Abstract
A number of studies showed that the development and the lifespan of Caenorhabditis elegans is dependent on mitochondrial function. In this study, we addressed the role of mitochondrial DNA levels and mtDNA maintenance in development of C. elegans by analyzing deletion mutants for mitochondrial polymerase gamma (polg-1(ok1548)). Surprisingly, even though previous studies in other model organisms showed necessity of polymerase gamma for embryonic development, homozygous polg-1(ok1548) mutants had normal development and reached adulthood without any morphological defects. However, polg-1 deficient animals have a seriously compromised gonadal function as a result of severe mitochondrial depletion, leading to sterility and shortened lifespan. Our results indicate that the gonad is the primary site of mtDNA replication, whilst the mtDNA of adult somatic tissues mainly stems from the developing embryo. Furthermore, we show that the mtDNA copy number shows great plasticity as it can be almost tripled as a response to the environmental stimuli. Finally, we show that the mtDNA copy number is an essential limiting factor for the worm development and therefore, a number of mechanisms set to maintain mtDNA levels exist, ensuring a normal development of C. elegans even in the absence of the mitochondrial replicase.
INTRODUCTION
Mitochondrial energy production is critically dependent upon the structural integrity of the mitochondrial genome (mtDNA). Individual mammalian cells have between 1000–10 000 mtDNA copies, with the more energy consuming tissues, such as heart, having proportionally higher amounts. Mature mammalian oocytes contain at least 100 000 copies of mtDNA, needed for normal development because mtDNA does not undergo replication through the early stages of embryogenesis. Mitochondrial DNA polymerase gamma (Polg) is the sole DNA polymerase, responsible for all replication and repair reactions within mitochondria (1). We recently described a knock-in mouse model that expresses an error-prone version of POLG (mtDNA mutator mice) (2). Abolished exonuclease activity leads to a 3- to 5-fold increase in somatic mtDNA mutations that, in turn, causes a progressive respiratory chain deficiency and premature aging phenotypes (2).
Over 100 different pathogenic mutations affecting almost every exon of the human POLG gene have been described over last years (3,4). These mutations are causing diseases characterized by a number of mtDNA maintenance defects like mtDNA depletion, multiple mtDNA deletions or multiple point mutations of mtDNA in the affected tissues (4). Clinically, POLG mutations can present from early neonatal life to late middle age, causing a spectrum of phenotypes that include autosomal progressive external ophthalmoplegia (adPEO), childhood encephalomyopathy with liver failure (Alpers–Huttenlocher syndrome), adult onset spinocerebellar ataxia (SANDO) and a number of other isolated clinical symptoms including premature ovarian failure, fatigue, muscle weakness and muscle pain (3,4). POLG is today recognized as a major human disease gene, possibly accounting for up to 25% of all patients with mitochondrial diseases (3,4).
Until now it has been shown that POLG is indispensable for development of various species ranging from yeast to mammals. A mutation in MIP, a gene coding for the catalytic subunit of yeast POLG, results in mtDNA depletion and formation of petite rho0 cells (5). Flies deficient in POLG activity (tam mutants) are weak and uncoordinated and grow significantly slower than wild-type flies, with noticeable defects in the development of the adult visual system (6). Due to a defect in locomotion, these flies also failed to undergo the behavioral changes characteristic of the wandering stage and died as a late third instar larvae (6). We have previously shown that POLG is absolutely essential for mammalian embryonic development and mtDNA maintenance. Homozygous disruption of the mouse POLG gene leads to embryonic death at late gastrulation and before early organogenesis (7). Loss of POLG coincides with a dramatic decrease in mtDNA levels, with POLG null embryos having around 2–5% of normal mtDNA level at embryonic day 8.5 (E8.5). In addition, POLG null embryos at E8.5 are much smaller than wild-type embryos and have a severe respiratory chain deficiency (7).
In the present study, we investigated the role of the Caenorhabditis elegans ortholog of mtDNA polymerase (POLG-1) in development by analyzing polg-1 deficient worms. Surprisingly, loss of polg-1 that resulted in animals having 25 times lower mtDNA levels than normal, did not affect the embryonic and larval development of C. elegans. Polg-1 deficient worms had normal developmental rates and even managed to produce a small number of embryos that arrested in the early embryonic stages due to extremely low mtDNA content. We could observe severe morphological and functional defects in the gonad as a result of serious mitochondrial depletion leading to sterility of polg-1 deficient animals. Marked depletion of mtDNA content in adult worms also leads to a shortened lifespan that could be extended almost to the normal level by prevention of egg laying, which otherwise leads to an early fatal gonadal protrusion. Furthermore, our results indicated that the mtDNA content in somatic tissues originates mainly from the embryo and is maintained on a high level without active replication during development and early adulthood. In the late adulthood, we have detected some signs of mitochondrial dysfunction in somatic tissues, presented as increasingly fused mitochondria and upregulation of the mitochondrial number in certain tissues, indicating increased mitochondrial biogenesis possibly as a compensatory mechanism for mtDNA depletion and mitochondrial turnover. Finally, our results clearly show that the adult gonad is the primary site of mtDNA replication in worms, probably due to the massive requirement for mitochondrial biogenesis during oogenesis, and that the mtDNA content in the embryo is the critical limiting factor for normal worm development. Consequently, our results show that high mtDNA levels are essential for C. elegans development, and that worms have created a mechanism ensuring normal development independent of mtDNA replication by providing embryos with mtDNA levels that correspond to the one found in human oocytes, set to create an organism with billions of cells.
MATERIALS AND METHODS
Strains
The following strains were used: N2 (wild type, Bristol), VC1224 (polg-1(ok1548)/mT1 II; +/mT1 [dpy-10 (e128)] II), DR2078 (mIn1[dpy10(e128)mIs14]/bli-2 (e768) unc-4(e120)II]), polg-1(ok1548)/mIn[dpy10(e128)mIs14]II (referred to as polg-1(ok1548)/+), polg-1(ok1548)II [referred to as polg-1(ok1548)], AA1058 Ex462(myo-3p::MTS::GFP + plasmid 8938) referred to as myo-3::MTS::GFP), Ex(myo-3p::GFP + plasmid 8938); polg-1(ok1548)II [referred to as polg-1(ok1548); myo-3::MTS::GFP], Is[HIS-72:: mCherry::unc-54 3′UTR]X (referred to as HIS-72::mCherry), polg-1(ok1548)II; Is[HIS-72::mCherry::unc-54 3′UTR]X [referred to as polg-1(ok1548), HIS-72::mCherry], SS104 (glp-4(bn2)I) [referred to as glp-4(bn2)], polg-1(ok1548)/mIn [dpy10(e128)mIs14]II, glp-4(bn2)I [referred to as polg-1(ok1548), glp-4(bn2)]. The strains were maintained at 20°C and all experiments were performed at 20°C, unless indicated differently.
Construction of mutants
polg-1(ok1548)/mIn[dpy10(e128)mIs14]II
polg-1(ok1548)II males were outcrossed 10 times to N2 hermaphrodites and then crossed to (mIn1[dpy10(e128)mIs14])/+ II hermaphrodites. The outcross was always done by a single male cross to a single N2 hermaphrodite. The presence of the ok1548 allele was confirmed by PCR.
