Abstract
Translation termination in eukaryotes is completed by two interacting factors eRF1 and eRF3. In Saccharomyces cerevisiae, these proteins are encoded by the genes SUP45 and SUP35, respectively. The eRF1 protein interacts directly with the stop codon at the ribosomal A-site, whereas eRF3—a GTPase protein—probably acts as a proofreading factor, coupling stop codon recognition to polypeptide chain release. We performed random PCR mutagenesis of SUP45 and screened the library for mutations resulting in increased eRF1 activity. These mutations led to the identification of two new pockets in domain 1 (P1 and P2) involved in the regulation of eRF1 activity. Furthermore, we identified novel mutations located in domains 2 and 3, which confer stop codon specificity to eRF1. Our findings are consistent with the model of a closed-active conformation of eRF1 and shed light on two new functional regions of the protein.
INTRODUCTION
Translation termination occurs when one of the three stop codons, UAA, UGA and UAG, enters the ribosomal A-site. In eukaryotes, two release factors eRF1 and eRF3 mediate translation termination. Either full or partial X-ray structures are available for both proteins (1,2). In Saccharomyces cerevisiae, eRF1 is encoded by the gene SUP45. This protein recognizes all three stop codons through an extended surface including the NIKS motif in its N-terminal domain (3–5) and triggers peptidyl–tRNA hydrolysis by activating the peptidyl transferase center of the ribosome through the highly conserved GGQ motif in the central domain of eRF1. The C-terminal domain of eRF1 is involved in binding eRF3, which is essential for in vivo termination (6). The precise molecular mechanisms underlying this process remain unclear, but it is possible that the binding of eRF1 to eRF3 induces a conformational change in eRF3 to stabilize the binding of GTP to eRF3 (7). The S. cerevisiae eRF3 protein, encoded by SUP35 gene, is composed of two different regions. The N-terminal region (called NM) specific to S. cerevisiae is not necessary for termination activity but is involved in the formation of prion-like polymers known as [PSI+]. The C-terminal region is highly conserved and is involved both in GTP and eRF1 binding. eRF1 binding to eRF3 is required for GTP binding. The ternary complex eRF1:eRF3:GTP may bind the ribosomal A-site, but the binding of GTP to eRF3 prevents eRF1 from catalyzing termination. If a stop codon is located in the A-site, a conformational rearrangement would occur and stimulate eRF3 GTPase activity (4). GTP hydrolysis allows proper positioning of the eRF1 GGQ motif in the peptidyl transferase center to catalyze peptidyl–tRNA cleavage (8,9).
Many studies have investigated eRF1 specificity for stop codons. However, most of these studies screened for eRF1 mutants with reduced termination efficiency (3,5,10–14). We developed a new approach based on the use of a weak stop codon to determine the role of the various eRF1 domains. Such a stop codon is easily suppressed by natural tRNAs, allowing a high rate of stop codon readthrough in a wild-type genetic background (15,16). We screened a random PCR mutagenesis library of the SUP45 gene to identify hyperactive mutants, using a method previously developed in our laboratory (17) (Figure 1 and Figure S1). Our study demonstrates that all the domains of the protein are involved in regulating termination efficiency. Moreover, we identified novel mutations in domains 2 and 3, which specifically improve termination at the UAA and UAG codons but have no effect on the UGA codon.
Figure 1.
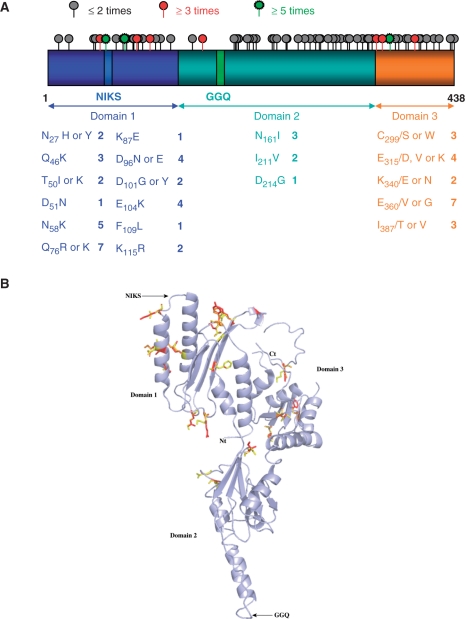
Distribution of the mutations in the SUP45 gene and eRF1 protein. (A) Schematic representation of the SUP45 gene divided into three functional domains. The mutations identified in this study are represented alongside the gene by a gray circle for positions at which mutations were found once or twice, a red circle for mutations found three to five times, and a green circle for mutations found at least five times. All mutants studied are shown below, with the number of times they were found. Mutations found only once are indicated when they are the only mutation identified in the gene. (B) Schematic 3D representation of the eRF1 protein using 1DT9 pdb file. Amino- and carboxy-terminal extremities and each domain are indicated. Functional motifs (GGQ and NIKS) are also shown. Mutated residues are shown in stick representation in yellow and mutations are displayed in red or orange.
MATERIALS AND METHODS
Yeast strains and media
The S. cerevisiae strains used in this study are listed in Table SI. The MT557/3b strain carries sup45–2 (SAL4) and shows a growth defect at a restrictive temperature (37°C). The modified FS1-TMV strain used for the screen was constructed as described in Supplementary Data. We generated an FS1 strain with deletion of the sup45 gene (FS1ΔS) as indicated in Supplementary Data. Strains were grown in minimal media supplemented with the appropriate amino acids for maintenance of the different plasmids after transformation. Yeast transformations were performed using the lithium acetate method (18).
