Abstract
Centromeres play a pivotal role in the life of a eukaryote cell, perform an essential and conserved function, but this has not led to a standard centromere structure. It remains currently unclear, how the centromeric function is achieved by widely differing structures. Since centromeres are often large and consist mainly of repetitive sequences they have only been analyzed in great detail in a handful of organisms. The genome of Dictyostelium discoideum, a valuable model organism, was described a few years ago but its centromere organization remained largely unclear. Using available sequence information we reconstructed the putative centromere organization in three of the six chromosomes of D. discoideum. They mainly consist of one type of transposons that is confined to centromeric regions. Centromeres are dynamic due to transposon integration, but an optimal centromere size seems to exist in D. discoideum. One centromere probably has expanded recently, whereas another underwent major rearrangements.
In addition to insights into the centromere organization and dynamics of a protist eukaryote, this work also provides a starting point for the analysis of the evolution of centromere structures in social amoebas by comparative genomics.
INTRODUCTION
Genomes of all organisms consist of chromosomes that are duplicated and split between daughter cells. Bacterial genomes are relatively simple structures containing one, at most two chromosomes and a number of plasmids. In most cases these genomic elements are circular with one origin of replication but linear chromosomes also exist (1). In contrast, eukaryote genomes are more structured and diverse: they consist of a variable number of chromosomes ranging from approximately 100 kb to many Mb. They are always linear and therefore must contain essential elements to enable maintenance and proper propagation to the progeny. The chromosome ends must be capped by telomeres, which protect the core chromosome from degradation and are responsible for the length maintenance during each replication cycle (2). Centromeres are required as attachment points for the spindle apparatus to attach during cell division so that the chromosomes can be exactly divided up between daughter cells.
Surprisingly, neither centromeres nor telomeres have a common organizational principal in all eukaryotes. Though most eukaryotic telomeres make use of short sequences (TTAGGG and variations thereof), which are synthesized at the ends of the chromosomes by a telomerase to form chromosome ends, others do not. In some insect groups, e.g. Drosophila, special transposon species are dedicated to form the chromosome ends and ensure sequential elongation of the chromosomes by transposition. Therefore, a commonly used principle for chromosome end maintenance is not essential, only the sequential elongation of telomeres is critical.
Centromeres seem to be even more variable than telomeres. Saccharomyces cerevisiae chromosomes contain a well-defined sequence motif of 125 bases, which is the only requirement for proper centromere function in this species (3). However, in most organisms centromeres are not defined by a certain sequence motif. For example, human centromeres are comprised of so-called alpha satellites, where no specific motif mediates the function of a centromere but rather a certain repetitiveness and size is required (4). In extreme cases like Caenorhabditis elegans, the centromere function cannot be assigned to a certain region of the chromosome, so no centromeres exist and chromosomes are holocentric in this organism (5). Other organisms like maize have centromeres containing transposons (6).
Dictyostelium discoideum belongs to the group of social amoebas in the evolutionary branch of Amoebozoa (7). It is a long-standing valuable model organism to study cell signaling, cytoskeleton and development, however its centromere structure is not well understood (8). Its genome is relatively small with 34 Mb (9) but contains a considerable fraction of repetitive elements including usual and unusual LTR and nonLTR transposons, DNA transposons, and not classified repetitive elements (10). Some of these transposon classes have a remarkable insertion preference: while tRNA gene-targeted retroelement (TRE) target exclusively the 3′- or 5′-end of tRNA genes, the Dictyostelium Intermediate Repeat Sequence (DIRS) elements are found preferentially to be integrated in another DIRS element although no specific target sequence could be defined so far making this insertion preference to an unresolved question.
