Abstract
It has been stipulated that repair of clustered DNA lesions may be compromised, possibly leading to the formation of double-strand breaks (DSB) and, thus, to deleterious events. Using a variety of model multiply damaged sites (MDS), we investigated parameters that govern the formation of DSB during the processing of MDS. Duplexes carrying MDS were inserted into replicative or integrative vectors, and used to transform yeast Saccharomyces cerevisiae. Formation of DSB was assessed by a relevant plasmid survival assay. Kinetics of excision/incision and DSB formation at MDS was explored using yeast cell extracts. We show that MDS composed of two uracils or abasic sites, were rapidly incised and readily converted into DSB in yeast cells. In marked contrast, none of the MDS carrying opposed oG and hU separated by 3–8 bp gave rise to DSB, despite the fact that some of them contained preexisting single-strand break (a 1-nt gap). Interestingly, the absence of DSB formation in this case correlated with slow excision/incision rates of lesions. We propose that the kinetics of the initial repair steps at MDS is a major parameter that direct towards the conversion of MDS into DSB. Data provides clues to the biological consequences of MDS in eukaryotic cells.
INTRODUCTION
Spatial distribution and reparability of damage to DNA are key parameters in triggering lethal or mutagenic event in cells. Multiply damaged sites (MDS) consist of closely spaced single lesions within one or two helical turns. Those induced by ionizing radiation typically include oxidized purines and pyrimidines, abasic (AP) sites and single strand breaks (SSB) (1–6). However, other genotoxic agents, such as UVC, UVA, radiomimetics and more recently alkylating agents have been reported to form closely spaced lesions (1,7,8). MDS are thought to be of high biological risk because their repair may be compromised and lead to the formation of double-strand break (DSB), the most deleterious type of DNA damage (8–10).
A number of studies using purified repair enzymes or cell extracts have established that the recognition and processing of a lesion may be affected by the presence of opposite neighboring lesions (11–19). Repair impairment largely depends on the interlesion distance and on the nature of the lesions present in the clusters. More specifically, the presence of an abasic (AP) site or a SSB drastically reduces the excision/incision efficiency of base damage or AP site situated 1–5 bp apart, contributing to limit DSB formation. A hierarchy in repair of closely spaced lesions exists. Such repair-resistant or repair-delayed clusters may persist for a substantial time in cells and lead to enhanced mutagenesis in comparison with single lesions, as shown for 8-oxoguanine (oG) containing clusters in Escherichia coli (20–23).
Alternatively, the fact that lesions at MDS can be partially processed or repaired at reduced rate (11–19) may favor the generation of DSB as repair intermediates. Indeed, the formation of DSB when MDS are processed in vitro also largely depends on the interlesion distance, on the nature of the lesions present within the cluster (11–19) and on the repair enzyme availability (19). In general, if two opposed base lesions are situated > 3 bp apart, the excision/incision step of base excision repair (BER) results in a DSB (9). DSB that could result from attempted simultaneous repair of closely spaced lesions has also been observed in bacteria and mammalian cells. For example, inactivation of the three major oxidative DNA N-glycosylases/AP-lyases in E. coli was correlated with a significant radioresistance of the triple mutant compared to wild-type cells, and with a reduced formation of DSB at radiation-induced clustered lesion (24). Overexpressing either hOGG1 or hNTH1, the two major BER DNA N-glycosylases/AP-lyases, in human lymphoblastoid cells resulted in a significant increase in DSB, which correlated with an elevated sensitivity to γ-rays and an enhanced induced mutagenesis. Down regulation of hOGG1 led to radioresistance and reduced DSB formation (25,26). Clustered lesions composed of two opposing uracil residues or AP sites situated ≤ 7 bp apart were readily converted into DSB in E. coli (27,28), in agreement with the 10-fold increase in deletion observed at uracil cluster compared to single uracil residue (29). Meanwhile, two clustered 8-oxoguanine (oG) or an oG and 5,6-dihydrothymine (DHT) resulted neither in DSB nor in deletion in E. coli, but in enhanced point mutation frequency, relative to single lesion (20–23). However, in HeLa cells, two opposing uracils did not seem to result in DSB formation, while AP sites did (30,31).
Using more complex MDS than those studied to date, our previous in vitro analysis demonstrated that the excision/incision efficiency of a lesion within MDS and the DSB formation not only depends on the nature and the distribution of the lesions, but also on the availability of repair proteins (18,19). This finding implies a hierarchy in the processing of the various lesions within a complex MDS. We wished to challenge our data to in vivo situation. The present work is meant to determine (i) if the processing of complex MDS leads to the formation of DSB in yeast Saccharomyces cerevisiae, and (ii) what are the parameters required for promoting or inhibiting DSB formation from a damage cluster. Our complex MDS previously studied, plus other less complex MDS composed of 2–3 base damage, were inserted into yeast replicative and integrative plasmids that served to transform yeast cells deficient or not in DNA repair. Plasmid viability was determined to be an appropriate assay to estimate the formation of DSB during the processing of MDS by cellular repair enzymes. The kinetics of excision/incision of base damage by yeast extracts were also investigated on both strands. We show that complex MDS carrying a 1-nt gap did not lead to DSB formation, whereas MDS composed of uracil or AP sites, which are extremely rapidly excised, did. The kinetics of the first steps of BER appears to be a key parameter that determines the fate of MDS. This work sheds light on the processing of MDS in eukaryotic cells and, moreover, provides clues to the biological consequences of such lesions.
MATERIALS AND METHODS
Oligonucleotides and plasmids
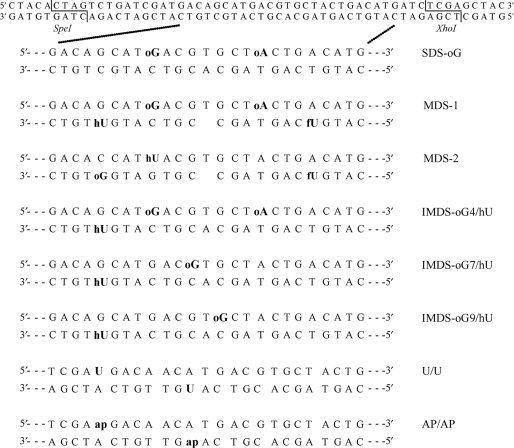
Unmodified oligonucleotides and oligonucleotides carrying uracil were purchased from Sigma-Proligo (France). Oligonucleotides carrying 8-oxoguanine (oG), 8-oxoadenine (oA), 5-hydroxyuracil (hU) and 5-formyluracil (fU) were synthesized as previously described (18). With these oligonucleotides, we built various MDS constructs of different orientation and complexities, as depicted in Figure 1. Duplexes carrying abasic (AP) sites were obtained by hybridization of single-stranded uracil-containing oligonucleotides digested with UDG protein (see below preparation of radiolabeled oligonucleotides). All duplexes harbor SpeI and XhoI restriction sites (Figure 1). Low-copy centromeric plasmid pRS415 (Stratagene) and integrative vector YIplac204-LPG (constructed as described in Supplementary Material; see Supplementary Figure S1a for plasmid maps) were used for introduction of oligonucleotides into yeast.
