/input values were obtained from the ratio of band intensities of immune-recovered relative to input sample, which was normalized against the ratios obtained with GAL1-10 promoter (GAL) (B and C) or wild-type strain (D). GAL, GAL1-10 promoter. VIR, right end of chromosome VI. WT, wild-type; HA, anti-HA antibody control.
Next, we investigated the effects of Sem1p and Ubp6p on the association of Sir2p to the telomere. Deletion of both SEM1 and UBP6 was observed to severely disrupt Sir2p association with telomere (Figure 2D). Therefore, Sem1p and Ubp6p are required for the physical interaction between the silencing complex and telomere. Parallel ChIP assay, however, revealed a manifold increase in the association of Sir2p at HMR and rDNA loci in mutant cells (Figure 2D). These combined observations presented a direct evidence for relocation of silencing factors from telomere to HM loci and rDNA array in the absence of Sem1p and Ubp6p.
Dynamic equilibrium of histone H2B ubiquitination and deubiquitination is critical for telomeric silencing
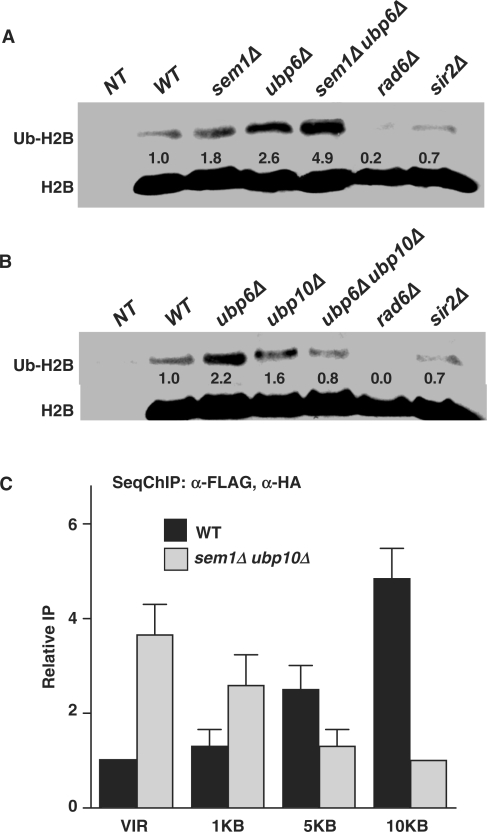
Our data, showing that (i) ubp6Δ suppresses ubp10Δ-induced defects in telomeric silencing (Figure 1C) and (ii) the enrichment of Sem1p and Ubp6p at telomeres, presuppose that Sem1p and Ubp6 along with Ubp10p could be implicated in regulating H2B ubiquitination at telomeres. To test this likelihood, we determined the cellular levels of ubiquitinated-H2B (ub-H2B) in the presence and absence of Sem1p and Ubp6p. Comparison of ub-H2B levels in wild-type and mutant strains by western blotting analysis, using anti-Flag M5 monoclonal antibody (Sigma) which specifically recognizes the Flag tag attached to the N-terminus of H2B, showed a 1.8- and a 2.6-fold increase in ub-H2B in sem1Δ and ubp6Δ mutants, respectively (Figure 3A). Moreover, simultaneous deletion of SEM1 and UBP6 resulted in about 5-fold increase in ub-H2B (Figure 3A). In contrast, transformation of sem1Δ ubp6Δ mutant with plasmids carrying wild-type SEM1 and UBP6, exhibited a reversal of ub-H2B to the wild-type level (data not shown). These results demonstrated that the loss of Sem1p and Ubp6p is clearly responsible for the increased levels of ub-H2B in sem1Δ ubp6Δ mutant. As a control, the band representing ub-H2B completely disappeared in the rad6Δ mutant (Figure 3A and B). Furthermore, Sir2p is known to function downstream of the core histone modification step. Accordingly, we also did not see any change in the ub-H2B level in the sir2Δ strain (Figure 3A and B). Interestingly, the increased ub-H2B level seen in either the ubp6Δ or the ubp10Δ mutants was restored back to the wild-type levels in the ubp6Δ ubp10Δ mutant (Figure 3B). The simultaneous loss of Ubp6p and Ubp10p might trigger the recruitment of another, yet unknown, deubiquitinating enzyme to the chromatin, thus lowering ub-H2B to the wild-type level. This observation is consistent with the phenotypic suppression of ubp10Δ-indcued silencing defects by ubp6Δ (Figure 1C), and suggested that Ubp6p and Ubp10p-dependent H2B deubiquitination are performing complementary functions. Taken together, these data support the important role of Sem1p, Ubp6p and Ubp10p-mediated histone H2B deubiquitination in maintaining telomere silencing. It can be surmised that Ubp6p- and Ubp10-dependent H2B deubiquitination process spawns a dynamic balance between H2B ubiquitination and deubiquitination that is instrumental in regulating telomeric silencing. However, a detailed mechanism of this process remains to be elucidated.