The genome is divided into six chromosomes, all of which harbor large repetitive regions at one tip. DIRS LTR transposons (11) are restricted almost exclusively to these regions. The only DIRS element not located in such a region is associated with a large duplication in chromosome 2 and thought to be the remainder of an unsuccessful attempt to establish a new centromere (9). The only experimental evidence that these regions perform centromeric functions is derived from the observation that these chromosome ends cluster in interphase and metaphase cells. Further striking evidence is that the DIRS repeat regions are the only obvious common feature of all chromosomes and are confined to one chromosomal area each. Furthermore, as in other organisms (12) these regions produce small RNAs, which might be involved in the regulation of centromere functions (13). In the following we will refer to these regions as centromere regions, albeit their function is not yet proven. Before this study our knowledge of the centromere structure in D. discoideum was scarce. Reconstruction of the centromere region from chromosome 1 was possible by using reads from the chromosome 1 enriched library together with the read pair information and unique sites in and at the borders of the repetitive units (9). This analysis showed that most but not all sections of this region are composed of transposons, mainly of the DIRS type. With only information on one such region it is impossible to generate broad conclusions about centromere structure and evolution in this organism. To enable a comparative analysis of centromeric regions of D. discoideum and infer possible modes of their generation, we reconstructed more centromeric regions using the same technique as with that of chromosome 1.
MATERIALS AND METHODS
Whole chromosome assemblies were obtained from dictybase (http://dictybase.org) (14). A BLAST database was created containing all reads from the genome shotgun sequencing project. All raw sequencing reads were blasted against these sequences using stringent BLAST parameters (95% identity over 100 bases). Using BLAST again, the remaining reads were checked for the presence of previously defined repetitive elements (10). The naming convention implemented in the genome project enabled the construction of chromosome-specific bins. As a result, the reads containing tentatively unambigous sequences were then assembled separately for each chromosome using the GAP assembler (http://staden.sourceforge.net/). With this method, we obtained unique sequence contigs not present in the current D. discoideum assembly. These ‘seed’ contigs were then used for a stepwise completion of the assembly. Several rounds of the following assembly steps were performed until no further sequences could be added: (i) incorporating the corresponding reads from the other end of the clone (read pair) enabled extension of the contigs. Contigs indicated by read pair information to be adjacent were joined; (ii) the resulting contigs were manually checked and wrongly incorporated reads were removed; (iii) polymorphisms, indels, truncation regions and unique sequences were then characterized; and (iv) used to find similar reads in the raw sequencing read database via BLAST (15).
Since in the final assembly, reads were derived from different chromosome-enriched libraries the individual contigs were assigned to specific chromosomes using a method described earlier (16). Sequencing gaps were filled by primer walking on the connecting clones. The sequence coverage of the individual centromere regions was comparable with their respective chromosomes. Only a few gaps that could not be closed, at most four, remained in the individual centromere regions. No read pair support exists for these gaps and a PCR approach in repetitive regions to obtain missing sequences is not feasible. These gaps could be spanned using the consensus sequence of transposons, which matched two contig ends, so the contigs were ordered according to these overlaps yielding the most probable centromere consensus. However, we cannot totally exclude that additional sequences reside in the gaps or the real order of the contigs in the centromeres is different from that presented here.
Identification of repetitive elements
The known transposon sequences were put in a BLAST database. Further putative repeated sequences were found during the assembly process of the centromeric regions. These sequences were added to the database of transposons to yield a comprehensive sequence repository for repetitive elements (Supplementary Material). A Blast analysis of the assembled centromeric contigs using this database was performed to identify all repetitive regions. Furthermore, tRNAscan (17) was used to search for tRNA genes. Repeat positions were transformed to a gff file format for each centromere. A graphical representation of the centromeres was obtained using gff2ps (18) on these gff-files.
Phylogenetic tree reconstruction
From the BLAST analysis, we identified 34 full-length or nearly full-length DIRS elements on all centromeres. These elements were aligned using clustalX (19). The resulting alignment was manually corrected. For tree reconstruction we used puzzle (20) and distance and maximum likelihood methods implemented in PHYLIP (21). Molecular clock and relative rate analyses were performed with HYPHY (22). Both analyses used the general reversible model and the DIRS element C2o 14 was used as the outgroup for the relative rate test.