Figure 1.
Duplexes carrying damaged sites used in this study. Oligonucleotides carried 8-oxoguanine (oG), 8-oxoadenine (oA), 5-hydroxyuracil (hU), 5-formyluracil (fU), uracil (U) or abasic site (AP) (obtained by conversion of uracil to AP site by UDG). Duplexes containing a U and an AP site in the two orientations were also used. oG was located at 4 bp from hU in MDS-1, MDS-2 and IMDS-oG4/hU, at 7 and 9 bp from hU in IMDS-oG7/hU and IMDS-oG9/hU, respectively. MDS-1 and MDS-2 carried a 1-nt gap terminated by 3′-OH and 5′-OH. Undamaged (upper) and damaged duplexes are 56-bp long and harbored SpeI and XhoI restriction sites at each extremity.
Media
Complete YEPD medium and synthetic complete medium SC that does not contain uracil (SC-URA), leucine (SC-LEU), leucine and tryptophane (SC-LEU-TRP) were used (32). Uracil auxotrophs were selected on the medium containing 5-fluoroorotic acid (5-FOA) (32). For drug-resistance selection, YEPD medium was supplemented either with 200 μg/ml of geneticin/G418 (YEPD + G418) or 350 μg/ml of hygromycin B (YEPD + HygB). For selection of ampicillin-resistant E. coli colonies, Luria broth medium was supplemented with 100 μg/ml of ampicillin (LB + Ap) (33).
Yeast strains
All yeast strains used in this study were derived from strain FF18733 (MATa his7-2 leu2-3,112 lys1-1 ura3-52 trp1-289) (F. Fabre, CNRS-CEA, Fontenay-aux-Roses, France), and obtained from Dr S. Boiteux (CNRS-CEA, Fontenay-aux-Roses, France). The following set of FF18733 derivatives was used: FF181134 (rev3Δ::URA3), BG300 (rad14Δ::kanMX6), CD182 (ntg1Δ::URA3 ntg2Δ::TRP1), BG310 (ntg1Δ::URA3 ntg2Δ::TRP1 rad14Δ::kanMX6) (29), BG3 (apn1::URA3, apn2::kanMX6). FF258 (rad51Δ::URA3) and FF742 (rad52Δ::URA3) were from Dr R. Chanet (CNRS, Institut Curie, Orsay, France). DGD39 (ung1Δ ntg1Δ ntg2Δ::kanMX6 ogg1Δ::URA3 mag1Δ::hphMX4) was constructed in the present study (as described in Supplementary Material).
Yeast transformation
Plasmids pRS415 and YIplac204-LPG were double digested either with SpeI and XhoI or with NheI and XhoI, respectively (Supplementary Figure S1a). Linearized vectors were purified by agarose gel electrophoresis using Geneclean Turbo Kit (Q-BIOgene). A total of 100 pmol of single-stranded oligonucleotides were annealed in 1 × restriction buffer #2 (New England Biolabs) to form the appropriate duplexes and digested simultaneously with SpeI and XhoI during 1 h at 37°C. Then oligonucleotides were purified using QIAquick Nucleotide Removal Kit (Qiagen). Concentrations of vectors and of oligonucleotides were quantified by UV absorbance.
For each transformation experiment, three types of ligation reaction (double cut vector + MDS-carrying duplexes, double cut vector + undamaged duplex and double cut vector without any oligonucleotide) were performed in parallel using the same stock solutions of vectors and oligonucleotides. For each ligation reaction, 200 ng of linearized pRS415 with or without 10 ng of SpeI/XhoI-digested damaged or undamaged duplex, or 7.2 μg of linearized YIplac204-LPG with or without 300 ng of SpeI/XhoI-digested MDS-1 or undamaged duplex, were used. Ligations were performed using T4 DNA ligase (New England Biolabs) in supplied buffer during 16 h at 16°C. YIplac204-LPG-based ligation mixtures were purified using Geneclean Turbo Kit and digested simultaneously with BsgI (to direct integration into the chromosomal trp1 gene) and NheI (to cut the fraction of nondigested vector molecules that may be present in the ligation mixture) during 1 h at 37°C. Restriction endonucleases were heat-inactivated at 65°C for 15 min.
Whole amount of each sample was used to transform yeast according to the LiAc procedure (32). Yeast cells were plated on SC-LEU plates (in the case of pRS415) or SC-LEU-TRP plates (in the case of YIplac204-LPG). Appropriate cell dilutions were plated on YEPD plates to determine the total number of cells used for transformation. The frequency of transformants was determined as the ratio of the number of colonies counted on SC-LEU or SC-LEU-TRP plates to the number of colonies counted on the YEPD plates. Leu+Trp+ clones formed after transformation with YIplac204-LPG ligation mixtures were colony-purified on SC-LEU-TRP plates. Single colonies were grown in liquid SC-LEU-TRP medium, and genomic DNA was isolated and analyzed by PCR as described in Supplementary Material.
Preparation of radiolabeled oligonucleotides
Oligonucleotides were 5′-32P-end-labeled as previously described (18). Unincorporated nucleotides were removed using a ProbeQuant G-50 Micro Columns (GE Healthcare). After phenol/chloroform extraction the radiolabeled oligonucleotides were hybridized with equimolar quantity of radiolabeled complementary strand or 1.75-excess of the nonradiolabeled complementary strand in hybridization buffer [140 mM NaCl, 10 mM Tris–HCl (pH 8), 1 mM EDTA (pH 8)] by heating 5 min at 95°C and slow cooling to room temperature. The hybridization efficiency was verified by migration of DNA samples on native 12% polyacrylamide gel [19:1, acrylamide/bisacrylamide (w/w), 100 mM Tris borate, 1 mM EDTA (pH 8)].
To produce an AP-site the single-stranded radiolabeled oligonucleotides (20 pmol) that contained a uracil residue was treated with 2U of uracil DNA glycosylase (UDG) (Trevigen) in 45 µl of buffer [10 mM Tris–HCl (pH 7.9), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT] for 1 h 30 min at 37°C. After the phenol/chloroform extraction, the efficacy of the reaction was checked. Hundred percent of oligonucleotides carried AP sites, as revealed by PAGE after cleavage at AP sites by 0.1 N NaOH. AP-containing oligonucleotides were hybridized as described above. Hybridization produced a weak cleavage at AP sites which was taken into account in cleavage efficiencies.
Preparation of yeast cell extract
The cell-free extract was prepared from 200 ml wild-type yeast exponential culture (∼10−7 cells/ml). After washing with cold water, yeast cells were pelleted and resuspended in 500 µl of lysis buffer [25 mM Tris–HCl (pH 7.5), 5 mM EDTA, 250 mM NaCl, and 0.6 mM PMSF]. A 2 ml of glass beads (size 425–600 μm) were added, and cell suspension was vortexed five times during 1 min (between vortexing cycles extract was kept on ice). Then, another 500 µl of lysis buffer were added and vortexed three more times during 1 min. After removing the glass beads by centrifugation, cell extract was ultracentrifuged for 30 min at 30 000 r.p.m. at 4°C in Beckman-Coulter Optima MAX Ultracentrifuge, using TLA-55 rotor. Then, glycerol was added (20% vol. final) and total protein concentration in the extract was measured using Bradford reagent (Biorad). The cell extract was aliquoted and stored at –20°C.