Plasmids and molecular biology methods
Mutagenesis
Random mutagenesis was carried out by PCR on a pUC19 vector with the SUP45 gene cloned at the SmaI site as indicated in Supplementary Data.
Western blot
Protein samples were separated on SDS–polyacrylamide gels and electrophoretically transferred to nitrocellulose. Antibodies raised against eRF1 or actin (MAB1501R, clone C4, Chemicon) were used.
Quantification of readthrough efficiency
Clones transformed with pHS-Sup45 constructs were then transformed with a dual reporter plasmid to quantify termination efficiency at each stop codon. Luciferase and β-galactosidase activities were assayed in the same crude extract as previously described (19). All reported values represent the median of at least five independent measurements, with 20% of standard error. The significance of variation was evaluated using a non-parametric statistical test to calculate a P-value (Mann–Whitney).
Computational analysis of eRF1 structure
Structural pictures were visualized with Pymol from DeLano (http://www.pymol.org) using either the surface or cartoon visualization mode. Hydrogen bonds were modeled using the tool, ‘find polar contacts’. Electrostatic views were generated using APBS Pymol plugging within a range of −5 kT (red) to +5 kT (blue).
RESULTS
Screening for hyperactive eRF1 mutants
We used a screen based on the ability to monitor the termination efficiency of a stop codon cloned into the ADE2 gene by assessing the color of yeast colonies (Figure S1). ADE2 encodes the P-ribosyl-amino–imidazole carboxylase (EC 4.1.1.21), responsible for the degradation of the red pigment amino–imidazole ribotide. We previously show that more than 10% of readthrough at the ADE2 gene is sufficient to switch the color from red to white (17). The screen is performed in a strain carrying the wild-type SUP45 gene and a chromosomal copy of the ade2 gene interrupted by an in-frame UAG stop codon at a weak termination site promoting 30% of readthrough (see Material and methods section). We carried out three independent random PCR mutagenesis experiments with different conditions on the wild-type SUP45 gene. These PCR products were inserted into the pFL44L plasmid (URA3) by gap repair and transformed directly into the reporter strain. Transformants were plated onto minimal medium supplemented with CSM containing a minimum quantity of adenine to allow healthy growth with optimization of the red color.
After 5 days at 30°C, we selected 46 red-colored clones among a large number of white transformants. Plasmids were counter-selected on 5-FOA medium. All but one clone reversed the phenotype indicating they were antisuppressor sup45 mutants.
Identification of sup45 mutations
We sequenced the SUP45 coding sequence of the candidates, to identify mutations present in the gene. One hundred and thirty-one mutations were identified equally distributed along the gene (Figure 1). Interestingly, however, there were no mutations in the ‘GGQ’ domain. Mutations resulting in a change in the amino-acid composition of eRF1 are presented in Table SII. The average number of all mutations in each coding sequence was 2.9, including 30 silent mutations. We focused our study on candidates presenting either a low number of mutations or mutations identified several times during the screen (Table 1). Most mutations were found in domain 1 (1–140), which interacts directly with the stop codon. Among the 34 mutations found in domain 1, 20 modified the charge of the amino acid, replacing a negative or uncharged amino acid by a positively charged one (R, K or H). Among the five mutations found in domain 2 (141–275), four mutations (N261, I211, D214, I248) were at the interface with the domain 1. In the only X-ray structure of eRF1 currently available (PDB 1DT9), the structure of domain 3 (276–437) is incomplete; thus, we were unable to localize all mutations in the 3D structure (Figure 1). Taking into account mutations that were unique or found in at least two independent clones, 12 mutated positions in domain 1, three positions in domain 2 and five positions in domain 3 were found.
Table 1.
Mutations identified during the screening and further analyzed
| Sup45 allele | Amino-acid substitution | Nucleotide change | Comments |
|---|---|---|---|
| C11 | N58K | AAT⇨AAA | NIKS motif in the domain 1 |
| B1 | Q76R | CAA⇨CGA | P2 |
| C6 | Q76K | CAA⇨AAA | P2 |
| A8 | E104K | GAA⇨AAA | P1 |
| A2 | E360V | GAA⇨GTA | Only domain 3 |
| B3 | E360G | GAA⇨GGA | Only domain 3 |
| A4 | N27H, K331N | AAT⇨CAT, AAA⇨AAT | P1 |
| A5 | Q46K, K115R | CAA⇨AAA, AAA⇨AGA | P2 |
| A19 | T50K, N379Y | ACA⇨AAA, GTT⇨GTA | P2 |
| A13 | Q76K, K340E | CAA⇨AAA, AAA⇨GAA | P2 |
| C1 | D96E, E360G | GAT⇨GAA, GAA⇨GGA | P1 |
| A16 | K276E, E345V | AAG⇨GAG, GAA⇨GTA | Only domain 3 |
| C12 | L21I, N209K, E315V | ATA⇨TTA, AAT⇨AAA, GAA⇨GTA | Domains 2 + 3 |
| A18 | D96E, N161I, I211V | GAT⇨GAA, AAT⇨ATT, ATT⇨GTT | P1 |
| C15 | E104K, C299S, I326N | GAA⇨AAA, TGT⇨AGT, ATC⇨AAC | P1 |
| C3 | D149N, Y287S, I387V | GAC⇨AAC, TAT⇨TCT, ATC⇨GTC | Domains 2 + 3 |
| C2 | I248V, E360G, I387T | ATT⇨GTT, GAA⇨GGA, ATC⇨ACC | Only domain 3 |
The various types of nucleotide transition and transversion are indicated, together with the resulting amino-acid changes. Mutants are listed in order of the corresponding mutations, from N-terminus to C-terminus residues. The column ‘comments’ correspond to the annotation of the bold residue (if more than one mutation) indicated in the second column. The complete list of mutations identified during the screening can be found in Supplementary Table SII.