RESULTS AND DISCUSSION
Centromere assembly
From a previous study (10), it was known that the D. discoideum genome harbors up to 10% transposon-derived sequences. The unique portion of the genome including shorter repetitive parts and a manually reconstructed centromere region from chromosome 1 was published in 2005 (9). In this assembly most raw sequencing reads were incorporated, but not all repetitive regions could be resolved. Automated assemblers tend to assign ambiguous positions in the genome to reads from repetitive regions if polymorphisms are scarce or exclude them totally from the final assembly. Thus, a considerable number of reads (>5% of all reads) remained unassembled due to repetitiveness or bad quality. The assembly of repetitive regions larger than 5 kb was especially hampered. This limitation by size is caused mainly by the fact that with current technology cloning of extremely AT nucleotide-rich sequences is not possible (23) and therefore clone resources for the D. discoideum genome are restricted to short insert libraries. We reasoned that much of this unassembled remainder should comprise sequencing reads from centromeric regions.
The genome sequence was built mainly from chromosome-enriched libraries. Therefore, it was possible to assign raw sequencing reads according to a tentative chromosome location and to whether or not the specific read occurred in the final assembly. Since the reads not present in the final assembly contained both the repetitive elements that are believed to be part of centromere and unwanted low-quality reads, these reads were assembled using stringent parameters in a chromosome-specific manner. This excluded the low-quality reads and ensured a correct assembly albeit resulting in only small contigs of up to 2.5 kb. An analysis of these contigs revealed that it would be impossible to assign contigs correctly to either chromosome 4 or 5. This is due to the fact that these two chromosomes have almost the same size and cannot be properly separated in pulsed field gels. Thus, the chromosome-enriched libraries overlapped too much to yield a stringent sorting parameter. Additionally, sequencing reads from chromosome 6 are almost completely derived from only one clone direction meaning that read pair information is not available for this chromosome. Since read pair information is indispensable for the assembly of large repetitive sequences, it was also not possible to reconstruct the sequence of the centromere from chromosome 6.
The final assembly of the centromeric regions of chromosomes 2 and 3 yielded five contiguous sequences for each region. These contigs were then oriented towards each other according to the transposon sequences they harbored at their ends (see Materials and Methods section). The centromere of chromosome 3 can be directly attached to the chromosome 3 unique core regions via two independent clone bridges. As a result, we estimate that this region is complete and correctly assembled. In case of the chromosome 2 centromeric region, two final contiguous sequences were obtained. Presumably the shorter one resides at the chromosomal duplication breakpoint (see below). In the larger sequence (360 kb) we designed HAPPY Map markers (24) which covered the entire region. These markers (DH5499, DH3377, DH3286, DH2916, DH2922 and DH2664) could be linked to the chromosome 2 core sequence indicating a correct assembly of the chromosome 2 centromeric region.
Centromere structure
All centromeres together occupy 3.8% of the three chromosomes (Table 1). Each individual functional centromeric region is longer than 170 kb. The smallest assembled region is located in the middle of chromosome 2. Since this region is probably only an artifact of the duplication event, it probably retained no function and cannot be counted as a proper centromeric region. The observed sizes of the true centromeric regions (171–361 kb) increase with the sizes of the chromosomes, although not linearly (Table 1). Since this analysis is restricted to only three chromosomes this observed nonproportional size increase remains a circumstantial evidence and might not be important for centromere function.
Table 1.
Centromere composition: overview
| Source | Chromosome length (Mb) | Length (bases) | Percentage of chromosome | Repetitive |
||
|---|---|---|---|---|---|---|
| Bases | Contigous fragments | Repetitive percentage | ||||
| Chr1 | 4.92 | 173 921 | 3.5 | 150 503 | 136 | 86.5 |
| Chr2 outer | 8.88 | 361 820 | 4.1 | 310 016 | 316 | 85.7 |
| Chr2 inner | 36 067 | 0.4 | 34 578 | 26 | 95.9 | |
| Chr3 | 6.55 | 191 571 | 2.9 | 163 766 | 154 | 85.5 |
| Alla | 20.35 | 763 379 | 3.8 | 658 863 | 632 | 86.3 |
aAll refers to the summation of all centromere sequence.