Figure 3.
Sem1p and Ubp6p regulate histone H2B ubiquitination. (A) The sem1Δ ubp6Δ mutant shows increased level of histone H2B ubiquitination. (B) Deletion of UBP10 in the ubp6Δ background restores the level of ub-H2B back to that in the wild-type cells. A plasmid copy of HTA1-FLAG-HTB1 was introduced into cells with both HTA1-HTB1 and HTA2-HTB2 deleted from the genome. (C) Sem1p and Ubp6p are involved in the regulation of H2B ubiquitination at telomere. Whole cell extracts were prepared from 25-ml cultures in mid-log phase and 25 µg protein were analyzed by western blotting. To calculate the fold-change, the density of wild-type strain was designated as 1.0, which was reset from the value obtained by dividing the density of each band with the density of the corresponding input band (H2B). SeqChIP was performed with anti-FLAG M5 and anti-HA (Covance) antibodies. Relative IP represents the IP signal normalized to the respective input signal and then to the respective background signal obtained from the yeast strain with K123R mutation. Densities of protein and DNA bands were quantitated as described in ‘Materials and methods’ section. Standard deviations were calculated from three replicates. Genotypes of yeast strains are indicated on the top. NT, non-tagged.
We next examined the effect on histone H2B ubiquitination induced by the loss of Sem1p and Ubp6p through SeqChIP. In the wild-type cells, ub-H2B levels were lowest at telomeric ends and showed a gradual increase at the ChIP primer sites away from telomere (Figure 3C). For instance at 10 kb from telomere, ub-H2B level was more than three times higher than at the ends. This wild-type pattern of H2B ubiquitination is consistent with previously published reports that have shown the requirement of Ubp10p for maintaining ub-H2B at low levels in the proximity of telomeres (13). Loss of Ubp10p disrupted this specific pattern of H2B ubiquitination. Similarly, simultaneous loss of Sem1p and Ubp6p was also shown here to disrupt the low ub-H2B levels in telomere. In contrast to wild-type cells, the sem1Δ ubp6Δ mutant had much higher levels of ub-H2B in the telomere compared to the 10-kb distance from the telomeric end, consistent with the results obtained in the ubp10Δ mutant (13). This reversed pattern of telomeric H2B ubiquitination induced by the loss of Sem1p and Ubp6p strongly suggests that Sem1p and Ubp6p together with Ubp10p are intimately associated in the maintenance of histone H2B ubiquitination at the telomeric ends. In essence, a dynamic equilibrium exists between ubiquitination catalyzed by Rad6p/Bre1p and deubiquitination catalyzed by Ubp6p/Sem1p and Ubp10p. These precisely regulated ub-H2B levels in turn aid in tethering essential factors at telomeres to orchestrate gene silencing.