Hyperactive sup45 mutants can complement the deletion of wild-type SUP45
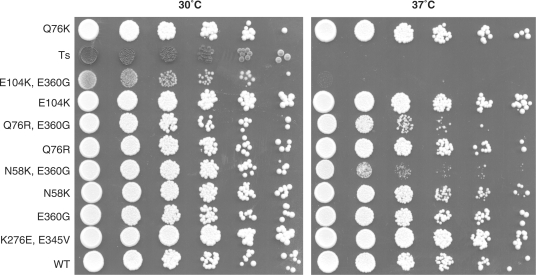
We carried out our genetic screen in the presence of the wild-type SUP45 gene. Translational elongation mutants with an elevated level of translational accuracy interfere with cell viability (20). Termination mutants with an elevated level of accuracy may thus severely affect yeast growth rate. To determine the potential effects of these mutant proteins on growth, we constructed a yeast strain deleted for SUP45 (FS1ΔS) rescued by a URA3 plasmid bearing wild-type SUP45. We selected a subset of mutants carrying only a single mutation, or a combination of these single mutations, for this analysis. The FS1ΔS strain was then transformed with the mutant constructs and the wild-type SUP45 gene was shuffled. Our results (Figure 2) clearly demonstrated that all the mutant cells were viable at 30°C. However, all three double mutants were thermosensitive.
Figure 2.
Hyperactive eRF1 mutants can complement the deletion of the SUP45 gene. The FS1ΔS strain was transformed with a plasmid carrying sup45 mutants and plated on selective media for this plasmid. Rescue plasmid carrying wild-type SUP45 gene was shuffled by plating the colonies on media containing 5FOA. The growing cells were transferred to a selective liquid culture until they reached 1 OD600 and were serially diluted. Plates were incubated either at 30 °C or 37 °C to test for thermosensitivity. Ts denotes the MT557/3b strain carrying a thermosensitive sup45 allele; E104K, E360G; Q76R, E360G; N58K, E360G; are double mutants created by directed mutagenesis, combining mutations identified independently during the screen within the same molecule.
Variation in the expression of sup45 mutants does not account for the phenotypes observed
A change in the nature of an amino acid can modify the activity or the stability of a protein. It has been previously reported that a depletion in the level of eRF1 causes a reduction in termination efficiency, indicating that the amount of eRF1 is important for ensuring efficient termination in yeast (21). In our study, an increase of eRF1 stability would result in a higher concentration of protein available for termination in the cytoplasm. This could explain the anti-suppressor phenotype observed.
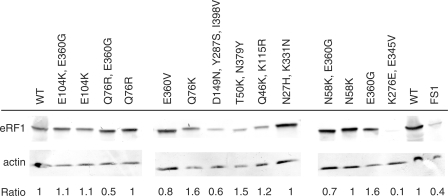
We quantified the levels of the 14 most interesting mutant proteins in the FS1ΔS strain deleted for the chromosomal copy of SUP45, by western blot. All the mutants but A16 were equally stable (Figure 3). Both FS1 and A16 strains display a significant variation of the eRF1 level. This was expected for the FS1 strain carrying only the chromosomal copy of the SUP45 gene. It is worth to note that increase in eRF1 level due to its overexpression (column WT in Figure 3) does not affect stop codon readthrough as previously published (22). However, results for the mutant A16 (K276E, E345V) are puzzling. Despite the low level of eRF1, the mutant is viable and does not show any obvious phenotype. One would expect an increase of stop codon readthrough in this strain due to the low level of eRF1, but the opposite occurs with a 2-fold reduction readthrough efficiency. This suggests that the mutant is very active and the effect on termination is underestimated due to the low level of eRF1 in the cell.
Figure 3.
Analysis of the expression of eRF1 variant alleles. Yeast cells carrying the various eRF1 alleles were grown to mid log phase. Equivalent amounts of proteins were incubated with antibodies directed against either eRF1 or actin. Ratio of eRF1 to actin proteins is indicated for each mutant. WT denotes the FS1ΔS strain carrying the SUP45wt gene. FS1 denotes the WT strain without the deletion of the SUP45 gene. All the mutants are named by their mutations. The ratios are equivalent for all the mutants, but K276E, E345V which is 10-fold lower than in the wild-type. Despite this low level of eRF1 this mutant still displays a strong termination phenotype.