All centromeres are highly repetitive with a total of 86% identifiable repetitive elements overall. The majority of this repetitiveness is achieved by transposons. However, we detected also small multiplied regions without defined transposons (Supplementary Material). These small sequence motifs have no obvious coding capacity and no significant G/C content difference to their surroundings. They are found exclusively in centromeric regions and may represent rare DNA transposable elements or could be derived from recombination events. Additionally, in the centromeres of chromosomes 1 and 2, but not 3, there are two tRNA genes each. Only one of these tRNA genes seems to be functional, since the other three were marked as pseudogenes (see Materials and Methods section).
The different transposon species occur at approximately the same percentage in each centromere (Table 2). DIRS elements comprise a total of 48.8% of all assembled centromere structures (Tables 1 and 2). These elements are restricted to only centromeric regions on the chromosome. The observed percentage of DIRS elements (1.8% of the total chromosomes) matches relatively closely the previously estimated percentage (3.2%) for the whole genome (10). The difference between calculated and observed values is probably due to the cloning bias towards A/T poorer sequences, which favors the cloning of DIRS element-derived sequences. Based on the observed and estimated numbers for DIRS elements in chromosomes 1, 2 and 3 and in the whole genome, we can estimate the total amount of DIRS sequences in chromosomes 4, 5 and 6 to be between 260 kb and 490 kb. Dictyostelium DNA Transposon (DDT) elements and the retrotransposon skipper contribute also 20% and 10%, respectively, to the overall length of the centromeres and therefore are enriched there compared with the rest of the genome (where they are present at only 1% and 0.9%, respectively). In contrast, the DNA transposons (Tdd elements) are only slightly overrepresented in this region, whereas nonLTR elements (TRE) are almost completely missing from centromeric regions. The only TRE element which is clearly located in a DIRS element containing region is that of the unfunctional chromosome 2 inner DIRS region.
Table 2.
Centromere composition: detailed repeat composition
| DIRS |
DDT |
Tdd |
Skipper |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Bases | cf (%) | >4 kb | Bases | cf (%) | Bases | cf (%) | Bases | cf (%) | |
| Chr1 | 80 920 | 30 (46.5) | 8 | 38 294 | 35 (22.0) | 1470 | 3 (0.9) | 19 098 | 4 (11.0) |
| Chr2 outer | 187 275 | 71 (51.8) | 17 | 69 510 | 78 (19.2) | 7404 | 5 (2.1) | 21 397 | 15 (5.9) |
| Chr2 inner | 15 254 | 4 (42.3) | 2 | 6421 | 6 (17.8) | 4640 | 2 (12.9) | 4930 | 2 (13.7) |
| Chr3 | 89 365 | 39 (46.7) | 7 | 33 966 | 32 (17.7) | 8383 | 4 (4.4) | 27 702 | 13 (14.5) |
| All | 372 814 | 144 (48.8) | 34 | 148 191 | 151 (19.4) | 21 897 | 14 (2.9) | 73 127 | 34 (9.6) |
cf, contigous fragments. % refers to the percentage of the centromere region that consists of the given repeat.
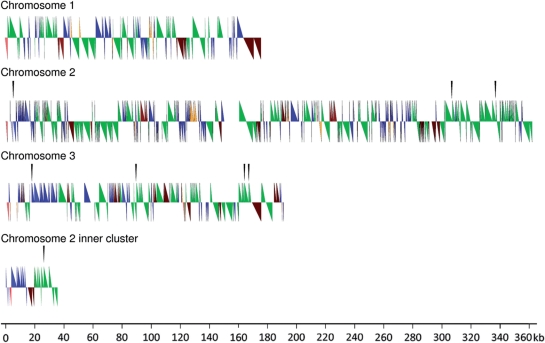
Most elements are highly fragmented (Table 2), mainly due to subsequent insertions of further transposable elements. DIRS elements and DDT/Tdd elements show a pronounced preference to be located in regions enriched for the respective elements. For example in the centromere of chromosome 3 the region from 17 kb to 35 kb is occupied by DDT and Tdd elements only, whereas the region from 85 kb to 100 kb is comprised solely of DIRS elements (Figure 1). Skipper and Dictyostelium Gypsy like Transposon (DGLT) elements apparently show no such preference.
Figure 1.