Sem1p and Ubp6p participate in telomeric silencing by regulating histone H3 K14/K23 acetylation
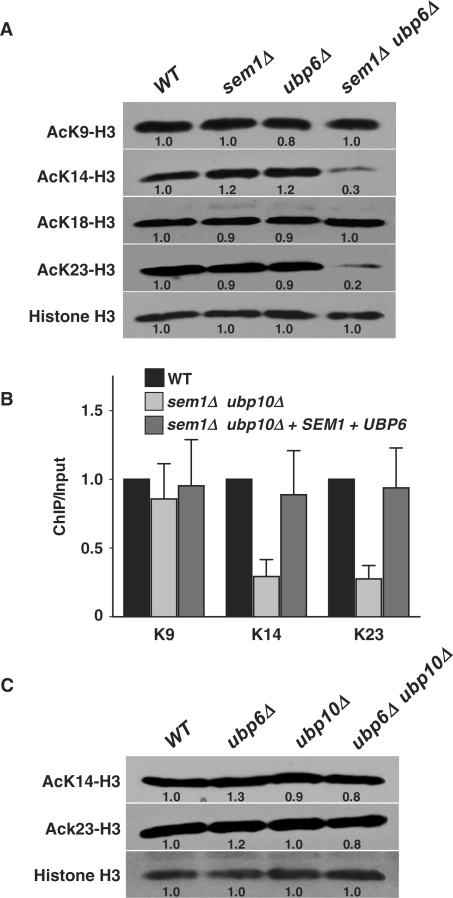
Acetylation of H3K14 and H3K23 is known to be a critical factor in telomeric silencing (17). Therefore, we investigated if the loss of Sem1p and Ubp6p affects acetylation by specific western blot analysis of a battery of acetylated forms of histones H3 and H4. Compared to wild-type cells, >3-fold decrease was seen in the acetylation of H3K14 and H3K23 in the sem1Δ ubp6Δ mutant (Figure 4A). On the other hand, acetylation levels of H3K9 and H3K18 remained unchanged. We next examined H3K14 and H3K23 acetylation in the sem1Δ ubp6Δ mutant carrying a plasmid containing the wild-type gene SEM1 and UBP6, and found that H3K14 and H3K23 are acetylated in the mutant as efficiently as the wild-type (data not shown). Analysis of acetylation of lysine residues in the N-terminal tail of histone H4, which is also known to impact telomeric silencing, did not show any significant changes at K5, K8, K12 and K16 moieties (data not shown). Thus, Sem1p and Ubp6p mediate their effects by altering acetylation at specific sites of histone H3, but not histone H4.
Figure 4.
Sem1p and Ubp6p mediate histone H3 acetylation. (A) Loss of Sem1p and Ubp6p results in reduced acetylation of K14 and K23 on histone H3. (B) Deletion of SEM1 and UBP6 causes reduction of H3 K14 and K23 acetylation in the telomere. (C) The ubp6Δ ubp10Δ mutant is able to acetylate K14 and K23. Global steady-state levels of acetylation of different residues were determined using site-specific antibodies. Western blotting analysis and quantitation were performed as described for Figure 3. The ChIP assay was performed as described for Figure 2. Standard deviations were calculated from three replicates. Genotypes of yeast strains are indicated on the top. VIR, right end of chromosome VI; WT, wild-type.
To further confirm that acetylation of H3K14 and H3K23 is exclusively occurring at the telomeric histones, and thus, is required for silencing at telomeres, we analyzed the acetylation pattern of H3K14 and H3K23 at telomeres via the ChIP assay. The results showed that acetylated-H3K14 and H3K23 were reduced >3-fold at the right telomeric end of chromosome VI in the sem1Δ ubp6Δ mutant (Figure 4B). However, there was no significant change in H3K9 acetylation in the sem1Δ ubp6Δ mutant in comparison to wild-type cells. These results are fully consistent with western blot analysis (Figure 4A), and confirmed that Sem1p and Ubp6p are required for acetylation of H3K14 and H3K23 in telomere. Finally, consistent with the results of Figures 1C and 3B, the ubp6Δ ubp10Δ mutant acetylated H3K14 and H3K23 as efficiently as the wild-type cells (Figure 4C). Therefore, histone H3 acetylation mediated by Sem1p and Ubp6p participates in telomeric silencing, and functions in a different pathway than that of the Ubp10 ubiquitin protease.