Sup45 mutants display significantly increased translation termination efficiency
To determine whether translation termination rate was actually increased in these mutants, we quantified stop codon readthrough in the FS1ΔS strain using the pAC reporter plasmids bearing a leaky UAA, UAG or UGA stop codon at the lacZ-luc junction (19,23). A stronger termination efficiency can be observed by a decrease of stop codon readthrough efficiency. The readthrough level was quantified and compared to that obtained in the presence of the pFL44L vector bearing the SUP45 wild-type gene. We measured the effect of 17 mutants on translation termination fidelity using the three different stop codons (Table 2). These results show that termination efficiency was significantly higher for all the mutants. The readthrough levels for UAA, UAG and UGA codons in the presence of the SUP45 WT gene were 16%, 34% and 22%, respectively. These values are consistent with previously published values (17,24). The two clones A8 and C11 carry mutations in domain 1 at positions 104 and 58, respectively. C11 carries a change at the first amino acid of the stop codon recognition sequence NIKS (N58K). These mutated sequences were able to confer a substantial increase in translational termination efficiency at all three stop codons; the rate of stop codon readthrough was between three and six times lower than that of wild-type Sup45p. The mutant C15 carries the same mutation as A8, together with mutations C299S and I326N. These two mutants had approximately the same effect, suggesting that the mutation E104K was responsible for the observed phenotype. Both B1 and C6 had mutations at position 76, replacing Gln by Arg or Lys, respectively. Stop codon readthrough was four times lower in B1, and six to eight times lower in C6 than in wild-type (Table 2). These two mutants carry a positively charged residue rather than the uncharged Gln. In comparison, the presence of both the Q76K and K340E mutations in the A13 clone had a weaker effect than the single mutation Q76K alone (C6) (Table 2). These data suggest that Q76K alone contributes to improved termination efficiency. The two other single mutants tested were A2 and B3, bearing E360V and E360G mutations, respectively. A strong effect was observed for UAA and UGA in A2, their rates of readthrough being five times lower than in wild-type. Similar patterns of anti-suppression were observed for all three stop codons in B3, with a 3-fold reduction in readthrough. This mutation was found in mutants C1 and C2, both of which also carried additional mutations. Unlike the mutant with only a single mutation, these two mutants did not display the same behavior for each of the different stop codons (Table 2). These different effects may thus be a result of the additional mutations. A 2–3-fold reduction in readthrough efficiency was observed for the mutant A4, which carries mutations in both domain 1 (N27H) and domain 3 (K331N). Among mutants with more significant reductions, C3 had a readthrough efficiency four to six times lower than that of wild-type (Table 2). This mutant carried three different mutations, one in domain 1 and two in domain 3. A cooperative effect of these three mutations may underlie the strong effect observed. Alternatively, given that we found the mutation I387V independently in three other mutants, this mutation could be the main determinant of hyperactivity in this mutant. A19 showed the largest reduction in readthrough efficiency in our screen. Readthrough of UAA and UGA codons was 10 and 14 times lower, respectively, in this mutant than in wild-type. This mutant had mutations at two positions; but only one (position 50) was identifed in another mutant (A9, Table SII). A16 and C12, both carrying mutations in domains 3 and 2, were of particular interest because they displayed unusual stop codon specificity. Both mutants had increased termination efficiency at UAA and UAG, but not at UGA.
Table 2.
Quantification of the effect of eRF1 mutants on the efficiency of stop codon readthrough
| UAA |
UAG |
UGA |
|||||
|---|---|---|---|---|---|---|---|
| Readthrough efficiency (%) | Decrease fold | Readthrough efficiency (%) | Decrease fold | Readthrough efficiency (%) | Decrease fold | ||
| WT | 16.3 | 34.3 | 22.4 | ||||
| C11 | N58K | 2.8 | ×6 | 7.9 | ×4 | 4.7 | ×5 |
| B1 | Q76R | 3.7 | ×4 | 9.5 | ×4 | 5.9 | ×4 |
| C6 | Q76K | 2.4 | ×7 | 5.8 | ×6 | 2.8 | ×8 |
| A8 | E104K | 3.6 | ×5 | 10.4 | ×3 | 3.9 | ×6 |
| A2 | E360V | 3.6 | ×5 | 15.9 | ×2 | 4.8 | ×5 |
| B3 | E360G | 4.8 | ×3 | 11.9 | ×3 | 6.9 | ×3 |
| A4 | N27H, K331N | 5.1 | ×3 | 12.1 | ×3 | 13.5 | ×2 |
| A5 | Q46K, K115R | 6.8 | ×2 | 12.1 | ×3 | 3.4 | ×7 |
| A19 | T50K, N379Y | 1.7 | ×10 | 7 | ×5 | 1.6 | ×14 |
| A13 | Q76K, K340E | 4.6 | ×4 | 11.7 | ×3 | 4.8 | ×5 |
| C1 | D96E, E360G | 3 | ×5 | 8.1 | ×4 | 5.7 | ×4 |
| A16 | K276E, E345V | 6.9 | ×2 | 20.1 | ×2 | 20.2 | ×1 |
| C12 | L21I, N209K, E315V | 11.1 | ×1.5 | 13.5 | ×3 | 17.5 | ×1 |
| A18 | D96E, N161I, I211V | 6.5 | ×2 | 13.9 | ×2 | 3.1 | ×7 |
| C15 | E104K, C299S, I326N | 4.3 | ×4 | 7.2 | ×5 | 3.2 | ×7 |
| C3 | D149N, Y287S, I387V | 2.6 | ×6 | 9 | ×4 | 3.7 | ×6 |
| C2 | I248V, E360G, I387T | 5.7 | ×3 | 11.5 | ×3 | 4.5 | ×5 |
Stop codon readthrough efficiency was quantified as described in the Material and Methods section. The values indicated are the median of at least five independent experiments. The fold decrease of readthrough efficiency relative to the value measured with the wild-type SUP45 gene is indicated.
Mutations found in domain 1 can be divided into two main groups, the pocket 1 (P1) and the pocket 2 (P2) (Table 1). To strengthen our data regarding the importance of these pockets we constructed by direct mutagenesis two single mutations (D96E in P1; T50K in P2) originally identified in double mutants. Results presented in Table 3 indicate that both single mutants increase termination at all stop codons. This clearly confirms our finding that P1 and P2 play an essential role in eRF1 function. However, the effects of the single mutants are weaker than the one observed in the initial double mutants. This clearly indicated that the second mutation also affects termination. This was already known for the E360G mutation found together with D96E, as this mutant is also identified alone, but not for N379Y (identified with T50K).