Centromere structures. Symbols above each chromosome's center line show features oriented towards the core of the chromosome, features below the line are oriented towards the upper end of the chromosome. The DIRS elements are depicted as green half-arrows. DNA transposons are in blue and skipper and DGLT elements in brown. The red triangles show the rDNA palindrome sequence whereas the orange hue indicates nonunique sequences not derived from transposons. Black triangles above the figures indicate gap locations.
rDNA Palindrome sequences as caps at centromeric ends
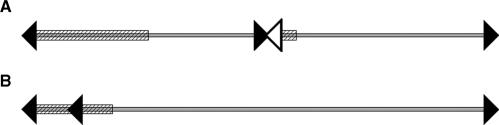
In the previous analysis of the whole D. discoideum genome, it was postulated that all chromosomes are capped at both ends with sequences derived from the rDNA palindrome (9). This kind of protection from slow chromosomal decay is thus different from that of most other eukaryotes. The previous finding was based on the observation that for each chromosome two junctions between palindrome and either transposons or unique chromosome sequence could be found. In each case, the palindrome and genome sequence were ordered in the following order: chromosome, junction, palindrome, palindrome end (Figure 2A). However, the position of these junctions on the chromosomes was only tentatively assigned. Chromosome ends adjacent to the centromere were especially difficult to analyze at that point, since only centromere sequences from chromosome 1 were available. We now can confirm that all centromeres analyzed are bordered by rDNA sequences at one side in the same order as was proposed (Figure 1). This fact proves that all chromosomes are indeed telocentric having only a q arm as previously suggested (25).
Figure 2.
Schematic overview of the centromeric rearrangements in centromeres 2 (A) and 3 (B) Arrowheads indicate rDNA palindrome sequences. The arrowhead direction gives the sequence direction from center to end of the rDNA palindrome. The rDNA palindrome sequence represented as empty arrowhead is only supported by theoretical evidence. Light gray line: unique chromosome sequence. The diagonal streaked rectangles depict the regions occupied by DIRS elements.
Scars from healing chromosome wounds
Chromosome breaks and rearrangements can also occur in centromeric regions. In the centromeres of chromosomes 2 and 3 we found potential marks of such events (Figure 2B).
Chromosome 3 centromeric fusion or extension mark: within the chromosome 3 centromere at 122 kb from the very end of the chromosome, we identified a short sequence derived from the rDNA palindrome. This sequence is currently the only case in the whole genome where we found a junction between the 3′-end of the palindrome and further repetitive structures. Most likely the innermost palindrome segment once built the former end of the chromosome until it was buried in further transposon-derived sequences and a further rDNA palindrome end was added. Possibly this event took place when the larger centromere was created that is now in the same size range as the chromosome 1 centromere.
The chromosome 2 duplication occurred in the laboratory strain AX4 ∼30 years ago and comprises more than 700 kb. The duplication event may have led to major additional changes in the chromosome. One of these likely changes is the presence of rDNA palindrome and DIRS sequences at the distal border of the duplication. The order of contigs in this part of the chromosome could only be confirmed by linking HAPPY map markers, leaving gaps between the contigs. The chromosome 2 centromere organization is supported by the HAPPY markers and the remaining 36 kb fragment does not fit at either position in this centromere region, so it very likely resides in this gap and extends the previously found DIRS containing sequence (Figure 2B). However, the junction between the rDNA palindrome sequence and the next DIRS elements could not be found in the raw sequencing reads nor are there bridging clones. Since the telomere would be nonfunctional at this position in the chromosome and, as previously explained, the centromeric sequence is likely nonfunctional, these junction sequences might have been destroyed and are thus no longer detectable. It is tempting to speculate that the knockout of this nascent centromere was caused by an overload of Tdd elements and/or the introduction of a TRE element since both features set this contig apart from the other centromeric regions. On the other hand, one less Tdd integration event would have caused a ‘normal’ transposon number distribution in this short centromere sequence piece. There should be a potential for TRE elements to integrate into centromeric regions since tRNAs, the targets of this integration, can reside there. Despite this fact, the only TRE element I found adjacent to DIRS elements is not associated with a tRNA hinting that this tRNA was deleted after the integration.