Cleavage of the lesions-containing duplexes by yeast cell extract
The cleavage assay mixtures (14 µl final volume) contained 200 fmol of radiolabeled double-stranded oligonucleotides and yeast cell extract (20 μg of proteins) in the incubation buffer [20 mM Tris–HCl (pH 7.6), 140 mM NaCl, 4 mM EDTA (pH 8), 8% glycerol]. The reactions were performed at 37°C for various time (as indicated in figures) and stopped by addition of stop-buffer [0.5% SDS and 50 mM EDTA (pH 8)]. Proteinase K (Eurobio) was added at 0.8 mg/ml, followed by incubation for 1 h at 37°C. After phenol/chloroform extraction and ethanol precipitation, samples were loaded on a 12% denaturing polyacrylamide gel [19:1, acrylamide/bisacrylamide (w/w), 7 M urea, 100 mM Tris borate, 1 mM EDTA (pH 8)]. Gels electrophoresis was conducted for 1 h at room temperature at 18.75 V/cm, dried, and exposed to an Amersham Biosciences PhosphorImager screen. The reaction products were visualized and quantified using Molecular Dynamics Storm 820 PhosphorImager and ImageQuant 5.2 software (Molecular Dynamics). The cleavage efficiency was expressed as the percentage of the amount of cleaved molecules to the total amount of (cleaved plus uncleaved) molecules.
RESULTS
The viability of replicative and integrative vector carrying MDS-1 indicates that this complex MDS is processed without formation of DSB
Using a biochemical approach, we previously showed that the formation of DSB during the repair process of complex MDS was limited due to impaired base excision, and that it largely depends on the distribution of base damage and SSB or gap (18,19). To gain insight into the processing of such complex damage in cells, we introduced MDS-1-containing DNA duplex (Figure 1) into either a centromeric plasmid or a nonreplicative (integrative) vector, and the resulting constructs were used to transform wild-type and DNA repair deficient yeast cells. Yeasts were then grown on selective medium to examine the ‘viability’ of these constructs in cells.
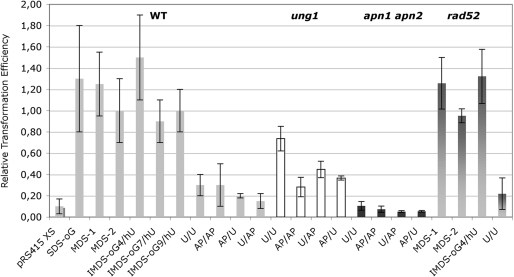
Wild-type yeast strain FF18733 and its isogenic derivatives defective in BER (ung1 ntg1 ntg2 ogg1 mag1 lacking all yeast DNA glycosylases of BER, and ntg1 ntg2), in NER (rad14), in both BER and NER (rad14 ntg1 ntg2) and in translesion DNA synthesis (rev3) were transformed with centromeric plasmid pRS415 carrying either MDS-1-containing oligonucleotide or undamaged oligonucleotide (Supplementary Figure S1a). Since the transformation efficiency by linear vector is very low, we assumed that the transformation efficiencies with vectors carrying the MDS would reflect the extent of DSB formed as repair intermediate. Surprisingly, we found that the transformation efficiencies with MDS-1-containing plasmid were similar to that with plasmid carrying undamaged oligonucleotide, in all tested strains (Table 1). This result may indicate that the repair process of MDS-1 did not generate a DSB that could lead to plasmid loss. Moreover, normal plasmid survival in rad51 or rad52 mutants (Table 1 and Figure 2) suggests that the major pathway of DSB repair in yeast, homologous recombination, is not involved in plasmid maintenance.
Table 1.
Relative transformation efficiency (RTEa) of pRS415 replicative plasmid harboring MDS-1-containing oligonucleotide
| Strain | Relevant genotype | RTEa |
|---|---|---|
| FF18733 | Wild-type | 1.25 [0.7–2.0] |
| DGD39 | ung1 ntg1 ntg2 ogg1 mag1 | 1.25 [0.6–1.4] |
| CD182 | ntg1 ntg2 | 1.25 [1.0–1.7] |
| BG310 | rad14 ntg1 ntg2 | 1.11 [0.7–2.0] |
| BG300 | rad14 | 1.25 [1.0–2.5] |
| FF181134 FF258 | rev3 rad51 | 1.25 [0.6–1.7] 1.25 [0.6–1.4] |
aRTE is calculated as the frequency of transformants with MDS-1-containing vector divided by the frequency of transformants with vector containing undamaged oligonucleotide. Median RTE values from 6 to 8 experiments with 95% confidential intervals are given. Typically, in all experiments, the frequency of transformants with linear (double cleaved) vector (control ligation) was no > 7% of that of transformants with vector ligated with oligonucleotides (see Materials and methods section).
Figure 2.
RTE of wild-type and repair deficient cells by replicative vector carrying diverse damaged duplex. Wild-type cells or ung1, apn1 apn2 or rad52 cells were transformed with pRS415 plasmid linearized by cleavage with SpeI and XhoI restriction enzymes (pRS415 XS, control ligation), or carrying SDS-oG, MDS-1, MDS-2, IMDS-oG4/hU, IMDS-oG7/hU, IMDS-oG9/hU, U/U, AP/AP, AP/U, U/AP constructs or undamaged oligonucleotide. RTE was calculated as the ratio of the transformation efficiency with pRS415 XS or with vector carrying a damaged duplex to the transformation efficiency with vector carrying undamaged oligonucleotide. Error bars represent the SD for at least three independent experiments. For each strain, in each independent experiment, the same stock of competent cells was transformed with all the tested MDS.
The normal survival of the MDS-1-containing replicative plasmid observed in our experiments may indicate that simultaneous repair of the closed lesions at the MDS, which could generate lethal DSB, does not occur. On the other hand, replication of plasmid occurring prior to repair of the MDS, might also prevent formation of DSB, and consequently lead to plasmid maintenance. To minimize the effect of replication, we examined MDS-1 within the nonreplicative vector YIplac204-LPG that, once transferred into yeast, must integrate into the chromosome in order to be replicated. This vector, linearized at BsgI site inside the TRP1 gene, can integrate into chromosome by homologous recombination with trp1 allele, restoring Trp+Leu+ phenotype of the trp1 leu2 (FF18733) host strain (see Materials and methods section and Supplementary Figure S1a and b). In this case, formation of a DSB at MDS-1 after integration into yeast genome, should result in chromosome loss or, at least, in loss of the MDS-1 sequence in the genome (34,35). This can be monitored by a decrease in transformation efficiency, as well as by reduced frequency of clones harboring oligonucleotide sequence in genome (relative to the undamaged control).