Table 3.
Values for single mutants constructed by point mutagenesis and compared to multiple mutants found during the screening
| UAA |
UAG |
UGA |
|||||
|---|---|---|---|---|---|---|---|
| Readthrough efficiency (%) | Fold WT | Readthrough efficiency (%) | Fold WT | Readthrough efficiency (%) | Fold WT | ||
| D96E | 7.3 | ×2 | 10.7 | ×2 | 12.3 | ×2 | |
| A2 | E360G | 4.8 | ×3 | 11.9 | ×3 | 6.9 | ×3 |
| A8 | E104K | 3.6 | ×5 | 10.4 | ×3 | 3.9 | ×6 |
| EE/KG | E104K, E360G | 3.2 | ×5 | 12.9 | ×3 | 5 | ×4 |
| B1 | Q76R | 3.7 | ×4 | 9.5 | ×4 | 5.9 | ×4 |
| QE/RG | Q76R, E360G | 2.4 | ×7 | 9.2 | ×4 | 2.2 | ×10 |
| T50K | 2.9 | ×6 | 5.3 | ×6 | 3.8 | ×6 | |
| C11 | N58K | 2.8 | ×6 | 7.9 | ×4 | 4.7 | ×5 |
| NE/KG | N58K, E360G | 2.2 | ×7 | 5.6 | ×6 | 2.7 | ×8 |
Same legend as for Table 2. The name of the allele is indicated in the left column for the mutants identified during the screening, and for the double mutants.
It remains to be determined whether mutations in the different domains increase termination efficiency through different mechanisms. If this was the case, these mutants would be expected to display increased efficiency at different stages of termination; such mutations may then have a cumulative effect. We constructed a set of double mutants all containing the mutation E360G in domain 3 (B3) with each of three mutations identified in different regions of domain 1, E104K, Q76R or N58K. These new mutants were called sup45-EE/KG, sup45-QE/RG and sup45-NE/KG, accordingly. The effect observed for sup45-EE/KG did not differ from that observed for the two single mutants (Table 3). The mutant sup45-QE/RG displayed a cumulative effect on termination at the UAA codon (7-fold reduction in readthrough) and a multiplicative effect at UGA (10-fold), whereas no effect was observed for the UAG codon. Cumulative effects on both UAG (6-fold) and UGA (8-fold) codons, but not on the UAA codon, were observed for the mutant sup45-NE/KG (Table 3).
Mutations are present in highly conserved domains
The eRF1 family is highly conserved; however, subtle changes are found throughout the proteins and several of the mutations affect highly conserved residues (Figure S2). An asparagine at position 27 in domain 1 is present in all sequences with the exception of Tetrahymena thermophila (yeast numbering). This Asn27 is at 6 Å from Asn126 in the adjacent Tyr122-Phe128 loop, which is essential for stop codon discrimination (11) (Figure S3A). Moreover, Asn27, together with residues Ser30-Asp96-Ile98-Gly102-Glu104, form a pocket 1 (P1) that lies at the top of domain 1 (Figure S3B). Replacing this Asn by either a His or Tyr would significantly affect the structure of the loop and create a deeper pocket. Moreover the importance of the Asn27 loop is reinforced by the fact that GTS residues immediately downstream from Asn27 are conserved in all eRF1 proteins (Figure 1). Position 51 is occupied by negatively charged residues only and is located in the external side of the helix 2 (Figure S3A). This part of the protein is thought to be in close interaction with both the ribosomal RNA and mRNA, the replacement of this polar amino acid by either a positively charged or aromatic amino acid is likely to strengthen the interactions between eRF1 and the ribosome. The Asn58 lies within the NIKSR loop, which contacts the uridine of the stop codon, and is crucial for eRF1 activity (25) (Figure S3A). Modification of this motif affects stop codon recognition (5,13). To our knowledge, substitution of Asn58 by a lysine has not been previously reported. It is possible that the interaction of the negatively charged mRNA with the positively charged lysine residue will be stronger than with the Asn residue. The residence time of eRF1 in the A-site would then be extended and termination significantly increased. Positions 101 and 104 are mostly occupied by acidic residues in the different species. These amino acids are localized at the surface of the protein. Notably, Asp101 lies close to the P1 domain. We identified mutations at each of the three residues of P1 (Asn27, Asp96 and Glu104), highlighting the importance of this domain in the translation termination mechanism.
All three mutations identified in domain 2 (N161I, I211V and D214G) affect highly conserved positions. Alignment of 14 wild-type eRF1 sequences revealed variation at these positions in only one sequence in each case (Figure S2). I211 and D214 are highly conserved, despite lying in the most variable region of domain 2.
Among the five mutations identified in domain 3, two were at highly conserved positions. The Val, Lys or Asp residues replacing Glu at position 315, which is conserved across all the species considered, have not been found to occur naturally at this location. Furthermore, we identified mutations in seven different clones at position 360, replacing Glu by neutral amino acids Val or Gly, despite this position being occupied by a negatively charged amino acid in all wild-type sequences. Notably, both positions 315 and 360 lie in the most variable region domain 3. This variability may indicate that this domain has co-evolved with variable regions of eRF3, which binds eRF1 specifically at this domain.