While Chromosome 2 is slightly larger than the other chromosomes analyzed here, its centromeric region is proportionally much larger, almost double the size of the others. Concurrently this centromere carries a large 10 kb unique region at position 150–160 kb (Figure 1). This is the largest unique region observed in all three centromeres. While the whole centromeric region has an A/T content of 68%, this region exhibits 78.5% A and T nucleotides indicating a lack of any coding capacity. It is possible that the chromosome 2 centromere is derived from a duplicated centromere and the unique region connects both. This could have been caused by increased repair activities after the chromosome break and the subsequent duplication event. This suggests duplicated centromeres are tolerated, while the rDNA mark on chromosome 3 suggests that short centromeres are not tolerated over longer evolutionary periods. The marks of wound healing at centromeres detected in this strain indicate that not only frequent duplication events (26) occur in different strains but also centromere reorganization is possible.
DIRS phylogeny and mode of centromere expansion
The availability of three centromeric regions makes it possible for the first time to analyze the mechanisms of centromere plasticity in D. discoideum. DIRS elements comprise the largest part of the centromeres and are restricted to these regions. Therefore, they most likely are the constitutive parts of this region. The other transposable elements may be functionally dispensable.
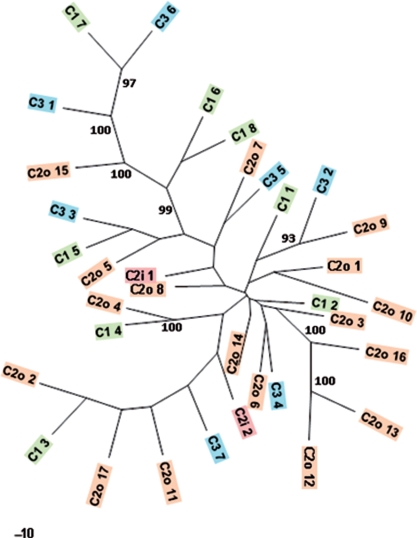
To further understand DIRS's role in centromeres and possible modes of integration preferences, we performed a phylogenetic analysis of these elements. On the three analyzed chromosomes 34 DIRS structures are located, which are longer than 4 kb and therefore are nearly complete elements. We assumed that these elements are the least decayed and therefore represent (besides possible insertions of truncated elements) the most recent DIRS element insertions. Indeed, polymorphisms between these 34 elements are scarce, under 100 in the aligned sequence. We reconstructed a phylogenetic tree of these elements (Figure 3). Due to the rare polymorphisms, the tree is only poorly resolved with weak bootstrap support for the main branches. Nevertheless, the relationship of the outermost branching DIRS elements is well supported. The tree shows clearly that there is no polymorphism restricted to only one chromosome. Instead, closely related DIRS elements are on different chromosomes suggesting that integration of DIRS elements occurs in trans and cis via retrotransposition.
Figure 3.
Phylogenetic tree of complete DIRS elements on the different chromosomes. The location of the DIRS elements is indicated by color (chromosome 1 green, chromosome 2 orange, chromosome 3 blue). The numbers after the chromosome indicator (C1; C2; C3) are given in ascending order beginning from the telomeric end. Chromosome 2 origins are also indicated by o (outer centromere) and i (inner centromere like). Only bootstrap values above 90% are shown. The scale is shown as a black bar.
To further investigate the dynamics of DIRS elements a molecular clock model was tested and is rejected (P < 0.0001). To examine closely related DIRS elements, pairwise relative rate tests were performed on all four pairs of DIRS elements where the terminal node had over 90% bootstrap support. The pairs C2o 12/C2o 13 and C2o 9/C3 2 each have nonsignificant rate differences, but the pairs C1 7/C3 6 (P < 0.04) and C2o 4/C1 4 (P < 0.0001) each have significant rate differences. While it is difficult to generalize from these four pairs, it is possible to make two conclusions. First, trans-retrotransposition and existing on different chromosomes (C2o 9/C3 2) does not necessarily lead to an increased rate of change as compared with cis-retrotransposition and existing on the same chromosome (C2o 12/C2o 13), since both pairs have nonsignificant differences. Second, trans-retrotransposition and existing on different chromosomes (C1 7/C3 6 and C2o 4/C1 4) can lead to different rates of change. It cannot be determined at this point whether DIRS elements established by cis-retrotransposition often have different relative rates due to the small sample size. The lack of a molecular clock and the relative rates tests together indicate that DIRS elements can be subject to variable mutational or selective forces after establishment but not in every case.