We transformed strain FF18733 with BsgI-linearized YIplac204-LPG plasmid carrying either undamaged oligonucleotide or MDS-1-containing oligonucleotide. Table 2 shows that the frequencies of Trp+Leu+ colonies formed after transformation with vector carrying undamaged or MDS-containing oligonucleotide were similar, and 2- to 3-folds higher than observed in control transformation with linearized vector (NheI/XhoI/BsgI-digested vector, see Materials and Methods section). Analysis of Leu+Trp+ clones by PCR (as described in Supplementary Material) showed that integration events into trp1 locus were as frequent after transformation with plasmid carrying MDS-1-containing oligonucleotide as after transformation with plasmid carrying undamaged oligonucleotide (41 and 31% of the transformants, respectively, Table 2). In both cases, 100% of the clones carrying vector integrated into trp1 locus also carried oligonucleotide sequence at the defined position (Table 2 and Supplementary Figure S2a). Analysis of the others clones, that carried YIplac204-LPG integrated outside the trp1 locus, showed the presence of oligonucleotide in 78% of the cases after transformations with vector ligated with undamaged or with MDS-1-containing oligonucleotide (Table 2 and Supplementary Figure S2b). Altogether, our data show that the overall frequency of the clones bearing the oligonucleotide sequence integrated into the genome is very similar after transformation with MDS-1-containing vector or with vector carrying undamaged oligonucleotide (87 and 85%, respectively). This result is also consistent with the proposal that the processing of MDS-1 does not generate DSB in vivo.
Table 2.
Characterization of the transformants of the wild-type strain with integrative plasmid YIplac204-LPG carrying either undamaged or MDS-1-containing oligonucleotide
| Transformation with YIplac204-LPG carrying oligonucleotide | Undamaged | MDS-containing | Controla | |
|---|---|---|---|---|
| Frequency of Trp+Leu+ clones observed in two experimentsb | I | 6.1 × 10−6 | 7.2 × 10−6 | 2.6 × 10−6 |
| II | 10 × 10−6 | 7.6 × 10−6 | 3.8 × 10−6 | |
| Total clones analyzed | 26 | 39 | ||
| Total integrations into trp1 locus | 8 (31%) | 16 (41%) | ||
| Integrations into trp1 locus those that contain oligonucleotide | 8 (100%)c | 16 (100%)c | ||
| Total integrations outside of trp1 locus | 18 (69%)d | 23 (59%)e | ||
| Integrations outside of trp1 locus those that contain oligonucleotide | 14 (78%) | 18 (78%) | ||
| Total clones carrying oligonucleotide | 22 (85%) | 34 (87%) | NAf |
aTransformation with NheI/XhoI/BsgI-digested YIplac204-LPG (see Materials and methods section).
bThe mean RTE (see footnote to Table 1) from the two experiments is 0.98.
cAll samples yielded PCR product with either ES3/trp-R primers or gal-P/trp-R primers.
dThree samples yielded PCR product neither with ES3/trp-P primers, nor with gal-P/trp-P primers. One sample yielded PCR product with gal-P/trp-P primers, but not with ES3/trp-P primers.
eFour samples yielded PCR product neither with ES3/trp-P primers, nor with gal-P/trp-P primers. One sample yielded PCR product with gal-P/trp-P primers, but not with ES3/trp-P primers.
fTransformants were not analyzed by PCR.
Are other complex MDS processed without DSB formation?
In MDS-1, the formation of a DSB should arise from the oG excision and DNA incision by BER (Figure 1). In fact, we showed that cleavage at oG by mammalian cell-free extracts was drastically reduced due to the presence of the 1-nt gap and of the efficiently cleaved hU at 4 bp on the opposite strand (18). In this context, no > 2.5% of MDS-1 were converted into DSB. This may well explain the observed unchanged transformation efficiency of yeast cells by MDS-1-carrying vector relative to that with undamaged oligonucleotide-containing vector. From the literature, it can be emphasized that the formation of a DSB within a MDS composed of two opposed base damage depends on the distance between the lesions and on their orientation (13,27,28,36). We then constructed other MDS that should harbor higher probability to give rise to DSB (Figure 1). Cleavage at MDS-2 by mammalian cell extracts was shown to result in 55% of DSB (18). We also constructed IMDS-oG/hU-constructs in which oG was located at 4-, 7- and 9-nt away from hU on the opposite strand. These MDS were inserted into the replicative plasmid as described above. Wild-type yeast cells were transformed with centromeric plasmid pRS415 carrying either MDS-containing oligonucleotide, oligonucleotide bearing a single oG (SDS-oG) or undamaged oligonucleotide. The left part of Figure 2 shows that the transformation with vectors carrying any of these MDS or SDS-oG, was as efficient as that with vector carrying undamaged oligonucleotide. For comparison, the relative transformation efficiency (RTE, see footnote to Table 1) with linearized vector was very low (circa 0.1 compared to 0.9–1.5 for the other lesions). Sequencing of transformants did not reveal deletion which might be formed, due to DSB, but instead point mutations were observed (Kozmin & Sage, manuscript in preparation).
To confirm that our assay was sensitive enough to detect the formation of DSB during the processing of MDS by the cellular machinery, we then built a duplex carrying two opposed uracil separated by 6 bp (Figure 1). Such clustered lesions have been reported to give rise to DSB and to deletions in bacteria (27,29). Indeed, Figure 2 shows that, in wild-type cells, the transformation efficiency with vector carrying U/U-construct was low, compared to that with vector carrying undamaged oligonucleotide, and it was close to that of linearized plasmid (RTE of 0.3 and 0.1, respectively). Similar relative transformation efficiencies (about 0.2–0.3) were found with vector carrying abasic sites instead of uracil (AP/AP, AP/U or U/AP constructs). This demonstrates that excision of uracils and incision at AP sites, probably, occurred simultaneously on both strands, resulting in formation of DSB, which linearized the vector. In marked contrast, and as expected, relative transformation efficiency with vector carrying U/U-construct, but not AP/AP-construct, significantly increased in ung1 cells (Figure 2). In these cells, uracil clusters are not readily excised and do not lead to extensive DSB. Apn1 and Apn2 are the two major AP endonucleases, but the DNA N-glycosylases/AP-lyases Ntg1 and Ntg2 can also incise AP sites. We found that, in apn1 apn2 cells, the relative transformation efficiencies of vector carrying U/U, AP/AP, U/AP or AP/U constructs were extremely low (Figure 2). This likely reflects the very high toxicity of either AP sites or 3'-blocked SSB left by AP lyases (37,38), leading to collapse of replication fork and thus preventing the plasmid to replicate. In WT cells, we cannot entirely rule out the possibility of breakage at AP sites by replication. Finally, Figure 2 also shows that the relative transformation efficiencies for vectors carrying MDS-1, MDS-2, IMDS-oG4/hU or U/U oligonucleotides were identical in wild-type cells and in cells deficient in homologous recombination and single strand annealing (rad52 strain), thus eliminating any recombinational events to repair potentially generated DSB in our assay.