Is[HIS-72::mCherry::unc-54 3′UTR]X
The strain was made by microinjecting (8) the reporter construct [his-72 (promotor + ORF)::mCherry::unc-54 3′UTR], i.e. the his-72 promotor and open reading frame fused to mCherry and the unc-54 3′-UTR, into N2 hermaphrodite animals. The final DNA concentration for microinjection was 171 ng/µl, consisting of 46 ng/µl of reporter array DNA and 125 ng/µl N2 genomic DNA, for faciliation of array formation, in water. The primer binding sites used for the amplification of his-72 were 5′-CTCTGTGTCGTCACGTGTCTTCGTGTTGG-3′ and 5′-AGCACGTTCTCCGCGGATGCGTCTG-3′. The fragments were fused by stitching PCR. Integration of the resulting extrachromosomal array was induced by 40 Gy gamma irradiation of L4/young adult animals. A line displaying early and at the same time low-level expression line was selected and then crossed to N2 at least four times. In the course of the experiments, the array was continuously transferred to a fresh wild-type background by crossing.
AA1058 (dhEx462(myo-3p::MTS::GFP))
The strain was made by microinjection of the plasmid pPD96.52 (plasmid 1608:L2534, Addgene, Fire Vector Library) that was co-injected with the coelomocyte marker (plasmid 8938: coel::RFP, Addgene, Fire Vector Library) at a concentration of 20 ng/μl and 80 ng/μl, respectively, in the N2 animals as previously described (8). The coelomocyte marker was used for identification of transformants.
glp-4(bn2)I, polg-1(ok1548)/mIn[dpy10(e128)mIs14]II
The strain was made by crossing (mIn1[dpy10(e128)mIs14]/+II males to glp-4(bn2)I hermaphrodites; then glp-4(bn2)/+ I;(mIn1[dpy10(e128)mIs14])/+II males were crossed to polg-1(ok1548)/+II hermaphrodites at 15°C. The presence of the ok1548 and bn2 alleles in strains cultivated at 15°C and 25°C were always confirmed by PCR (ok1548) and DIC (Nomarski) microscopy (bn2).
Brood size and lifespan assay
Singled L4 staged hermaphrodites (n = 5) were placed on NGM plates seeded with Escherichia coli OP50 bacteria and the number of hatched embryos was counted over the next 4 days. Brood size was determined at 20°C, 25°C and 27°C.
Synchronized L4 stage hermaphrodites were used for lifespan measurements. Worms were transferred every second day on NGM plates seeded with E. coli OP50 and examined every day for probe-provoked movement and pharyngeal pumping until death. Worms that died due to internally hatched eggs, desiccation or due to crawling out of the plate were censored. The cumulative survival rate was determined according to Kaplan and Meier (9). The log-rank (Mantel–Cox) test was used for comparing significant distributions between different groups in the lifespan assays. All calculations and plots were performed with Microsoft Office Excel 2003, Octave 2.9.12 (version 2.x and 3.x, http://www.gnu.org/software/octave/), Gnuplot (version 4.2, http://www.gnuplot.info/) or R (version 2.7, http://www.r-project.org/).
RNAi assay
RNAi assay was done following a standard protocol (10). Worms feeding on bacteria carrying the empty vector (L4440) were used as a control whereas worms feeding on bacteria carrying JA:Y57A10A.m (II-8O11) vector was used for RNAi analysis.
Rhythmic behavior analysis
Defecation was scored as previously described (11). In each experiment, at least 13 adult hermaphrodites were scored for posterior body wall muscle contraction (pBoc) and expulsion (Exp) cycle over 10 min at day 1, day 4 and day 6 of adulthood. All counts were carried out at 20°C under the dissecting scope. For statistical analysis, we first calculated the mean defecation cycle period for each animal and then the mean and standard deviation of these results.
Determination of the mtDNA copy number
The mtDNA copy number was measured by quantitative PCR. Worms at the respective developmental stage were singled and lyzed by standard protocol (12). Primers for NADH dehydrogenase subunit 1 (nd1) and actin-3 (act-3) were used in determination of mtDNA copy number. The act-3 forward primer 5′-TGCGACATTGATATCCGTAAGG-3′ and reverse primer 5′-GGTGGTTCCTCCGGAAAGAA-3′. nd1 forward primer 5′-AGCGTCATTTATTGGGAAGAAGAC-3′ and reverse primer 5′-AAGCTTGTGCTAATCCCATAAATGT-3′. Real-time PCR conditions were 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Amplified products were detected with SYBR Green (Platinum SYBR Green qPCR Super Mix-UDG with ROX, Invitrogen) and fluorescent signal intensities were determined by ABI Prism 7700 Sequence Detector System (Applied Biosystems) by software SDS (version 1.91). The CT values within the linear exponential phase were used to measure the mtDNA copy numbers from a standard curve generated either by using plasmid containing cloned target sequence (nd1) into pCR2.1 plasmid (TOPO kit, Invitrogen) in order to determine absolute values or by five 10-fold dilutions of nuclear DNA (act-3) in order to measure the nuclear DNA copy number. All reactions were checked on the agarose gel after performing quantitative PCR and as a control for amplification accuracy we measured mtDNA copy number from few embryos or worms in one reaction. Quantitative PCR was performed for each initial template copy number at least four times and the results were reproducible.
Measurement of transcript levels
Transcript levels of cytochrome b (ctb-1), polg-1 and MTCE.35 (NADH dehydrogenase subunit 5, referred to as nd5) genes were analyzed in N2 and polg-1(ok1548) adult hermaphrodite animals by quantitative real-time PCR. Primers for ctb-1: forward 5′-TTCCAATTTGAGGGCCAACT-3′ and reverse 5′-AACTAGAATAGCTCACGGCAATAAAAA-3′; for nd5 forward 5′-TTAGCAAGTTTGGTCGAAGAAGATT-3′ and reverse 5′-GGCCCAAAGTAACTATTGAAAAACC-3′ and for polg-1 forward 5′-CTGCCTAATACCGTTGCCTTCTT-3′ and reverse primer 5′-TTGGAGCCGTCCGGATT-3′. Total RNA from 20 worms was isolated from N2 and polg-1(ok1548) strain with Trizol (Invitrogen). Amplification was performed with the ABI Prism 7700 Sequence Detector (Applied Biosystems) with the following PCR conditions: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Amplified products were detected with SYBR Green (Platinum SYBR Green qPCR Super Mix-UDG with ROX, Invitrogen). Relative quantification was performed using the standard curve method. The standard curve was constructed by using five 10-fold dilutions of the standard sample. Standards were run in triplicate with act-3 as endogenous control.
Analysis of mitochondrial morphology
Mitochondrial morphology in the body wall muscle was examined by expressing GFP under the myo-3 promoter. Worms were mounted in four times diluted egg salts and observed with a Zeiss Axioplan 2 imaging microscope. All pixels representing mitochondria were annotated. The number of connected pixels was used as a measure of individual mitochondrial size. Image analysis was performed in ImageJ (ver. 1.40 http://rsbweb.nih.gov/ij/) and Matlab (ver.R2007, The MathWorks).