We also determined if the acetylation of H3K14 and H3K23 is the direct result of Sem1p and Ubp6p activities in yeast. For this purpose, the sem1Δ ubp6Δ mutant cells were transformed with two plasmids expressing the wild-type Sem1p and Ubp6p and their extracts were analyzed by western blot and ChIP assays. Introduction of the two activities into the sem1Δ ubp6Δ mutant restored the H3 K14 and K23 acetylation back to the wild-type level (Figure 4B). These results confirmed that the defect in H3 acetylation is directly dependent on the loss of Sem1p and Ubp6p function. Collectively, our data reveal that enhanced H2B ubiquitination and accompanying reduction in H3K14 and H3K23 acetylation levels account for the silencing defects in the absence of Sem1p and Ubp6p at telomere. At this point, however, it is not entirely clear how H2B deubiquitination signals histone H3K14 and H3K23 acetylation, but both these events require participation of Sem1p and Ubp6p.
Sem1p and Ubp6p maintain telomeric silencing independently of the proteolytic degradation
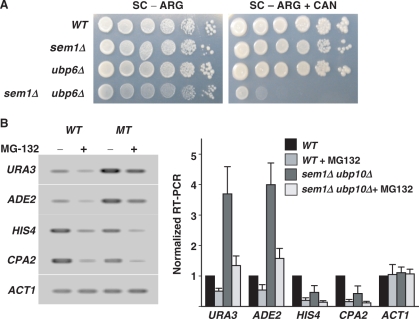
As functional components of the proteasome, Sem1p and Ubp6p could impart their effect on gene expression independently or through proteasome-dependent proteolysis. To resolve this, we first determined the involvement of these two proteins in the protein degradation pathway. Thus, arginine in the synthetic media was replaced with canavanine, a nonproteinogenic analog of arginine. In this medium, cells are forced to take up canavanine instead of arginine to support their protein synthesis. However, the incorporation of canavanine into the polypeptide chain results in synthesis of abnormal protein products, which can accumulate if the ubiquitin-proteasome pathway is rendered defective. Consequently, accumulation of indigestible protein products prevents normal cellular growth and causes cell death. Both sem1Δ and ubp6Δ are known to induce sensitivity to high concentrations of canavanine (27,29). However, under our assay conditions using a lower (1 μg/ml) concentration, neither sem1Δ nor ubp6Δ showed any discernable sensitivity to canavanine. Yet, at this low concentration, simultaneous deletion of SEM1 and UBP6 caused a dramatic growth defect in the presence of canavanine (Figure 5A). This suggests that Sem1p and Ubp6p function cooperatively to undertake efficient proteolysis for sustaining cell survival.
Figure 5.
Sem1p and Ubp6p are involved in the proteolytic function of the proteasome. (A) Sensitivity to canavanine was done by spotting 10-fold serial dilutions of cells onto synthetic complete plates with arginine dropped out (SC-ARG) and SC-ARG plates containing 1 μg/ml canavanine (SC-ARG+CAN). Plates were photographed after 3–5-day growth at 30°C. Genotypes of yeast strains are indicated at the left. (A) Sem1p and Ubp6p regulate telomeric silencing independently of the proteolytic function of the proteasome. Indicated strains were treated with MG-132 (50 μM) or DMSO for 30 min and processed for RT-PCR All assays were repeated three times. WT, wild-type.