Collectively, our data demonstrate that when oG or hU are located opposite a 1-nt gap within MDS, this does not lead to substantial formation of DSB during repair in yeast. Our results also demonstrate that when oG and hU are located on opposite strands and separated by 3–8 bp, this also does not give rise to DSB formation. In contrast, the repair process by BER of MDS composed of opposed U and/or AP sites leads to extensive formation of DSB. The literature (21) also reported that the processing of two opposed oG, or oG opposed uracil, does not result in the production of DSB in bacteria, regardless of the interlesion distance (thus not investigated in the present study). What could be the driven force that promotes or prevents the formation of DSB?
The kinetics of cleavage at MDS and of DSB formation by yeast cell extract differ drastically, depending on the type of damage
It has been shown using mammalian cell nuclear extracts that opposing oG and AP site present within a cluster are processed sequentially, limiting the formation of DSB, whereas two opposed AP sites are incised simultaneously, giving rise to DSB (36). This report also suggests that excision of AP site is fast. On the other hand, we showed, using mammalian whole-cell extracts, that the initial rate of cleavage at hU is higher than that at oG, resulting in a hierarchy in the repair of lesions within complex MDS (18,19). It appears that the kinetics of DNA cleavage at the various lesions present within a MDS may be that limiting parameter which leads to or prevents the formation of DSB.
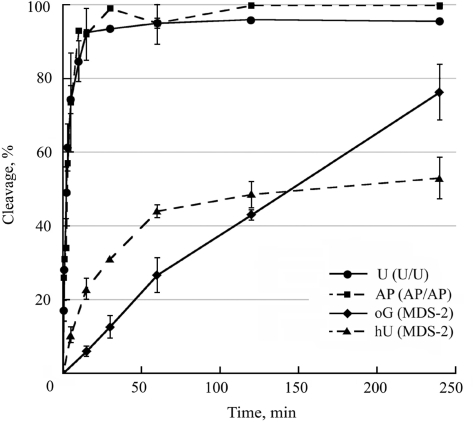
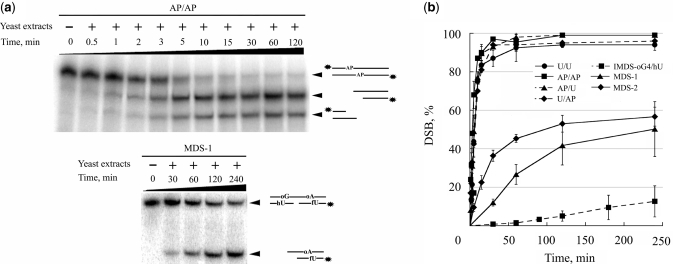
Using yeast cell-free extracts, we analyzed the rate of cleavage at uracil, AP site, hU and oG in our damaged duplex on denaturing gels and also examined the rate of DSB formation by native gel electrophoresis. Figure 3 shows the rate of cleavage at uracil, AP, hU and oG in duplexes U/U, AP/AP and MDS-2. The results demonstrate that uracil and AP site were extremely quickly excised, in comparison with hU or oG. As observed with mammalian cell extracts, initial rate of cleavage at hU is higher than at oG for all MDS tested (Figure 3 and data not shown). In addition, the rates of cleavage at oG or hU observed for IMDS-oG4/hU and MDS-1 were similar to those shown in Figure 3 for MDS-2 (data not shown). The extent of cleavage at hU and oG could indicate that incision may occur on different molecules. The induction of DSB was thus checked. We observed (Figure 4a) that within 5 min a large part of the initial duplex containing AP sites was converted into cleaved products, while after 4 h incubation with the extract about half of MDS-1 remained uncleaved. Figure 4b reveals that most of DSB were produced within a few minutes in all duplexes containing uracil and/or AP sites, and that after 30 min of incubation about 95% of DSB were formed in these initial duplexes, whereas only 35% and 10% of DSB were formed in MDS-2 and MDS-1, respectively. The plateau for DSB formation reached virtually 100% for duplexes containing uracil and/or AP sites, while a maximum of 50% and 60% of substrate was converted into DSB for MDS-1 and MDS-2, respectively, under our experimental conditions. Interestingly, cleavage at hU and oG in IMDS-oG4/hU occurred mainly on separated molecules, as revealed by the very low extent of DSB formed (10% after 4 h incubation). These observations imply that in MDS-1 and MDS-2, the formation of DSB is largely favored by the presence of the 1-nt gap which cannot be repaired under our in vitro experimental conditions (18). Altogether, these data demonstrate that when excision of a damage is very fast, like for uracil and AP sites, excision occurs simultaneously on both strands leading to the formation of DSB (as long as there is no steric hindrance for two proteins at the binding site). In contrast, a lower excision rate protects from the formation of DSB, which may be mainly induced in the presence of pre-existing SSB, at least in vitro.
Figure 3.
Cleavage efficiency at uracil, AP sites, oG and hU by yeast whole-cell extracts. 32P-labeled duplexes U/U, AP/AP or MDS-2 were incubated with 20 μg of proteins from whole-cell extracts at 37°C for various periods of time and separated on 12% denaturing polyacrylamide gels. Two different cell extracts were used. Data represent the means of at least two independent experiments for each extract. Kinetics of cleavage at U or at AP were similar on the two strands and similar to that in AP/U and U/AP. Kinetics of cleavage at oG and hU are given for MDS-2, but they are similar to that in IMDS-oG4/hU or SDS-oG and SDS-hU.
Figure 4.
Assessment of DSB formation during repair. 32P-labeled (*) duplexes were incubated with 20 μg of proteins from whole cell extracts at 37°C for various period of time and separated on 12% nondenaturing polyacrylamide gels. (i) Representative polyacrylamide gels demonstrating DSB formation following incubation of duplexes AP/AP and MDS-1 with yeast whole-cell extract for various periods of time. (ii) rates of DSB induction by cell extract in duplexes U/U, AP/AP, AP/U, U/AP, MDS-2, MDS-1 and IMDS-oG4/hU. Two different cell extracts were used. Data represent the means of at least two independent experiments for each extract.
DISCUSSION
Clustered lesions were predicted to be the most deleterious DNA damage, due to the potential to generate DSBs. As a matter of fact, it has been observed that, at the genomic scale, abortive repair by BER pathway of radiation-induced clustered lesions is a significant source of lethal DSBs, at least in E. coli (24). On the other hand, the fact that excision of one lesion within a cluster may compromise the repair of another closely spaced lesion has been documented in a number of studies performed with defined MDS (see Introduction section). In the present study, we used model MDS of various complexity, carrying diverse types of base damage (oG, hU, U, AP site, 1-nt gap), to investigate the parameters that govern the formation of DSB during the processing of lesions within MDS. The interlesion spacings (3–8 bp) were chosen so that one lesion would not fully block the repair of another lesion located on the opposite strand, according to data from the literature (11–17, 20–23, 27–31, 36). The induction of DSB was assessed by the reduction of yeast transformation efficiency with plasmids carrying these MDS. The combined use of replicative and integrative plasmids bearing diverse MDS and of repair-deficient strains (in BER or homologous recombination) helped us to demonstrate that transformation efficiency assay was relevant to assess the formation of DSB during the processing of MDS carried on plasmid. This is the first extensive study on repair of MDS composed of diverse types of lesions in eukaryotic cells.