Transmission electron microscopy
Worms at different stages during adulthood and isolated gonads were fixed in 2% glutaraldehyde and 0.5% formaldehyde. Samples are embedded in gelatin and postfixed in 2% osmium tetra-oxide. All specimens were examined using a Philips 420, TECNAI 10 microscope and images were captured with the Mega View Soft Imaging System.
Morphology of the nuclei in the gonad
To localize nuclei in isolated gonads, Hoechst 33258 stain (1 µg/ml) was added to the mounting medium (M9) and the fluorescent staining was detected using a DAPI filter on a Zeiss Axioplan 2 Imaging microscope. After staining, the gonadal structure was analyzed with the software Endrov (ver.2.11.0) by 3D reconstruction of microscopic images and by annotation of stained nuclei.
Determination of stage of embryonic arrest
The stage in which embryos arrested was determined by inspection of embryonic corpses. In order to assess the stage of arrest more precisely, the transgene HIS-72::mCherry was crossed into the polg-1(ok1548)/+ mutant background. The transgene is expressed after eight-cell stage in somatic cells. The translational fusion protein is stable several hours after death of the animal, allowing stage analysis of embryos in which morphological analysis already fails due to autolysis. Photomicrographs of arrested embryos were visually compared with time-lapse recordings of the same transgene in wild-type background by three observers. The time point of the morphologically best matching stage was used to quantify the stage of arrest of the embryo.
Embryonic development—time-lapse microscopy
One- or two-cell stage embryos were dissected from adult hermaphrodites. In the case of the polg-1(ok1548) animals, young adults were used in order to isolate the first embryos produced by the gonad. Embryos were mounted on polylysine coated cover slides recorded on a custom-built video time-lapse microscopy setup. This setup is based on an Axioplan 2 imaging microscope (Zeiss) with a piezo stage (ASI) controlled by OpenLab (Improvision) and a custom made software (ORS, Endrov, Henriksson J. unpublished results). The microscope is equipped for DIC/Nomarski and epifluorescent imaging. Time-lapse recordings were processed and analyzed using the software Endrov (ver.2.11.0).
RESULTS
Polg-1 deficiency causes severe mtDNA depletion, but not a developmental arrest
Based on sequence similarities Y57A10A.15 has been annotated as the C. elegans ortholog of mitochondrial DNA polymerase (POLG). The amino acid sequence of Y57A10A.15 (termed polg-1) shows 26% identity and 45% similarity to the human POLG. The multiple alignment of the POLG amino acid sequences for human, murine, fly, yeast and worm protein indicates the presence of highly conserved regions in both the exonuclease and the polymerase domains (Figure S1A and B). In addition, an iPSORT (http://hc.ims.u-tokyo.ac.jp/iPSORT/), a TargetP (http://www.cbs.dtu.dk/services/TargetP/) and a MitoProt II (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html) analysis confirmed the presence of a mitochondrial targeting peptide at the N-terminus of polg-1.
To investigate the in vivo function of polg-1, we analyzed a deletion mutant polg-1(ok1548) carrying a 2149 bp deletion that removes exons 8–10 of the predicted polymerase domain of polg-1. The mutant strain VC1224 was outcrossed 10 times against the N2 background and then balanced. Quantitative PCR analysis confirmed that the polg-1 transcripts are extremely reduced in adult homozygous deletion mutants (Figure 1A). Importantly, the transcription efficiency of the downstream gene Y57A10A.14 is unaffected by this deletion (data not shown). The outcrossed homozygous polg-1(ok1548) deletion mutant animals were viable, born in Mendelian proportions and reached adulthood without any obvious morphological or behavioral defects.
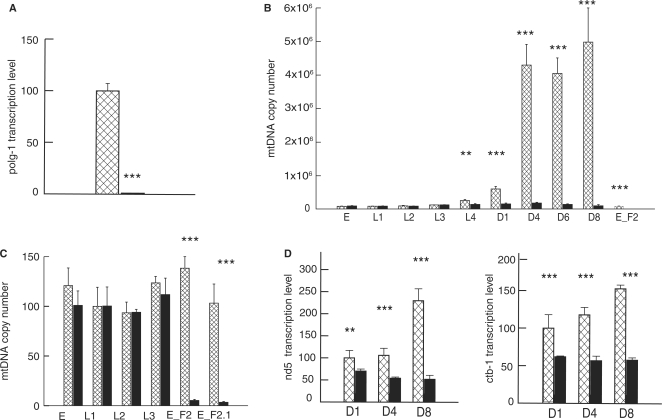
Figure 1.
Steady-state levels of mtDNA and mitochondrial transcripts in wild-type (N2, squared bars) and polg-1(ok1548) animals (black bars). (A) Relative transcript levels of polg-1 gene presented as a percentage of the wild type measured at day 1 of adulthood. (B, C) Steady-state mtDNA levels during worm development. Values represent the mtDNA copy number per animal (B) or per nuclear genome (C). (D) Quantification of transcript levels for mitochondrial-encoded nd5 and ctb-1 genes in polg-1(ok1548) animals at day 1 (D1), day 4 (D4) and day 8 (D8) of adulthood. Values are presented in the percentage relative to the wild-type levels at adult day 1. The error bars represent the SD. Asterisks show statistical significance (Student's t-test, **P < 0.01, ***P < 0.001).
We employed quantitative real-time PCR to estimate the mtDNA copy number in single embryos or adult animals of both wild-type (N2) and polg-1 deficient animals. Both strains have comparable mtDNA levels in the embryos (90 000–100 000) and that number remains essentially unchanged up to the L3 stage, indicating an absence of replication of the mitochondrial genome during this period (Figure 1B; Table 1). In order to further confirm the lack of mtDNA replication in early larval stages, we performed additional quantification relative to the amount of nuclear DNA that should be constant for a given developmental stage. For example, absolute mtDNA levels in all isolated embryos were the same but when we performed quantification of mtDNA molecules relative to nuclear DNA content, we observed that early embryos (E_F2, before comma stage) had 38% higher relative levels of mtDNA compared with late embryos (E_F2.1, folded), L1 and L2 larvae, corresponding to the lower cell number, but similar mtDNA content in these embryos (Figure 1C). Increase in the mtDNA copy number in N2 worms at 20°C starts around L3 stage and reaches a roughly 3-fold increase with the transition to L4 (from 90 000 to 250 000) (Table 1). The increase of the mtDNA levels closely mirrors the germ line proliferation in the late larval stages and the early adulthood. Transition to adult worms results in a tremendous increase of the mtDNA copy number that reaches around 4 millions mtDNA molecules per animal after adult day 1, coinciding with the high production of embryos (Figure 1B; Table 1).