DISCUSSION
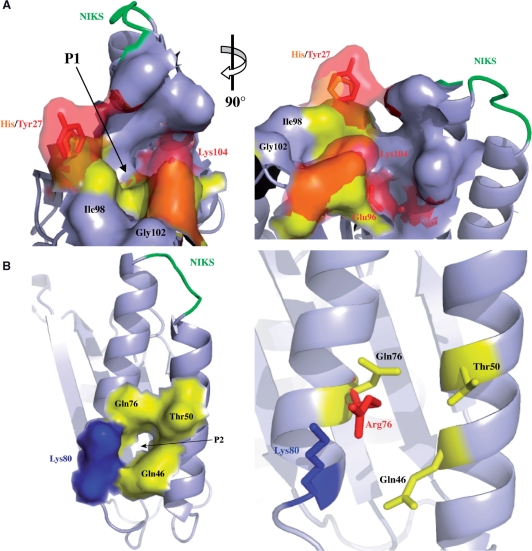
Both release factors eRF1 and eRF3 are key components of the termination process. The eRF1 protein can be divided into three functional domains. Domain 1 binds the stop codon directly in the ribosomal A-site. Domain 2 interacts with the peptidyl transferase center leading to the hydrolysis of the last peptidyl-tRNA bond through the GGQ motif (10,26). Little structural data is available for domain 3, however, in S. pombe, the last 11 residues are necessary for the binding of eRF3 (6,27–29). We developed an original approach to identify new mutants displaying an anti-suppressor phenotype in a wild-type genetic background by taking advantage of the weak termination efficiency of stop codons present in readthrough sequences. Using this screen, we identified 131 mutations distributed throughout the SUP45 gene. All single mutants were viable at either 30°C or 37°C in a strain carrying deletion of SUP45 and did not display any obvious phenotype. Thus, the increase in termination efficiency had no deleterious effect on yeast viability. However, these mutants were not viable in the absence of eRF3, demonstrating that these mutations do not compensate for loss of eRF3 by directly activating eRF1 (data not shown). We demonstrated most of the mutants had an equal amount of eRF1 protein and showed a substantial increase in termination efficiency, demonstrating a direct effect on termination mechanisms. Moreover, we showed that the combination of single mutations, identified separately in domains 1 and 3, affects termination efficiency differently, depending on the mutations and stop codon involved. These findings suggest that mutations in the various domains modify termination efficiency through their effects on different mechanisms. We were able to localize these mutations on the 3D structure of the human eRF1 using Pymol software (see Materials and methods section). All mutations found in domains 1 and 2 appear to modify the surface of the protein, whereas the mutations found in domain 3 have negligible effects on the surface of the protein (Figure S4). Mutations located in domain 1 are subdivided into three distinct groups (Figure 4). The first group (affecting Asn27, Asp96 and Glu104) constitutes a pocket 1 (P1) at the top of this domain (Figure 4A), lying very close to the regions directly involved in the stop codon recognition. A ‘pocket model’ has been proposed for stop codon recognition by eRF1 (4,11). An important feature of this model is the H-bond formed between residues Glu55 and Tyr122, which would maintain the two remote regions of domain 1 (the two NIKS domain-containing helices, and the beta-strand on the opposite side, represented in dark blue in Figure S3) in close proximity to each other. The new region P1 is continuous with this functional area. Both these regions could together accommodate the mRNA for proper orientation of eRF1 in the A-site. As observed in Figure 4, the introduction of either His or Tyr at position 27 should exert a similar structural bulk effect on the protein surface. These mutations would deepen pocket 1 and probably strengthen interactions with the helix domain.
Figure 4.
Structural model of mutations identified in eRF1. (A) The left panel shows P1 in the same orientation as shown in Supplementary Figure S3B. Wild-type amino acids are displayed in yellow, whereas mutated residues are shown in red and orange. The changes induced by these mutations at the surface of the protein are shown in transparency surface mode. The right panel is rotated as indicated to give a more detailed view. (B) The left panel is a surface representation of P2 with labeling of the four residues creating the pocket 2. The three residues found mutated are shown in yellow; the residue with no associated mutation is shown in blue. The right panel is a magnification of the image showing P2 with the same orientation. The residue Arg76 found in our screen is shown in red.
The residues affected by group 2 mutations (Gln46, Thr50, Gln76), together with Lys80, define a second pocket (P2) (Figure 4B). A direct role of P2 in stop codon recognition is unlikely as the residues are located too far away from the eRF1 mRNA-binding site. However, we found mutations involving three of four residues constituting P2. In particular, A19 (T50K, N379Y), for which we observed the strongest effect on readthrough, carried a mutation at position T50 of P2. Moreover mutations T50K and Q76K alone induced a 6-fold and 6- to 8-fold reduction in readthrough respectively, which are the strongest effects observed for single mutations. This highlights the crucial role of this region in termination efficiency (Figure 4B). It is worth noting that an arginine residue at position 76 creates a steric clash with Lys80 (Figure 4B), probably inducing a structural change of this part of domain 1. Furthermore, all the mutations found in P2 modify the charge of the residues by adding a positive charge (Q46K, T50K, Q76R/K). This strongly suggests that the electrostatics charges of this region are important for the observed improvement in eRF1 activity. When eRF1 is in the A-site, it probably interacts with the rRNA (30–32). We proposed that this electropositive side of domain 1 (Figure S5) interacts with the rRNA and that the P2 region plays a central role in these interactions. It would then follow that hyperactive mutations in P2 increase the residence time of eRF1 in the A-site and thus promote termination, whereas mutations in P1 probably affect the binding or recognition of the stop codon itself.