We observed that, in wild-type cells, whether or not a DSB is formed at MDS largely depends on the type of base damage present in the cluster. None of the MDS carrying oG and hU situated on opposite strand and separated by 3–8 bp gave rise to DSB, despite that MDS-1 and MDS-2 contained a SSB (1-nt gap) (Figures 1 and 2). In contrast, DSB were readily induced at clusters composed of uracil and/or AP sites separated by 6 bp. Analyzing the kinetics of excision/incision at base damage and formation of DSB on the same MDS by yeast cell extracts, we found that an extensive in-cellulo formation of DSB correlates with fast in vitro excision/incision of damage, and a contrario, MDS carrying base damage associated with slow excision/incision rates are not converted into DSB.
The initial excision/incision rate of damage could be related to the amount of protein available. In exponentially growing yeast cells, the transcript level of APN1, encoding the major AP endonuclease in this organism, is 3–4 folds higher than that of OGG1, NTG2 or APN2 (39). Anyhow, the differences in repair rates of single oG, U or AP site by OGG1, UNG and APE/HAP1 proteins, respectively, found in mammalian cells (oG≪ U < AP) only partly depend on levels of initiating enzymes (40,41). The comparison of the kinetic parameters may be more relevant to explain our observation. The affinities of these proteins for their respective substrates (Km) are in the same order of magnitude. However, the turnover (Kcat) and catalytic efficiency (Kcat/Km) are much higher for yeast Apn1p than for yeast Ogg1 or Ngt 1 and 2 proteins (42–44).
Our findings have several implications. First, in yeast, uracils in U/U-construct, AP sites in AP/AP-construct, or uracil and AP site in AP/U and U/AP constructs were probably cleaved simultaneously (Figure 2). Indeed, we could not see any significant difference in the excision/incision velocity by yeast extracts between the two strands and among these duplexes (Figures 3 and 4). Furthermore, this simultaneous excision of closely opposed uracils and AP sites only occurs because of the fast and roughly identical kinetics. Our data are in very good agreement with in vitro data showing that two opposed AP sites situated 1–5 bp to each other are incised simultaneously by mammalian cell extracts and by purified HAP1, while less efficiently in clusters separated by 1 or 3 bp in the 3′ orientation (17,36,45). Our data are also in accord with the reported conversion of clustered uracils or AP sites into DSB in E. coli (27–29). In human cells, it appears that two closely opposed uracils introduced within luciferase gene did not lead to luciferase inactivation (30), suggesting that the majority of the clustered lesions were not converted into DSB and that mammalian cells evolved mechanisms to avoid the accumulation of SSB as repair intermediates. However, recently Malyarchuk et al. (31) were able to detect luciferase inactivation by the processing of two opposed furan-type of AP site. Using a more sensitive plasmid survival assay, they observed plasmid loss which was increased in the absence of active nonhomologous end joining (NHEJ), the major DSB repair pathway in mammalian cells (31). This indicates the generation of DSB during repair of clustered AP sites in mammalian cells. Our data also point out the high toxicity of unrepaired or misrepaired clustered AP sites (Figure 2).
In contrast to the situation with clustered uracils and AP sites, excision/incision of oxidative base damage such as oG and hU in vitro was much slower and IMDS-oG/hU, MDS-1 and MDS-2 were not converted into DSB in yeast. In particular, in the case of IMDS-oG/hU, in vitro cleavage at oG and hU occurs predominantly on different molecules, emphasizing that, in cells, oG and hU are repaired sequentially. The initial rate of cleavage at hU is higher than at oG both in yeast (this work) and mammalian extracts (19), implying that, in vivo, hU should be cleaved first. Shikazono et al. (23) came to similar conclusion, which is that an oxidized or reduced pyrimidine [like dihydrothymine in (23) or hU in the present work], cleaved by Nth and Nei proteins in E. coli or by Ntg1p or Ntg2p in yeast, is repaired first, then repair of oG (cleaved by Fpg in E. coli and Ogg1p in yeast) can occur. Furthermore, using mutational and repair assays, Malyarchuk et al. (21) showed that DSB repair intermediates and deletions were not formed during repair of MDS consisting of two opposing oG or oG opposite uracil in E. coli. Altogether, these data suggest that, as long as MDS comprises a base damage with slow excision rate by DNA N-glycosylases/AP-lyases, the removal of the lesions is sequential, and moreover, full repair of the first excised lesion occurs before the second lesion is excised and then repaired. The case of MDS-1 and MDS-2 was more complex because of the pre-existing 1-nt gap. The formation of DSB at MDS-1 and MDS-2 by yeast cell extracts was favored by the presence of the 1-nt gap nearby, which could not be processed under our experimental conditions (18). It should be noted that in MDS-1 context, oG was more efficiently cleaved by yeast than by rodent cell extracts (25% versus 5% of cleavage after 1 h incubation, this work and 19). The absence of detectable conversion of MDS-1 and MDS-2 into DSB in yeast may suggest that the 1-nt gap was repaired first. Anyhow, a sequential repair of hU and oG was likely to occur. The importance of kinetics of the initial repair step is further exemplified by the following observations. Overexpression of N-glycosylases/AP-lyases largely enhanced the initial excision rate at MDS by mammalian cell extract, leading to increased DSB formation (19). It also increased DSB-mediated lethality and mutagenesis in human cells exposed to ionizing radiation (25,26). The overexpression of N-glycosylases/AP-lyases may thus have partly overcome the slow excision/incision by these enzymes.
In the case of MDS composed of lesions with slow and different excision/incision kinetics (like our IMDS constructs), what prevents the first SSB created by BER to be transformed into a DSB by excision/incision of a second opposed lesion? DNA compaction proteins like the bacterial HUαβ may bind to the nick created by the first excision/incision event within an MDS and inhibit further endonuclease activity on the opposite strand. In fact, this protein has been shown to interact in vitro with a nick situated opposite a dihydrouracil and to prevent Nth protein from excising the base damage, without inhibiting repair synthesis and ligation (46). HU protein is very abundant in E. coli with regard to repair enzymes, it binds with high affinity to DNA containing nicks or 1- to 2-nt gap, or one base gap generated by DNA N-glycosylases (47), and exhibits a protective effect against lethal action of UV and ionizing radiation (48). It also binds efficiently to MDS containing a 1-nt gap (namely MDS-1 and MDS-2, E. Sage, unpublished data) and it may well be involved in the processing of MDS and protect from the generation of DSB, as suggested in ref. (46). Meanwhile, it appears that, in the case of fast excision/incision, like for clustered uracils and AP sites, protection by HUαβ does not operate efficiently, with regard to the high level of DSB generated during repair in E. coli (27–29). The eukaryotic orthologs of HUαβ, HMGB1 and HMGB2 proteins, may play a similar role. In mammalian cells, poly(ADP-ribose) polymerase (PARP1) which binds to SSB produced by ionizing radiation and is involved in BER (49,50), could also play a role similar to that of HUαβ. Interestingly, Ku70/80 complex could also bind SSB. Indeed, Hashimoto et al. (51) demonstrated that Ku70/80 complex inhibits the N-glycosylase/AP-lyase activity of bacterial N-th at a dihydrouracil residue located opposite an SSB, but also decrease the production of free DSB in vitro. In yeast, NHEJ is a minor pathway devoted to repair of DSB operating mainly during G1 phase, while DSB are essentially repaired by homologous recombination. In yeast, the reduced transformation efficiency of plasmid containing clustered uracils and AP sites suggests that the DSB, produced as repair intermediates, are probably not protected, like in E. coli. They are also not repaired by homologous recombination or single-strand annealing since the transformation efficiency relative to plasmid carrying undamaged oligonucleotide is similar in WT and rad52 mutant cells (Figure 2). In human cells, NHEJ protects only partially furan-type AP sites to be converted into DSB (31).