Table 1.
mtDNA copy number in polg-1 (ok1548) mutant at 20°C
| Developmental stages | N2 (wild type) ×104 | polg-1(ok1548) ×104 |
|---|---|---|
| Embryo (F1) (n = 4) | 8.96 ± 0.75 | 9.97 ± 0.82 |
| L1 (n = 4) | 9.04 ± 0.65 | 9.42 ± 0.86 |
| L2 (n = 4) | 10.15 ± 1.62 | 9.43 ± 0.55 |
| L3 (n = 4) | 12.94 ± 0.29 | 13.10 ± 0.52 |
| L4 (n = 6) | 25.96 ± 3.37 | 14.56 ± 2.15** |
| D1 (n = 6) | 60.42 ± 7.74 | 16.07 ± 2.47*** |
| D4 (n = 6) | 429.65 ± 61.89 | 18.62 ± 2.22*** |
| D6 (n = 6) | 404.79 ± 46.23 | 14.37 ± 1.90*** |
| D8 (n = 4) | 498.34 ± 127.35 | 10.43 ± 3.47*** |
| Embryo (F2) (n = 4) | 8.96 ± 0.75 | 0.33 ± 0.08*** |
Numbers represent absolute mtDNA levels/worm ± SEM. Level of statistical significance **P < 0.01, ***P < 0.001, Student's t-test). D1, D4, D6 and D8 refer to days 1, 4, 6 and 8 of adulthood, respectively.
Gonad as a primary site of mtDNA replication
The mtDNA copy number in polg-1(ok1548) animals remained constant during early development and was comparable with the N2 mtDNA levels. It seems that the maternal contribution of POLG-1 protein and/or polg-1 transcripts in the embryos is quite high as polg-1 deficient animals manage to partially respond to the initial request for upregulation of the mtDNA content during late larval and early adult stages (Figure 1B, Table 1). However, this roughly 2-fold increase is far from the 50-fold increase in the mtDNA levels observed in N2 animals and it was not sustained later in adult polg-1(ok1584) worms, probably due to the exhausted POLG-1 storages and mitochondrial turnover. In order to evaluate the somatic versus the germline component of mtDNA levels, we have measured the mtDNA copy number in a germline proliferation deficient glp-4 strain. The function of the glp-4 gene product is not known at the molecular level, however, the glp-4(bn2) allele limits the production of germline nuclei to 12, while the somatic gonad retains a wild-type morphology (13). The mtDNA copy number in glp-4(bn2) mutant is similar to the wild type at L4 stage, but the high increase that follows the transition to adulthood in N2 worms is lacking in the glp-4(bn2) mutant (Tables 1 and 2). Instead, a mild, <2-fold increase in the mtDNA levels is observed with a transition to adulthood in glp-4(bn2) mutant, indicating that the majority of the mtDNA replication occurring after the L4 stage accounts for germline proliferation and embryo production. Double mutants, glp-4(bn2), polg-1(ok1548) had comparable mtDNA levels during development, but failed to upregulate them during adulthood. Instead, we detected a significant reduction of mtDNA levels with transition to adulthood in polg-1(ok1548), glp-4(bn2) worms. In addition, environmental temperature both higher (25°C) and lower (15°C) than 20°C present quite a metabolic stress on worms, as we have detected that all different strains, including N2, glp-4(bn2), polg-1(ok1548) and polg-1(ok1548) glp-4(bn2) mutants, had highly increased mtDNA content when grown at those temperatures (Table 2). Worms cultured at low temperature (15°C) had 4–5 times increase in mtDNA levels, indicating a high metabolic demand probably due to an increased environmental stress. The main effect of a higher temperature (25°C), besides a moderate increase in mtDNA copy number, was in the increased mtDNA turnover that started already during larval development (Table 2).
Table 2.
mtDNA copy number in polg-1(ok1548); glp-4(bn2) mutant
| Developmental stages | 25°C |
15°C |
||||||
|---|---|---|---|---|---|---|---|---|
| N2 (wild type) ×104 | polg-1(ok1548) ×104 | glp-4(bn2) ×104 | polg-1(ok1548); glp-4(bn2) ×104 | N2 (wild type) ×104 | polg-1(ok1548) ×104 | glp-4(bn2) ×104 | polg-1(ok1548); glp-4(bn2) ×104 | |
| L3 (n = 3) | 31.28 ± 0.12 | 22.42 ± 2.35 | 24.28 ± 3.49 | 17.40 ± 2.29 | 69.99 ± 8.12 | 51.47 ± 3.46 | 64.18 ± 5.37 | 45.91 ± 0.81 |
| L4 (n = 3) | 191.19 ± 13.97 | 22.74 ± 0.45 | 28.45 ± 9.30 | 17.62 ± 1.48 | 192.09 ± 2.45 | 62.78 ± 2.28 | 108.41 ± 18.26 | 53.24 ± 4.16 |
| D1 (n = 3) | 388.70 ± 67.09 | 16.59 ± 2.60 | 37.56 ± 5.89 | 12.54 ± 1.82 | 499.38 ± 59.41 | 62.44 ± 0.27 | 289.82 ± 48.83 | 56.13 ± 6.17 |
| D4 (n = 3) | 692.05 ± 65.80 | 14.24 ± 1.12 | 43.20 ± 8.43 | 9.91 ± 1.98 | 849.49 ± 129.12 | 52.62 ± 1.12 | 745.62 ± 87.33 | 50.49 ± 2.36 |
Numbers represent absolute mtDNA levels/worm ± SEM. Level of statistical significance (**P<0.01, ***P<0.001, Student's t-test). D1 and D4 refers to a day 1 and day 4 of adulthood, respectively.
Mitochondrial transcript levels in polg-1 mutants
Mitochondrial transcript levels in polg-1 deficient worms are decreased 30–77% compared with that in wild-type worms at different days of adulthood (Figure 1D). Although the reduction in nd-5 and ctb-1 transcript levels was quite robust (30%, 49% and 77% decrease for nd-5 transcripts and 38%, 51% and 62% for ctb-1 transcripts at adult day 1, 4 and 6, respectively), it was still much smaller than the observed decrease in the mtDNA copy number (up to 35 times) (Figure 1C and D). This is consistent with previously described upregulation of mitochondrial RNA and protein stability as a result of mtDNA deficiency (8).
Mitochondrial phenotypes in polg-1 mutants
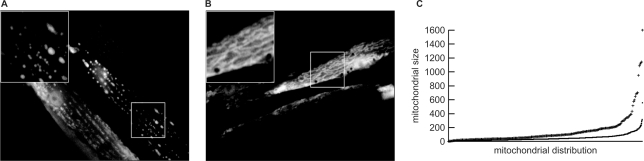
High mtDNA depletion as observed in adult polg-1 deficient worms may lead to mitochondrial dysfunction, predominantly observed in the high energy demanding, postmitotic tissues, e.g. muscle. Using a muscle-specific GFP fusion gene (myo-3::MTS::GFP) that targets mitochondria in live worms, we observed marked changes in the mitochondrial morphology of polg-1(ok1548) worms (Figure 2B). In control worms, the muscle mitochondria were well organized, running in parallel with the body axis (Figure 2A). In polg-1 deficient worms, the mitochondria were noticeably disorganized and fused (Figure 2B). After quantification of the mitochondrial length we verified that polg-1(ok1548) animals have a much higher incidence of long mitochondria than wild-type worms (Figure 2C). We propose that this increased mitochondrial fusion could be an adaptive response to the mtDNA depletion, allowing functional complementation of mtDNA-encoded proteins.