Preferential cis integration would have explained, at least partially, the observed clustering of DIRS elements at specific chromosomal positions. Since there is no preference for such cis integration, other mechanisms should be at work to establish specific integration. Without any clues on the presence of a specific target sequence it is tempting to speculate that regulation by small RNAs is the major force, which restricts DIRS elements to centromeric regions.
A further possibility for centromere expansion would be unequal crossing over via homologous recombination during meiosis. This would create tandemly repeated units not restricted to single transposon elements but rather crossing integration and truncation borders. We did not observe identical integration or truncation patterns on different regions of the same or different centromeres demonstrating that, if such unequal crossovers take place, they are rare. Furthermore, the traces of such events would quickly be erased by subsequent transposon integration events.
Comparison to centromeres of other model organisms
Only for a few organisms full chromosomal centromeres were described. Small defined sequence motifs as in S. cerevisiae seem to be rare, in most organisms centromeres are large, highly repetitive regions. Most analyses therefore yield only the major sequence motifs in addition to size estimates (27–30). A common feature of all described centromere structures is its repetitiveness. According to available data most centromeres consist of short repeated units (alpha satellites) sometimes interspersed with transposon species. Dictyostelium discoideum centromeres share this repetitiveness, but we observed no trace of alpha satellite sequences. Thus, while repetitiveness in large centromeres seems to be indispensable, small repeated sequences (alpha satellites) are not.
CONCLUSION
This analysis provides a first glimpse into the dynamics of centromere formation in D. discoideum offering the possibility to study centromeres and their evolutionary dynamics in this well-defined model system. A shotgun survey sequencing of related species showed that the transposon DIRS is present only in a few copies in other genomes of social amoebas. We also found no other transposon species, which could take over the role of DIRS elements in these amoebas (our unpublished data). This makes it unlikely that DIRS elements or other transposons play a major role in centromere establishment in other social amoebas. Possibly the large centromeres seen in D. discoideum are an evolutionary late invention and the ancestral state is a short centromere as in S. cerevisiae. Ongoing comparative genome analysis within social amoebas will help to further elucidate the dynamic evolution of centromeres.
ACCESSION NUMBERS
The sequences described here were deposited in GenBank with the Accession Numbers: Chromosome3 centromere: FJ387222; Chromosome2 centromere: FJ387223; Chromosome2 inner centromere: FJ387224
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG (Gl235/1-2, to A.H.). Funding for open access charge: DFG.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to all members of the Dictyostelium Sequencing Consortium for the provision of all raw sequencing reads. We also appreciate the very constructive contributions of the two anonymous reviewers.