Our data lead us to propose that, whatever is the complexity of MDS, we can distinguish two types of MDS whose deleterious effects will be different. The MDS containing lesions, like uracils and AP clusters, which are incised with high speed, generate DSB, and will be either lethal or be subjected to error-prone repair (29,31). This process does not prevent some AP clusters to persist in human cells, which may become lethal (17). In contrast, the MDS, carrying base damage removed by N-glycosylases/AP-lyases at slow speed, do not substantially generate DSB. The nick created by the first incision within these MDS would be protected by interaction with proteins that inhibit a second nicking and prevent DSB formation. In fact, both, the direct impairment of N-glycosylases/AP-lyases activities by a SSB nearby and the interaction of specific proteins with the first nick generated, avoid the accumulation of DSB at such MDS. The price to pay may be an increase of point mutations, as observed in E. coli (20–23). Indeed, preliminary data reveal an increase in point mutations targeted at MDS (Kozmin & Sage, manuscript in preparation). This model implies that excision/incision rate is the limiting step for generation of DSB, while repair synthesis and resealing steps are not. In conclusion, our work emphasizes the biological importance of a better understanding of the processing of clustered lesions, which may also be produced by DNA damaging agents such as UV radiation and alkylating agents, beside ionizing radiation (1–8).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique (CNRS) (to E.S. & S.K.); Institut Curie (to E.S., A.R. & S.K.); Centre National d'E;tudes Spatiales (CNES) (to E.S.); Electricité de France (EDF) (to E.S.), Association pour la Recherche sur le Cancer (to S.K.) and Institut National du Cancer (to Y.S.). Funding for open access charge: Electricité de France.
Conflict of interest statement. S.K. was recipient of post-doctoral fellowships from Institut Curie, Association pour la Recherche sur le Cancer and CNRS.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs S. Boiteux (CEA/CNRS), G. Faye (Institut Curie) and Y.I. Pavlov (University of Nebraska) for gift of yeast strains and plasmids. We thank Drs R. Chanet, P.M. Girard (Institut Curie) for stimulating discussions, and Katrina Mitchel (Duke University, USA) for careful reading of the manuscript.
REFERENCES
- 1.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 2.Ward JF. Biochemistry of DNA lesions. Radiat. Res. 1985;104:S103–S111. [PubMed] [Google Scholar]
- 3.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int. J. Radiat. Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 4.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered damages and total lesions induced in DNA by ionizing radiation: oxidized bases and strand breaks. Biochemistry. 2000;39:8026–8031. doi: 10.1021/bi9927989. [DOI] [PubMed] [Google Scholar]
- 5.Georgakilas AG, Bennett PV, Sutherland BM. High efficiency detection of bi-stranded abasic clusters in gamma-irradiated DNA by putrescine. Nucleic Acids Res. 2002;30:2800–2808. doi: 10.1093/nar/gkf393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulston M, Fulford J, Jenner T, de Lara C, O’Neill P. Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 2002;30:3464–3472. doi: 10.1093/nar/gkf467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song JM, Milligan JR, Sutherland BM. Bistranded oxidized purine damage clusters: induced in DNA by long-wavelength ultraviolet (290-400 nm) radiation? Biochemistry. 2002;41:8683–8688. doi: 10.1021/bi020137x. [DOI] [PubMed] [Google Scholar]
- 8.Ho ELY, Parent M, Satoh MS. Induction of base damages representing a high risk site for double strand break formation in genomic DNA by exposure of cells to DNA damaging agents. J. Biol. Chem. 2007;282:21913–21923. doi: 10.1074/jbc.M610651200. [DOI] [PubMed] [Google Scholar]
- 9.Blaisdell JO, Harrison L, Wallace SS. Base excision repair processing of radiation-induced clustered DNA lesions. Radiat. Prot. Dosimetry. 2001;97:25–31. doi: 10.1093/oxfordjournals.rpd.a006634. [DOI] [PubMed] [Google Scholar]
- 10.Georgakilas AG. Processing of DNA damage clusters in human cells: current status of knowledge. Mol. Biosyst. 2008;4:30–35. doi: 10.1039/b713178j. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhry MA, Weinfeld M. Reactivity of human apurinic/apyrimidinic endonuclease and Escherichia coli exonuclease III with bistranded abasic sites in DNA. J. Biol. Chem. 1997;272:15650–15655. doi: 10.1074/jbc.272.25.15650. [DOI] [PubMed] [Google Scholar]
- 12.Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 13.David-Cordonnier MH, Boiteux S, O’Neill P. Efficiency of excision of 8-oxo-guanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Biochemistry. 2001;40:11811–11818. doi: 10.1021/bi0112356. [DOI] [PubMed] [Google Scholar]
- 14.Budworth H, Dianova II, Podust VN, Dianov GL. Repair of clustered DNA lesions. Sequence-specific inhibition of long-patch base excision repair by 8-oxoguanine. J. Biol. Chem. 2002;277:21300–21305. doi: 10.1074/jbc.M201918200. [DOI] [PubMed] [Google Scholar]
- 15.David-Cordonnier MH, Cunniffe SM, Hickson ID, O’Neill P. Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry. 2002;41:634–642. doi: 10.1021/bi011682l. [DOI] [PubMed] [Google Scholar]
- 16.Lomax ME, Cunniffe S, O’Neill P. 8-OxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break, when present within a clustered DNA damage site. DNA Repair. 2004;3:289–299. doi: 10.1016/j.dnarep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Georgakilas AG, Bennett PV, Wilson DM, III, Sutherland BM. Processing of bistranded abasic DNA clusters in γ-irradiated human hematopoietic cells. Nucleic Acids Res. 2004;32:5609–5620. doi: 10.1093/nar/gkh871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eot-Houllier G, Eon-Marchais S, Gasparutto D, Sage E. Processing of a complex multiply damaged DNA site by human cell extracts and purified repair proteins. Nucleic Acids Res. 2005;33:260–271. doi: 10.1093/nar/gki165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eot-Houller G, Gonera M, Gasparutto D, Giustranti C, Sage E. Interplay between N-glycosylases/AP-lyases at multiply damaged sites and biological consequences. Nucleic Acids Res. 2007;35:3355–3366. doi: 10.1093/nar/gkm190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malyarchuk S, Youngblood R, Landry AM, Quillin E, Harrison L. The mutation frequency of 8-oxo-7,8-dihydroguanine (8-oxodG) situated in a multiply damaged site: comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Repair. 2003;2:695–705. doi: 10.1016/s1568-7864(03)00040-5. [DOI] [PubMed] [Google Scholar]