Figure 2.
Mitochondrial morphology in the body wall muscle of (A) control (myo-3::MTS::GFP) and (B) polg-1 deficient (polg-1(ok1548),myo-3::MTS::GFP) adult hermaphrodite animals. (C) Distribution of different mitochondrial lengths “+” stands for polg-1(ok1548) animals and “°” stands for N2 animals.
The integrity of mitochondria in adult polg-1(ok1548) worms was further investigated by transmission electron microscopy (TEM). Hypodermal cells, intestine and body wall muscle mitochondria of either wild-type or polg-1(ok1548) worms appeared normal, with numerous cristae and similar size (Figure S2A–C). We could observe a moderate upregulation of the number of mitochondria in the skin and body wall muscle of polg-1 deficient worms at day 6 of adulthood (Figure S2A and C).
mtDNA depletion causes change in physiological rhythms
To test if mtDNA depletion affects physiological rhythms, we examined the intervals of different stages of the defecation cycle (11). These rhythmic contractions represent an ultradian cycle in worms that repeats every 50–60 s (11) and we detected a progressive increase in the intervals with increasing age in wild-type worms (Figure S3). Defecation rates in polg-1 deficient worms basically remained on the same level, but later in life muscle contraction became very weak and defecation rates were much shorter in comparison with wild type worms (Figure S3). On the contrary, several long-lived mutants affecting mitochondrial energy metabolism (e.g. clk-1, tpk-1 and isp-1) have slowed rates of pharyngeal pumping and defecation when compared with wild-type worms (15).
Lack of mtDNA replication causes irregular gonadal development
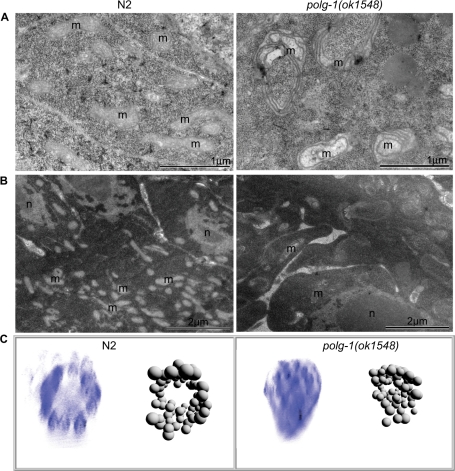
Next we analyzed mitochondria in the gonad, the only proliferating tissue in adult worms. Gonadal development in polg-1(ok1548) animals appears to be normal until the first embryos are produced. However, the number of mitochondria is drastically reduced in polg-1 deficient gonads and the remaining mitochondria appear to be enlarged and display a number of morphological abnormalities in young adults (Figure 3A). Later in adulthood the gonad seems to be seriously depleted of mitochondria, probably due to the production of a certain number of embryos and inability to replicate mtDNA (Figure 3B). These results are in agreement with our previous observation that the majority of the mtDNA replication appears to take place in proliferating gonads. In addition, adult polg-1(ok1548) hermaphrodites did not develop a rachis, the central core of cytoplasm that connects the developing oocytes in the syncytial gonad, which in wild-type worms contains a large number of mitochondria (Figure 3B). No obvious difference in the number of nuclei was observed in isolated polg-1(ok1548) gonads stained with Hoechst 33258 (data not shown).
Figure 3.
Gonad morphology of wild-type (N2) and polg-1(ok1548) animals. Electron micrographs of gonads isolated from worms at first (A) and sixth (B) day of adulthood; m: mitochondria, n: nuclei. (C) 3D representation of the gonad morphology in wild-type (N2) and polg-1(ok1548) mutant adult hermaphrodites. 3D reconstruction of microscopic images was performed after Hoechst staining of isolated gonads with the software Endrov (ver. 2.11.0).
The 3D reconstruction images of gonadal nuclei stained with Hoechst 33258 allowed us to further investigate gonadal morphology in polg-1(ok1548) hermaphrodites (Figure 3C). While wild-type gonads had a well-developed rachis with a number of surrounding nuclei, polg-1(ok1548) gonads lacked the rachis with nuclei being equally distributed through the gonad (Figure 3C).
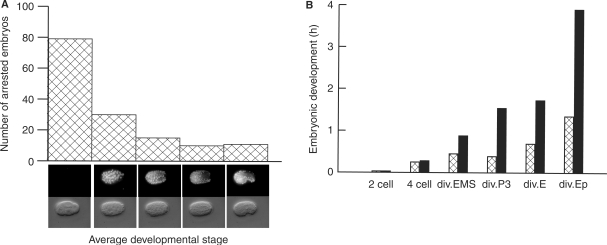
Progeny of polg-1 mutants arrests in the early development
Even though the development of polg-1(ok158) animals was rather normal, these animals could not produce viable progeny at any given temperature (Table 3). The brood size of polg-1(ok158/+) heterozygous animals is smaller than in N2, yet similar to the balancer background (see Materials and methods section) at different temperatures (Table 3). However, polg-1 deficient animals could produce a handful of eggs that arrested during embryonic development. We quantitatively characterized the embryonic arrest in polg-1(ok158) progeny by determining the approximate number of cells at the time of arrest. An integrated transgene expressing a mCherry tagged histone (HIS-72::mCherry), driven by its native promoter, allowed for accurate screening even in partially decayed embryos. The arrest occurred during all stages of embryogenesis, with most of the embryos arrested before the four-cell stage (Figure 4A). However, a small fraction of embryos reached the comma stage or even developed up to the 3-fold stage. Extremely rarely, the first embryo of a young adult hatched, but nevertheless, those larvae were not viable. These differences in the embryonic development are likely to be determined by the maternally inherited mtDNA levels; with the first ones produced probably having higher mtDNA levels than later ones. The 4D DIC time-lapse microscopy of polg-1(ok158) progeny showed that the speed of development is drastically reduced (Figure 4B, Movie S1 and S2), and it varies tremendously between individuals. The development of polg-1(ok158) progeny exponentially slows down between four cell and comma stage (Figure 4B). The rate of development most likely slows down due to mitochondrial dilution with each successive cell division leading to energy depletion in these embryos.
Table 3.
Brood size of polg-1 (ok1548) mutant at different temperatures
| Strains | 20°C | 25°C | 27°C |
|---|---|---|---|
| N2 (wild type) (n = 5) | 250 | 226 | 40 |
| polg-1(ok1548/+) (n = 5) | 160 | 187 | 19 |
| polg-1(ok1548) (n = 5) | 0 | 0 | 0 |
All values presented in the table are average values for the brood size of the total number of worms examined.
Figure 4.
Embryonic development of polg-1(ok1548) progeny. (A) Quantification of the number of embryos arrested on specific stages during embryonic development. (B) Time intervals needed for embryonic development of wild-type (N2, squared bars) and polg-1(ok1548) (black bars) progeny to reach the two-cell, four-cell stage, the division of the EMS cell (div.EMS), of the P3 cell (div.P3), of the E cell (div.E) and the Ep cell (div.Ep).