To ascertain the uniqueness of silencing effect and any impact of proteolysis, we investigated the changes of specific gene transcription in response to SEM1 and UBP6 deletion under conditions of inhibited proteasomal activity. For this purpose, wild-type and mutant strains were made sensitive to the uptake of Velcade analog MG-132 by deletion of their pdr5 gene. These sensitized strains were used for gene expression studies using RT-PCR assay (Figure 5B). As expected, transcription of a constitutive gene, ACT1, was not sensitive to either simultaneous deletion of SEM1 and UBP6 or the MG-132 treatment. Consistent with previous reports (24), transcription of HIS4 and CPA2 was significantly reduced in the presence of MG-132. In addition, loss of Sem1p and Ubp6p also negatively affected the HIS4 and CPA2 transcription, but to a lesser extent. The negative effects on HIS4 and CPA2 transcription induced by MG-132 treatment and simultaneous deletion of SEM1 and UBP6 were additive. However, expression of telomeric URA3 and ADE2 showed a >3-fold increase in response to the simultaneous loss of Sem1p and Ubp6p, albeit as seen with HIS4 and CPA2, their transcription was also reduced by MG-132 treatment. More importantly, a 2-fold decrease in telomeric URA3 and ADE2 expression induced by MG-132 could not completely reverse the strong de-repressing effect due to the loss of Sem1p and Ubp6p (Figure 5B). This differential response of telomeric URA3 and ADE2 suggests that gene silencing regulated by Sem1p and Ubp6p is independent from that of Sem1p and Ubp6p-dependent protein degradation.
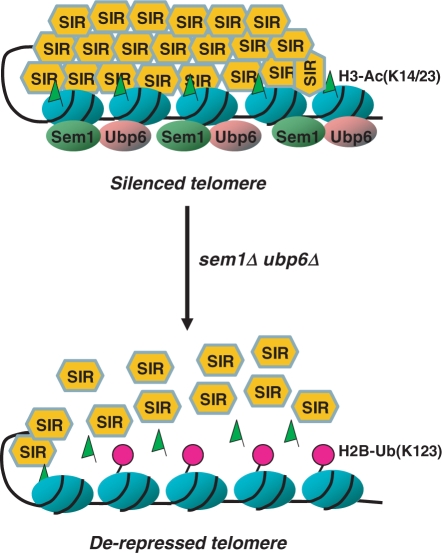
Based upon the overall results of this study, a simplified scheme is presented to illustrate the mechanics of Sem1p and Ubp6p participation in telomeric silencing (Figure 6). According to this model, Sem1p and Ubp6p regulate a major catalytic force that deubiquitinates histone H2B. Ubp6p is activated by interacting with Sem1p and its activation untangles a delicate equilibrium between ubiquitination and deubiquitination of H2B. This disequilibrium translates into a specific acetylation pattern of H3 K14 and K23, which is essential for the maintenance of silent chromatins at telomeres. Simultaneous deletion of SEM1 and UBP6 is shown to disrupt this tightly regulated equilibrium between H2B ubiquitination and deubiquitination, and compromise the acetylation pattern of H3 K14 and K23. These changes, in turn, result in reduced recruitment of silencing protein factors and disruption of silent chromatin at telomere. The silencing factors released from telomere promptly redistribute to other genomic regions, i.e. HM and rDNA loci, causing increased levels of gene silencing at these sites. Taken together, our results provide a new regulatory pathway of telomeric silencing by the proteasome-associated Ubp6p and Sem1p.
Figure 6.
Schematic models depicting the participation of Sem1p and Ubp6p in telomeric silencing. Ubp6p interacts with the 19S proteasome through its binding to Sem1p. Simultaneous deletion of SEM1 and UBP6 causes the blocking of histone H2B deubiquitination, and subsequent reduction of H3K14 and H3K23 acetylation. Such alteration of histone H2B ubiquitination and H3 K14/23 acetylation results in dissociation of the SIR complexes from the telomere, and consequently de-represses telomeric genes. ub, ubiquitin moiety; Ac, acetylation of histone H3; SIR, sir silencing complex.
FUNDING
Public Health Service grants ES2388, ES12991 and CA93413 (to A.A.W.); and a T32 oncology training fellowship (to S.Q.) by the Public Health Service grant CA09338-27 from the National Cancer Institute. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Drs Daniel Finley, Daniel Gottschling, Mary An Osley, Kurt Runge, Brain Strahl and Hideyoshi Yokosawa for providing yeast strains and plasmids. We are grateful to Dr Mark Parthun for critical reading of the manuscript.