- 21.Malyarchuk S, Brame KL, Youngblood R, Shi R, Harrison L. Two clustered 8-oxo-7,8-dihydroguanine (8-oxodG) lesions increase the point mutation frequency of 8-oxodG, but do not result in double strand breaks or deletions in Escherichia coli. Nucleic Acids Res. 2004;32:5721–5731. doi: 10.1093/nar/gkh911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson CG, Shikazono N, Thacker J, O’Neill P. Enhanced mutagenic potential of 8-oxo-7,8-dihydroguanine when present within a clustered DNA damage site. Nucleic Acids Res. 2004;32:263–270. doi: 10.1093/nar/gkh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shikazono N, Pearson C, O’Neill P, Thacker J. The roles of specific glycosylases in determining the mutagenic consequences of clustered DNA base damage. Nucleic Acids Res. 2006;34:3722–3730. doi: 10.1093/nar/gkl503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaisdell JO, Wallace SS. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang N, Galick H, Wallace SS. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA Repair. 2004;3:1323–1334. doi: 10.1016/j.dnarep.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 26.Yang N, Chaudhry MA, Wallace SS. Base excision repair by hNTH1 and hOGG1: a two edged sword in the processing of DNA damage in γ-irradiated human cells. DNA Repair. 2006;5:43–51. doi: 10.1016/j.dnarep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 27.D'Souza DI, Harrison L. Repair of clustered uracil DNA damages in Escherichia coli. Nucleic Acids Res. 2003;31:4573–4581. doi: 10.1093/nar/gkg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison L, Brame KL, Geltz LE, Landry AM. Closely opposed apurinic/apyrimidinic sites are converted to double strand breaks in Escherichia coli even in the absence of exonuclease III, endonuclease IV, nucleotide excision repair and AP lyase cleavage. DNA Repair. 2006;5:324–335. doi: 10.1016/j.dnarep.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dianov GL, Timchenko TV, Sinitsina OI, Kuzminov AV, Medvedev OA, Salganik RI. Repair of uracil residues closely spaced on the opposite strands of plasmid DNA results in double-strand break and deletion formation. Mol. Gen. Genet. 1991;225:448–452. doi: 10.1007/BF00261686. [DOI] [PubMed] [Google Scholar]
- 30.Malyarchuk S, Harrison L. DNA repair of clustered uracils in HeLa cells. J. Mol. Biol. 2005;345:731–743. doi: 10.1016/j.jmb.2004.10.079. [DOI] [PubMed] [Google Scholar]
- 31.Malyarchuk S, Castore R, Harrison L. DNA repair of clustered lesions in mammalian cells: involvement of non-homologous end-joining. Nucleic Acids Res. 2008;36:4872–4882. doi: 10.1093/nar/gkn450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose MD, Winston F, Hieter P. A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. Methods in Yeast Genetics. [Google Scholar]
- 33.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 34.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 35.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomax ME, Cunniffe S, O’Neill P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry. 2004;43:11017–11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 37.Guillet M, Boiteux S. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 2002;21:2833–2841. doi: 10.1093/emboj/21.11.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Michan C, Monje-Casas F, Pueyo C. Transcript copy number of genes for DNA repair and translesion synthesis in yeast: contribution of transcription rate and mRNA stability to the steady-state level of each mRNA along with growth in glucose-fermentative medium. DNA Repair. 2005;4:469–478. doi: 10.1016/j.dnarep.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Cappelli E, Degan P, Frosina G. Comparative repair of the endogenous lesions 8-oxo-7, 8-dihydroguanine (8-oxoG), uracil and abasic site by mammalian cell extracts: 8-oxoG is poorly repaired by human cell extracts. Carcinogenesis. 2000;21:1135–1141. [PubMed] [Google Scholar]
- 41.Cappelli E, Hazra T, Hill JW, Slupphaug G, Bogliolo M, Frosina G. Rates of base excision repair are not solely dependent on levels of initiating enzymes. Carcinogenesis. 2001;22:387–393. doi: 10.1093/carcin/22.3.387. [DOI] [PubMed] [Google Scholar]
- 42.Johnson AW, Demple B. Yeast DNA 3′-repair diesterase is the major cellular apurinic/apyrimidic endonuclease: substrate specificity and kinetics. J. Biol. Chem. 1988;263:18017–18022. [PubMed] [Google Scholar]
- 43.You HJ, Swanson RL, Doetsch P. Saccharomyces cerevisiae possesses two functional homologues of Escherichia coli endonuclease III. Biochemistry. 1998;37:6033–6040. doi: 10.1021/bi973042h. [DOI] [PubMed] [Google Scholar]
- 44.Guibourt N, Castaing B, Auffret van der Kemp P, Boiteux S. Catalytic and DNA binding properties of the Ogg1 protein of Saccharomyces cerevisiae: comparison between the wild type and the K241R and K241Q active-site mutant proteins. Biochemistry. 2000;39:1716–1724. doi: 10.1021/bi992262n. [DOI] [PubMed] [Google Scholar]
- 45.David-Cordonnier MH, Cunniffe SM, Hickson ID, O’Neill P. Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry. 2002;41:634–642. doi: 10.1021/bi011682l. [DOI] [PubMed] [Google Scholar]
- 46.Hashimoto M, Imhoff B, Ali MM, Kow YW. HU protein of Escherichia coli has a role in the repair of closely opposed lesions in DNA. J. Biol. Chem. 2003;278:28501–28507. doi: 10.1074/jbc.M303970200. [DOI] [PubMed] [Google Scholar]
- 47.Castaing B, Zelwer C, Laval J, Boiteux S. HU protein of Escherichia coli binds specifically to DNA that contains single-strand breaks or gaps. J. Biol. Chem. 1995;270:10291–10296. doi: 10.1074/jbc.270.17.10291. [DOI] [PubMed] [Google Scholar]
- 48.Boubrik F, Rouviere-Yaniv J. Increased sensitivity to gamma irradiation in bacteria lacking protein HU. Proc. Natl Acad. Sci. USA. 1995;92:3958–3962. doi: 10.1073/pnas.92.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 50.Parsons JL, Dianova II, Allinson SL, Dianov GL. Poly(ADP-ribose) polymerase-1 protects excessive DNA strand breaks from deterioration during repair in human cell extracts. FEBS J. 2005;272:2012–2021. doi: 10.1111/j.1742-4658.2005.04628.x. [DOI] [PubMed] [Google Scholar]
- 51.Hashimoto M, Donald CD, Yannone SM, Chen DJ, Roy R, Kow YW. A possible role of Ku in mediating sequential repair of closely opposed lesions. J. Biol. Chem. 2001;276:12827–12831. doi: 10.1074/jbc.M010906200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.