The mtDNA copy number in the progeny of polg-1(ok158) animals is reduced to only 3300 copies, around 30 times less mtDNA content than in normal wild-type embryos (Table 1). We tested whether the lack of viable progeny could be rescued by introducing a wild-type copy of polg-1 into the polg-1(ok158) embryos by mating mutant hermaphrodites with HIS-72::mCherry males. However, no viable offspring was obtained from this mating, indicating that a wild-type copy of polg-1 cannot rescue the phenotype due to very low levels of mtDNA present in these embryos (around 3300 copies) and the lack of mtDNA replication in the early embryonic and larval stages.
MtDNA depletion causes shortening of the lifespan
All polg-1(ok158) animals die after nearly complete gonadal and intestinal protrusion, impairing the movements of the animal (Figure S4A). The animals died within 1 or 2 days after protrusion, most likely due to starvation. The protrusion phenotype starts to occur around the sixth day of adulthood. The mean lifespan of polg-1 deficient animals was 10 ± 3 days, with a maximum of 15.3 days (Figure 5A, Table 4). Heterozygous animals carrying the balancer chromosome as well as animals homozygous for the balancer showed the same life span as N2 (Figure 5A, Table 4). However, previous RNAi-based genome-wide screening analysis did not reveal any phenotypes caused by downregulation of the polg-1 gene (16–18). We believe that RNAi over just one generation failed to decrease mtDNA levels under the critical threshold for obvious mitochondrial dysfunction. Therefore, we performed RNAi analysis by continuous feeding over several generations. The third generation of worms develops the same phenotype as polg-1(ok158) mutants, with gonad or intestinal protrusion and their progeny undergoes embryonic arrest after 72 h (Figure SB).
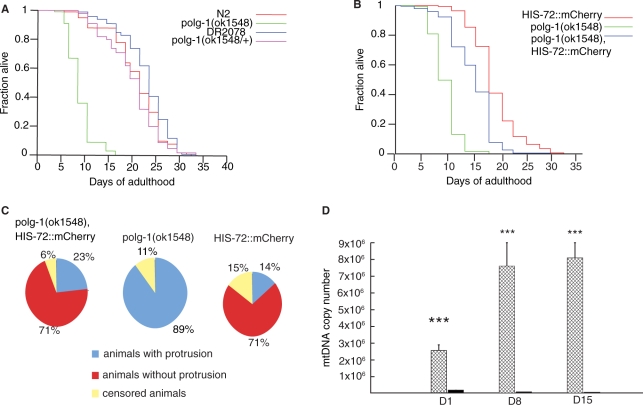
Figure 5.
Lifespan analysis of polg-1(ok1548) animals. (A) Lifespan of polg-1(ok1548) mutant was shown in comparison with DR2078, wild-type (N2) and polg-1(ok1548/+) animals. (B) Lifespan analysis of polg-1(ok1548), HIS-72::mCherry, with HIS-72::mCherry and polg-1(ok1548) animals used as controls. (C) Pie charts present percentage of animals that developed vulva protrusion during their lifespan. (D) mtDNA copy number in polg-1(ok1548), HIS-72::mCherry (black bars) and HIS-72::mCherry (squared bars) animals. The steady-state mtDNA levels are measured at day 1 (D1), day 8 (D8) and day 16 (D16). Values represent the mtDNA copy number per animal. The error bars represent the SD. Asterisks show statistical significance (Student's t-test, ***P < 0.001).
Table 4.
Statistical analysis of adult lifespan
| Genotype | Mean ± SEM | Maxa (days) | Number of animals that died/totalb |
|---|---|---|---|
| polg-1(ok1528) | 10 ± 3 | 15.3 | 313/365*** |
| wild type (N2) | 19 ± 7 | 30.2 | 334/370 |
| DR2078 | 22 ± 8 | 32.1 | 152/165*** |
| polg-1(ok1548/+) | 19 ± 6 | 28.5 | 119/165 |
| polg-1(ok1528),HIS72::mCherry | 17 ± 4 | 23.4 | 195/204***/c*** |
| HIS-72::mCherry | 21 ± 4 | 29.9 | 162/200 |
| wild type (N2)d | 9 ± 2 | 14.2 | 201/223 g*** |
| polg-1(ok1548)d | 8 ± 2 | 12 | 190/200 g*** |
| polg-1(ok1548), glp-4(bn2)d | 8 ± 3 | 15.4 | 186/200 g***/e* |
| glp-4(bn2)d | 10 ± 4 | 18.1 | 196/200 |
aMaximum lifespan shown is a median lifespan of the longest lived 10% of the animals assayed.
bThe total number of events equals the number of animals that died plus the number censored. For comparing significant distributions, statistical analysis of data was performed with the log-rank (Mantel–Cox) test. P-values were calculated for individual experiments consisting of control and experimental animals examined at the same time.
dAnimals were grown on 25°C. Comparisons are done either with wild type, (c) with HIS-72::mCherry animals, (g) with glp-4(bn2) and (e) with polg-1(ok1548) animals (*P<0.05, ***P<0.0001).
Decreased egg laying prolongs lifespan in polg-1 deficient worms
We propose that the gonadal protrusion that was observed in all polg-1 deficient animals was the result of mitochondrial dysfunction, probably due to the weakening of vulval muscle cell after repeated egg laying. To test if the protrusion phenotype of polg-1(ok158) animals could be suppressed we introduced an Egl (egg-laying deficient) phenotype by mating hermaphrodites heterozygous for polg-1(ok158) allele with HIS-72::mCherry males. Presence of the HIS-72::mCherry allele leads to spontaneous occurrence of phenotypes including partial Egl, Dpy or Unc. These phenotypes might arise due to mutagenic or epigenetic effects of the transgene product (Hench J., unpublished results). We found a line with a partial Egl phenotype that almost completely suppressed the protrusion phenotype observed in polg-1(ok158) animals (Figure 5C) and partially corrected for the shorter lifespan (Figure 5B). Nevertheless, the median and maximal lifespan of polg-1(ok158), HIS-72::mCherry worms was still significantly shorter than that of controls (17 ± 4 days and 23.4 days, respectively) (Table 4). Similarly, higher environmental temperature (25°C) used to promote glp-4(bn2) mutant phenotype also had an effect on polg-1(ok1548) mutants. At this temperature, polg-1 deficient worms did not lay any eggs and therefore did not developed gonadal protrusion which in turn resulted in a prolongation of their lifespan to the levels comparable with polg-1 deficient gonad less mutants (polg-1(ok1548), glp-4(bn2)) (Table 4).