REFERENCES
- 1.Glöckner G, Schulte-Spechtel U, Schilhabel M, Felder M, Sühnel J, Wilske B, Platzer M. Comparative genome analysis: selection pressure on the Borrelia vls cassettes is essential for infectivity. BMC Genomics. 2006;7:211. doi: 10.1186/1471-2164-7-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grandin N, Charbonneau M. Protection against chromosome degradation at the telomeres. Biochimie. 2008;90:41–59. doi: 10.1016/j.biochi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Cottarel G, Shero JH, Hieter P, Hegemann JH. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic. Mol. Cell Biol. 1989;9:3342–3349. doi: 10.1128/mcb.9.8.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Maddox PS, Oegema K, Desai A, Cheeseman IM. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- 6.Jin W, Lamb JC, Vega JM, Dawe RK, Birchler JA, Jiang J. Molecular and functional dissection of the maize B chromosome centromere. Plant Cell. 2005;17:1412–1423. doi: 10.1105/tpc.104.030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 8.Insall R. The Dictyostelium genome: the private life of a social model revealed? Genome Biol. 2005;6:222. doi: 10.1186/gb-2005-6-6-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichinger L, Pachebat JA, Glöckner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glöckner G, Szafranski K, Winckler T, Dingermann T, Quail MA, Cox E, Eichinger L, Noegel AA, Rosenthal A. The complex repeats of Dictyostelium discoideum. Genome Res. 2001;11:585–594. doi: 10.1101/gr.162201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cappello J, Handelsman K, Lodish HF. Sequence of Dictyostelium DIRS-1: an apparent retrotransposon with inverted. Cell. 1985;43:105–115. doi: 10.1016/0092-8674(85)90016-9. [DOI] [PubMed] [Google Scholar]
- 12.Li F, Sonbuchner L, Kyes SA, Epp C, Deitsch KW. Nuclear non-coding RNAs are transcribed from the centromeres of Plasmodium. J. Biol. Chem. 2008;283:5692–5698. doi: 10.1074/jbc.M707344200. [DOI] [PubMed] [Google Scholar]
- 13.Kuhlmann M, Borisova BE, Kaller M, Larsson P, Stach D, Na J, Eichinger L, Lyko F, Ambros V, Soderbom F, et al. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nucleic Acids Res. 2005;33:6405–6417. doi: 10.1093/nar/gki952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fey P, Gaudet P, Pilcher KE, Franke J, Chisholm RL. dictyBase and the Dicty Stock Center. Methods Mol. Biol. 2006;346:51–74. doi: 10.1385/1-59745-144-4:51. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Glöckner G, Eichinger L, Szafranski K, Pachebat JA, Bankier AT, Dear PH, Lehmann R, Baumgart C, Parra G, Abril JF, et al. Sequence and analysis of chromosome 2 of dictyostelium discoideum. Nature. 2002;418:79–85. doi: 10.1038/nature00847. [DOI] [PubMed] [Google Scholar]
- 17.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abril JF, Guigo R. gff2ps: visualizing genomic annotations. Bioinformatics. 2000;16:743–744. doi: 10.1093/bioinformatics/16.8.743. [DOI] [PubMed] [Google Scholar]
- 19.Thompson JD, Gibson TJ, Higgins DG. Curr. Protoc. Bioinformatics. 2002. Multiple sequence alignment using ClustalW and ClustalX. Chapter 2, Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 21.Retief JD. Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 2000;132:243–258. doi: 10.1385/1-59259-192-2:243. [DOI] [PubMed] [Google Scholar]
- 22.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 23.Glöckner G. Large scale sequencing and analysis of AT rich eukaryote genomes. Curr. Genomics. 2000;1:289–299. [Google Scholar]
- 24.Dear P, Cook PR. Happy mapping: linkage mapping using a physical analogue of meiosis. Nucleic Acids Res. 1993;21:13–20. doi: 10.1093/nar/21.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loomis WF, Welker D, Hughes J, Maghakian D, Kuspa A. Integrated maps of the chromosomes in Dictyostelium discoideum. Genetics. 1995;141:147–157. doi: 10.1093/genetics/141.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bloomfield G, Tanaka Y, Skelton J, Ivens A, Kay RR. Widespread duplications in the genomes of laboratory stocks of Dictyostelium. Genome Biol. 2008;9:R75. doi: 10.1186/gb-2008-9-4-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alkan C, Ventura M, Archidiacono N, Rocchi M, Sahinalp SC, Eichler EE. Organization and evolution of primate centromeric DNA from whole-genome shotgun. PLoS Comput. Biol. 2007;3:1807–1818. doi: 10.1371/journal.pcbi.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawabe A, Nasuda S. Structure and genomic organization of centromeric repeats in Arabidopsis species. Mol. Genet. Genomics. 2005;272:593–602. doi: 10.1007/s00438-004-1081-x. [DOI] [PubMed] [Google Scholar]
- 29.Sun X, Le HD, Wahlstrom JM, Karpen GH. Sequence analysis of a functional Drosophila centromere. Genome Res. 2003;13:182–194. doi: 10.1101/gr.681703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H. The size and sequence organization of the centromeric region of Arabidopsis. DNA Res. 2001;8:285–290. doi: 10.1093/dnares/8.6.285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.