Group 3 comprises two mutations located at the bottom of domain 1 opposite the top part of the domain 2. These two mutations (Lys115 and Lys87) both point in the direction of domain 2 (Figure S2B). However, the crystal structure reveals these residues to be too far away from other amino acids to form a hydrogen bond. This crystal structure of eRF1 is unlikely to represent the active structure of eRF1 bound to ribosome (2). Indeed, the NIKS and GGQ domains are separated by 100 Å in the crystal structure, whereas the distance between the codon in the A-site and the peptidyl center is around 73 Å. A conformational change has been modeled to create a closed form, which fits the range needed for functional binding of eRF1 within the ribosome (33). The closed conformation of eRF1 involves motion of domain 2 relative to other domains. This conformational reorganization would, for example, bring the Asp141 to within approximately 4 Å of the Lys115, whereas these residues are separated by 13 Å in the open conformation (Figure S3C). However, due to approximations in the modeled eRF1-closed conformation, these observations should be carefully interpreted. The distance between Lys and Arg in this new model is in the range allowing formation of hydrogen bonds. We propose that H-bond formation between the mutated residues and the hinge region (or the surface of domain 2) would stabilize the closed conformation, locking eRF1 in its active conformation. This would explain the increased translation termination efficiency observed for the single mutants. Similarly, the three mutations identified in domain 2 were all located on the same side of this domain, opposite domain 1. Nussinov and colleagues (33) also showed that neutralization of the electrostatic repulsions between domains 1 and 2 by histidine protonation is required for the closed conformation. Two of the three mutations change a charged residue to a neutral residue. These substitutions may therefore help to lower the repulsion between domains 1 and 2, promoting the transition from the inactive open conformation to the active closed conformation.
As mentioned above, structural analysis of the mutations in domain 3 is difficult because the available structure for domain 3 is incomplete. Indeed, mutations E360V/G, found in seven independent mutants, are located in one of the missing regions. All the remaining mutations do not appear to affect the surface of the protein, but probably affect the general conformation of the domain. For example, the replacement of a Cys by a Trp residue at position 299 creates a steric clash with helix 9. The resulting movement of either helix 9, or the Trp residue is likely to affect the overall structure of domain 3. Domain 3 is the binding site of eRF3; thus, mutations in this domain could promote termination by increasing the likelihood of eRF3 binding to eRF1. Alternatively, these mutations may allow stronger binding of eRF1 to the ribosome by altering the overall shape of the release factor. The two variant alleles expressed in A16 and C12 significantly increased termination efficiency at UAA and UAG codons but had no effect on the UGA codon. We identified several point mutations in these two mutants—K276E and E345V and L21I, N209K and E315V—respectively. Both mutations in A16 were located in domain 3. We found mutations at Glu315 in four different mutants. This mutation is therefore likely to be relevant for termination. However, we did not identify any single mutants with a mutation at position 315 only; so the effects on termination efficiency observed may have been due to the presence of several mutations. To the best of our knowledge, this is the first report of eRF1 mutations located outside domain 1 affecting the specificity of stop codon recognition. Biochemical analysis of these mutants will help to determine which step of the termination process is affected.
The comparison of the potential functional consequences between these various mutations is of particular interest. We created a series of double mutants combining mutations identified in three different areas of domain 1 with the most frequent mutation found in domain 3 (E360G). The first double mutant EE/KG, carrying both the E104K (P1) and E360G mutations, did not display any synergistic effect between the two mutations on termination efficiency at any of the stop codons. Thus, these two mutations probably affect a common step in termination. The second double mutant (sup45-QE/RG) carried the Q76R (from B1, P2) mutation associated with E360G (from B3) and demonstrated an additive effect of these two mutations compared to the single mutations. However, the effect observed on termination at the UAG codon was equivalent to that observed in the mutant with Q76R alone. Notably, the mutant T50K N379Y, showing the strongest effect observed for termination efficiency, showed a much less marked effect on the UAG stop codon than on the two other stop codons. Thus, different regions of eRF1 seem to be involved in termination, depending on the stop codon present in the A site. The third mutant (sup45-NE/KG) carried the N58K mutation (from C11) with E360G. This mutant showed a cumulative effect for both UAG and UGA stop codons. This suggests that mutations in domains 1 and 3 act on termination through different mechanisms. These findings provide support for the three mutations in domain 1 affecting different steps of the termination.
We exploited the use of a stop codon at a weak termination site to search for release factor 1 mutants with a greater activity than wild-type. These mutant proteins inevitably adopt a functional conformation, so their effects cannot be explained by prevention of correct folding of the protein. We identified and characterized two previously unknown regions involved in termination. This powerful approach can be easily extended to study proteins involved in other translational mechanisms such as frameshifting.
Our findings strengthen the previous study suggesting that eRF3 may act as a proofreading factor by coupling stop codon recognition by eRF1 to efficient polypeptide chain release (8). Moreover, our study suggests that the termination process is a multistep reaction. Understanding how these mutations affect the interactions between ribosome and eRF1 will be a major step in further characterization of the termination process in eukaryotes. This will help to design drugs that prevent eRF1 from acting on premature stop codons, allowing the re-expression of genes inactivated by a nonsense mutation (34). Atomic resolution structures have been solved for prokaryotic factors; it is hoped that similar structures of eRF1 in a complex with the ribosome will be solved in the future and will help to further elucidate the molecular mechanisms underlying termination.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by grants from the Agence Nationale pour la Recherche (grant ANR-06-BLAN-0391-01 to J.P.R.), the Association pour la Recherche sur le Cancer (grant 3849 to J.P.R.). ON was funded by a fellowship from the Association pour la Recherche sur le Cancer during part of this study. Funding for open access charge: ANR-06-BLAN-0391-01.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to acknowledge Ruth Nussinov and Buyon Ma for providing the PDB file of the closed-eRF1 simulated conformation. The authors also want to thank Valerie Heurgue for the kind gift of the eRF1 antibody. We are very grateful to M. Tuite for providing us with the MT557/3b strain carrying the thermosensitive SUP45 allele. English usage was corrected by Alex Edelman & Associates.