We believe that the shorter lifespan of polg-1(ok158), HIS-72::mCherry mutant hermophrodites was a consequence of even larger mtDNA depletion and this was confirmed by quantification of mtDNA levels. We detected an additional 40% decrease in the mtDNA levels between day 8 and 15 of adulthood (Figure 5D, Table 5). Also, the mtDNA levels in the control HIS-72::mCherry worms were almost two times higher than in N2 worms, indicating that deficiency in the egg laying does not affect mtDNA replication (Tables 1 and 5). On the contrary, the massive mtDNA levels imply that mtDNA replication is normal, but because these worms carry more embryos in the uterus, mtDNA content per worm is much higher. We have detected upregulation of mitochondrial mass in the hypodermis of polg-1(ok158), HIS-72::mCherry worms at day 8 of adulthood (Figure S5A), but not at day 1 (data not shown). At day 15, mitochondria in the hypodermis from both control (N2) and polg-1 deficient worms looked enlarged and swollen (Figure S5B), as previously reported for aged worms (19). We could observe an additional tissue distortion and a large number of membrane-like structures that resemble mitochondria in the process of mitophagy in polg-1(ok158), HIS-72::mCherry animals. Also, there was a marked increase of mitochondrial mass in other tissues, e.g. pharynx of polg-1(ok158), HIS-72::mCherry animals at this late time point (Figure S5C).
Table 5.
mtDNA copy number in polg-1(ok1548), HIS-72::mCherry mutants
| Developmental stages | HIS-72::mCherry (×104) | polg-1(ok1528), HIS-72::mCherry (×104) |
|---|---|---|
| D1 (n = 4) | 256.32 ± 33.78 | 19.66 ± 1.20*** |
| D8 (n = 4) | 760.11 ± 181.86 | 9.28 ± 0.49*** |
| D15 (n = 4) | 809.06 ± 148.44 | 5.72 ± 1.58*** |
Numbers represent absolute mtDNA levels/worm ± SEM. Level of statistical significance [***P<0.001, Student's t-test, (n = 4)]. mtDNA copy number was determined at day 1 (D1), day 8 (D8) and day 15 (D15) of adulthood.
DISCUSSION
Our results clearly show that although POLG is believed to be a necessary component of the normal developmental program in different species ranging from yeast to mammals (5–7), C. elegans development is not dependent on the presence of the polg-1 gene. This should not be mistaken for the lack of importance of mtDNA for worm development. On the contrary, we show that C. elegans embryos carry around 100 000 copies of the mitochondrial genome, a number that corresponds the one found in mammalian oocytes. And while a single mammalian oocyte will give rise to billions of cells, a C. elegans embryo will end up developing into adult worm with only around a thousand somatic cells. This very high number of mtDNA molecules in the developing embryo indicates the importance of the mtDNA for C. elegans development.
Others have reported that the mtDNA copy number range from 25 000 in early embryos to 800 000 in adult worms, but the pattern of increase was basically the same (20). Differences in the experimental approach could explain why we found a higher number of mtDNA molecules. We have performed all our experiments with DNA obtained from a single worm, while others have pooled 20 animals for DNA isolation (20).
Our results suggest that in addition to the high mtDNA levels, worms also have a high maternal contribution of POLG-1, which ensures normal development of polg-1 mutants. A high maternal contribution of proteins and/or mRNAs to the embryo appears to be conserved for many mitochondrial proteins and probably represents a safety mechanism ensuring normal maturation to the L4 stages when the aerobic respiration peaks (21). However, we cannot exclude the possibility of existence of additional DNA polymerases in the mitochondria, although we believe this to be very unlikely, because mtDNA levels were not sustained through the life. Instead mtDNA levels progressively decrease during the adult life, suggesting mitochondrial turnover and a lack of biogenesis.
The lack of active mtDNA replication in polg-1 deficient worms results in mitochondrial deficiency that could be partially compensated for by upregulation of the mitochondrial transcripts. A similar trend was observed in mice that had a moderate reduction of the mtDNA copy number, and yet presented normal levels of several mtDNA-encoded transcripts and proteins (14). Furthermore, studies of cell lines have shown that the inhibition of mtDNA expression dramatically increases the stability of mtDNA-encoded transcripts and proteins (22).
We propose that the increased mitochondrial fusion observed in somatic tissues of polg-1 deficient worms is another compensatory mechanism set to allow functional complementation of mtDNA-encoded proteins. There is a growing body of evidence that mitochondrial fusion, in addition to controlling the shape of mitochondria, is important for their bioenergetic function (23). When mitochondrial fusion is abolished in mammalian cells, a large fraction of the mitochondrial population loses mtDNA (24). In addition to mtDNA, it is also proposed that other components, such as substrates, metabolites or specific lipids, can be restored in defective mitochondria by fusion (25). In worms increased fusion seems to be a common response to a mitochondrial dysfunction as increasingly fused and disorganized mitochondria were reported in worms undergoing RNAi inactivation of the various electron-transport chain subunits, the mitochondrial ribosomal subunit and the cytochrome C heme lyase (26).
It seems that mitochondrial dysfunction could be compensated for by various mechanisms in somatic tissues of polg-1 deficient animals, but in the gonad where a constant urge for massive mtDNA replication exists, these mechanisms fail to assist. This results in the development of disorganized gonads that lacked the rachis and were seriously depleted of mitochondria. Ultimately, mtDNA depletion in polg-1 deficient animals precluded them from producing viable offspring, even if they produced a handful of embryos that arrested very early in development. Similar phenotypes were reported when levels of MTSSB-1, mitochondrial single-stranded DNA binding protein, were highly reduced after RNAi using a continuous feeding over generations (27). Results from other studies also suggest that mitochondria may play a role in the gonadal development. For example, high levels of uaDf5, an mtDNA deletion that removes nearly 25% of mitochondrial genome, have been associated with reduced rates of egg laying, defecation and sperm motility (28). RNAi for a C. elegans ortholog of mitochondrial ATP synthase b subunit (asb-1) causes 100% penetrant sterility (29). Furthermore, it has been shown that prohibitins, inner-mitochondrial membrane proteins that control cell proliferation, cristae morphogenesis and the functional integrity of mitochondria, are also required for somatic and germline differentiation in the larval gonad (30).
Although we have detected a number of changes in polg-1 deficient worms, we were rather surprised that mtDNA depletion leaving only around 4% of mtDNA content had a rather mild phenotype in comparison with other models, where similar levels of mtDNA depletion persistently resulted in the early developmental arrest (5–7,14,31). Previously, mtDNA depletion in worms has also reported to lead to a developmental arrest at L3 larval stage (20). However, this mtDNA depletion was evoked by treatment with ethidium bromide (EtBr) that predominantly results in a complete block of mtDNA replication, but after longer treatments could also have an effect on the maintenance of the nuclear genome (20). Similarly, large deletions in genes encoding essential subunits of the mitochondrial respiratory chain (MRC) complexes I and V result in developmental arrests at the third larval stage (L3) and the arrest of gonadal development in the second larval stage (L2), with impaired mobility, pharyngeal pumping and defecation (32). In addition, when MRC biogenesis is blocked by inhibitors of mitochondrial translation (chloramphenicol and doxycycline), a quantitative and homogeneous arrest as L3 larvae also takes place (32). These data demonstrate that C. elegans is dependent on mitochondria as the major source of ATP and that a functional MRC is a prerequisite for normal development. Here, we went a step further and showed that since mitochondria are so important for C. elegans development, a mechanism, providing embryos with a substantial amount of mtDNA, evolved to ensure that enough active MRC is present during development.