REFERENCES
- 1.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 2.Rudner AD, Hall BE, Ellenberger T, Moazed D. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell Biol. 2005;25:4514–4528. doi: 10.1128/MCB.25.11.4514-4528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emre NC, Berger SL. Histone post-translational modifications regulate transcription and silent chromatin in Saccharomyces cerevisiae. Ernst Schering Res. Found. Workshop. 2006;57:127–153. doi: 10.1007/3-540-37633-x_8. [DOI] [PubMed] [Google Scholar]
- 4.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Ann. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 5.Shukla A, Chaurasia P, Bhaumik SR. Cell Mol. Life Sci. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 7.Yamashita K, Shinohara M, Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc. Natl Acad. Sci. USA. 2004;101:11380–11385. doi: 10.1073/pnas.0400078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 9.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 10.Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 11.Tresaugues L, Dehe PM, Guerois R, Rodriguez-Gil A, Varlet I, Salah P, Pamblanco M, Luciano P, Quevillon-Cheruel S, Sollier J, et al. Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J. Mol. Biol. 2006;359:1170–1181. doi: 10.1016/j.jmb.2006.04.050. [DOI] [PubMed] [Google Scholar]
- 12.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emre NC, Ingvarsdottir K, Wyce A, Wood A, Krogan NJ, Henry KW, Li K, Marmorstein R, Greenblatt JF, Shilatifard A, et al. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol. Cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell Biol. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cockell MM, Perrod S, Gasser SM. Analysis of sir2p domains required for rDNA and telomeric silencing in saccharomyces cerevisiae. Genetics. 2000;155:2021. doi: 10.1093/genetics/155.4.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly TJ, Qin S, Gottschling DE, Parthun MR. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol. Cell Biol. 2000;20:7051–7058. doi: 10.1128/mcb.20.19.7051-7058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 19.Meijsing SH, Ehrenhofer-Murray AE. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 2001;15:3169–3182. doi: 10.1101/gad.929001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Guterman A, Glickman MH. Deubiquitinating enzymes are IN/(trinsic to proteasome function) Curr. Protein Pept. Sci. 2004;5:201–211. doi: 10.2174/1389203043379756. [DOI] [PubMed] [Google Scholar]
- 23.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 24.Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- 25.Qin S, Parthun MR. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell Biol. 2002;22:8353–8365. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz RT, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 27.Chernova TA, Allen KD, Wesoloski LM, Shanks JR, Chernoff YO, Wilkinson KD. Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J. Biol. Chem. 2003;278:52102–52115. doi: 10.1074/jbc.M310283200. [DOI] [PubMed] [Google Scholar]
- 28.Park KC, Woo SK, Yoo YJ, Wyndham AM, Baker RT, Chung CH. Purification and characterization of UBP6, a new ubiquitin-specific protease in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 1997;347:78–84. doi: 10.1006/abbi.1997.0311. [DOI] [PubMed] [Google Scholar]
- 29.Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 30.Bhaumik SR, Malik S. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit. Rev. Biochem. Mol. Biol. 2008;4:1. doi: 10.1080/10409230802605914. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 31.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Qin S, Parthun MR. Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol. Cell Biol. 2006;26:3649–3658. doi: 10.1128/MCB.26.9.3649-3658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardner RG, Hampton RY. A ‘distributed degron' allows regulated entry into the ER degradation pathway. EMBO J. 1999;18:5994–6004. doi: 10.1093/emboj/18.21.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 36.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 37.Ray A, Hector RE, Roy N, Song JH, Berkner KL, Runge KW. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat. Genet. 2003;33:522–526. doi: 10.1038/ng1132. [DOI] [PubMed] [Google Scholar]
- 38.Roy N, Runge KW. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol. 2000;10:111–114. doi: 10.1016/s0960-9822(00)00298-0. [DOI] [PubMed] [Google Scholar]
- 39.Smith JS, Brachmann CB, Pillus L, Boeke JD. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]