REFERENCES
- 1.Kong C, Ito K, Walsh MA, Wada M, Liu Y, Kumar S, Barford D, Nakamura Y, Song H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 2.Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1 – mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 3.Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, Thompson SR, Pestova TV, Bedwell DM. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol. Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seit-Nebi A, Frolova L, Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eurwilaichitr L, Graves FM, Stansfield I, Tuite MF. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 1999;32:485–496. doi: 10.1046/j.1365-2958.1999.01346.x. [DOI] [PubMed] [Google Scholar]
- 7.Pisareva VP, Pisarev AV, Hellen CU, Rodnina MV, Pestova TV. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J. Biol. Chem. 2006;281:40224–40235. doi: 10.1074/jbc.M607461200. [DOI] [PubMed] [Google Scholar]
- 8.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 10.Seit-Nebi A, Frolova L, Justesen J, Kisselev L. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 2001;29:3982–3987. doi: 10.1093/nar/29.19.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolosov P, Frolova L, Seit-Nebi A, Dubovaya V, Kononenko A, Oparina N, Justesen J, Efimov A, Kisselev L. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito K, Frolova L, Seit-Nebi A, Karamyshev A, Kisselev L, Nakamura Y. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc. Natl Acad. Sci. USA. 2002;99:8494–8499. doi: 10.1073/pnas.142690099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frolova L, Seit-Nebi A, Kisselev L. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002;8:129–136. doi: 10.1017/s1355838202013262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavatte L, Kervestin S, Favre A, Jean-Jean O. Stop codon selection in eukaryotic translation termination: comparison of the discriminating potential between human and ciliate eRF1s. EMBO J. 2003;22:1644–1653. doi: 10.1093/emboj/cdg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buckingham RH. Codon context and protein synthesis: enhancements of the genetic code. Biochimie. 1994;76:351–354. doi: 10.1016/0300-9084(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 16.Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29:4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Namy O, Hatin I, Rousset JP. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001;2:787–793. doi: 10.1093/embo-reports/kve176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stahl G, Bidou L, Rousset JP, Cassan M. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruusala T, Andersson D, Ehrenberg M, Kurland CG. Hyper-accurate ribosomes inhibit growth. EMBO J. 1984;3:2575–2580. doi: 10.1002/j.1460-2075.1984.tb02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stansfield I, Eurwilaichitr L, Akhmaloka, Tuite MF. Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol. Microbiol. 1996;20:1135–1143. doi: 10.1111/j.1365-2958.1996.tb02634.x. [DOI] [PubMed] [Google Scholar]
- 22.Hatin I, Fabret C, Namy O, Decatur WA, Rousset JP. Fine-tuning of translation termination efficiency in Saccharomyces cerevisiae involves two factors in close proximity to the exit tunnel of the ribosome. Genetics. 2007;177:1527–1537. doi: 10.1534/genetics.107.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Namy O, Hatin I, Stahl G, Liu H, Barnay S, Bidou L, Rousset JP. Gene overexpression as a tool for identifying new trans-acting factors involved in translation termination in Saccharomyces cerevisiae. Genetics. 2002;161:585–594. doi: 10.1093/genetics/161.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bidou L, Stahl G, Hatin I, Namy O, Rousset JP, Farabaugh PJ. Nonsense-mediated decay mutants do not affect programmed -1 frameshifting. RNA. 2000;6:952–961. doi: 10.1017/s1355838200000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chavatte L, Seit-Nebi A, Dubovaya V, Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito K, Ebihara K, Nakamura Y. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998;4:958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frolova LY, Merkulova TI, Kisselev LL. Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA. 2000;6:381–390. doi: 10.1017/s135583820099143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merkulova TI, Frolova LY, Lazar M, Camonis J, Kisselev LL. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 1999;443:41–47. doi: 10.1016/s0014-5793(98)01669-x. [DOI] [PubMed] [Google Scholar]
- 30.Liu R, Liebman SW. A translational fidelity mutation in the universally conserved sarcin/ricin domain of 25S yeast ribosomal RNA. RNA. 1996;2:254–263. [PMC free article] [PubMed] [Google Scholar]
- 31.Velichutina IV, Dresios J, Hong JY, Li C, Mankin A, Synetos D, Liebman SW. Mutations in helix 27 of the yeast Saccharomyces cerevisiae 18S rRNA affect the function of the decoding center of the ribosome. RNA. 2000;6:1174–1184. doi: 10.1017/s1355838200000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velichutina IV, Hong JY, Mesecar AD, Chernoff YO, Liebman SW. Genetic interaction between yeast Saccharomyces cerevisiae release factors and the decoding region of 18 S rRNA. J. Mol. Biol. 2001;305:715–727. doi: 10.1006/jmbi.2000.4329. [DOI] [PubMed] [Google Scholar]
- 33.Ma B, Nussinov R. Release factors eRF1 and RF2: a universal mechanism controls the large conformational changes. J. Biol. Chem. 2004;279:53875–53885. doi: 10.1074/jbc.M407412200. [DOI] [PubMed] [Google Scholar]
- 34.Keeling KM, Bedwell DM. Clinically relevant aminoglycosides can suppress disease-associated premature stop mutations in the IDUA and P53 cDNAs in a mammalian translation system. J. Mol. Med. 2002;80:367–376. doi: 10.1007/s00109-001-0317-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.