Our results show that the bulk of mtDNA replication is taking place in the gonad of adult worms and therefore the lack of mtDNA replication has a deleterious effect on the gonad of polg-1 deficient animals. It seems that polg-1 deficient animals distribute large amounts of the inherited mtDNA to the somatic cells and that this DNA is maintained throughout larval development, well into adulthood, when increased turnover in the absence of mtDNA replication could be detected. Also, it seems likely that somatic cells of polg-1 deficient animals inherit close to normal mitochondrial pools and that the majority of the mtDNA detected in adult worms (4% of normal) is actually present in somatic tissues. Hence, a suppression of deleterious organ protrusion phenotype, by partial egg laying deficiency, allowed polg-1 mutant animals to have a much longer lifespan and only when mitochondrial turnover in somatic tissues has depleted mtDNA levels under a critical threshold these animals die. This is in agreement with the observation that ∼35–40% reduction of mtDNA copy number in animals heterozygous for the mitochondrial transcription factor A null mutation (Tfam+/−) has minimal effects on mtDNA expression and mitochondrial function (14). Furthermore, mtDNA levels in gonad-less mutant, glp-4(bn2), when grown at the nonpermissive temperature, at the late larval stages are comparable with the one in polg-1 deficient animals, and around 10 times lower than in wild-type (N2) worms, indicating again, that the mtDNA levels present in the somatic tissues are much lower than in the gonadal ones. Only during adult life polg-1(ok1548) mutants failed to maintain the mtDNA levels due to the lack of replicase and increased mitochondrial turnover. This was even more apparent from the results of gonad-less polg-1 deficient animals polg-1(ok1548), glp-4(bn2) that had 4–5 times lower mtDNA copy number than gonad-less glp-4(bn2) mutants at adult day 4. Interestingly, the comparison between polg-1(ok1548) mutants with or without developed gonads suggests that most of the replication machinery that originates from embryos is dedicated to the gonad. We cannot, however exclude the possibility that glp-4(bn2) mutations have some kind of impact on the mtDNA replication and/or turnover.
In conclusion, this study emphasizes the role of polg-1 as a key player in the mtDNA maintenance in C. elegans. Our work also provides evidence that the mtDNA copy number is an essential limiting factor for C. elegans development and that a high maternal contribution of both, mtDNA and POLG-1 in the embryo ensures a normal development, even in the absence of polg-1 gene. Furthermore, our results demonstrate that the mtDNA copy number is highly regulated during development and can be adjusted to much higher levels than normal as a response to high energy demands caused by environmental stimuli. Finally, we believe that our results unraveled C. elegans as an excellent new model system to study mechanisms of mtDNA inheritance and maintenance in both postmitotic and proliferative tissues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council (to A.T. - 522-2005-7145 na to T.R.B. - 2003-3449); Åke Wiberg Foundation (to A.T. - 868332643); Loo and Hans Ostermans Foundation (to A.T. - 2006Oste0012); Marie Curie Early Stage Training Fellowship (to I.B.); Swedish Foundation for Strategic Research (to J.H. and T.R.B. - A3 04:160f) U.S. National Institutes on Health/National Institute on Aging (to A.A. - AG027498), Ellison Medical Foundation Senior Scholar Award in Aging (to A.A. - AG-SS-1920). Funding for open access charge: Swedish Research Council [522-2005-7145].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Peter Swoboda for helpful discussions, Kjell Hultenby and Ingrid Lindell for help with the TEM, Johan Dethlefsen for technical assistance and Lene Sorenson for help with the editing of the article. All strains not made by involved laboratories were kindly provided by the Caenorhabditis Genetics Center, University of Minnesota, USA.
REFERENCES
- 1.Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu. Rev. Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 2.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 3.Hudson G, Chinnery PF. Mitochondrial DNA polymerase-gamma and human disease. Hum. Mol. Genet. 2006;15:R244–R252. doi: 10.1093/hmg/ddl233. [DOI] [PubMed] [Google Scholar]
- 4.Longley MJ, Graziewicz MA, Bienstock RJ, Copeland WC. Consequences of mutations in human DNA polymerase gamma. Gene. 2005;354:125–131. doi: 10.1016/j.gene.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 5.Genga A, Bianchi L, Foury F. A nuclear mutant of Saccharomyces cerevisiae deficient in mitochondrial DNA replication and polymerase activity. J. Biol. Chem. 1986;261:9328–9332. [PubMed] [Google Scholar]
- 6.Iyengar B, Roote J, Campos AR. The tamas gene, identified as a mutation that disrupts larval behavior in Drosophila melanogaster, codes for the mitochondrial DNA polymerase catalytic subunit (DNApol-gamma125) Genetics. 1999;153:1809–1824. doi: 10.1093/genetics/153.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum. Mol. Genet. 2005;14:1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 8.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 9.Kaplan EL, Meier P. Nonparametric estimation from incomplete observation. J. Am. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- 10.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. research0002.1-0002.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu DW, Thomas JH. Regulation of a periodic motor program in C. elegans. J. Neurosci. 1994;14:1953–1962. doi: 10.1523/JNEUROSCI.14-04-01953.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barstead RJ, Kleiman L, Waterston RH. Cloning, sequencing, and mapping of an alpha-actinin gene from the nematode Caenorhabditis elegans. Cell Motil. Cytoskeleton. 1991;20:69–78. doi: 10.1002/cm.970200108. [DOI] [PubMed] [Google Scholar]
- 13.Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- 14.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 15.Branicky R, Hekimi S. What keeps C. elegans regular: the genetics of defecation. Trends Genet. 2006;22:571–579. doi: 10.1016/j.tig.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez AG, Gunsalus KC, Huang J, Chuang LS, Ying N, Liang HL, Tang C, Schetter AJ, Zegar C, Rual JF, et al. New genes with roles in the C. elegans embryo revealed using RNAi of ovary-enriched ORFeome clones. Genome Res. 2005;15:250–259. doi: 10.1101/gr.3194805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 19.Yasuda K, Ishii T, Suda H, Akatsuka A, Hartman PS, Goto S, Miyazawa M, Ishii N. Age-related changes of mitochondrial structure and function in Caenorhabditis elegans. Mech. Ageing Dev. 2006;127:763–770. doi: 10.1016/j.mad.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Tsang WY, Lemire BD. Mitochondrial genome content is regulated during nematode development. Biochem. Biophys. Res. Commun. 2002;291:8–16. doi: 10.1006/bbrc.2002.6394. [DOI] [PubMed] [Google Scholar]
- 21.Vanfleteren JR, De Vreese A. Rate of aerobic metabolism and superoxide production rate potential in the nematode Caenorhabditis elegans. J. Exp. Zool. 1996;274:93–100. doi: 10.1002/(SICI)1097-010X(19960201)274:2<93::AID-JEZ2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 22.England JM, Costantino P, Attardi G. Mitochondrial RNA and protein synthesis in enucleated African green monkey cells. J. Mol. Biol. 1978;119:455–462. doi: 10.1016/0022-2836(78)90226-7. [DOI] [PubMed] [Google Scholar]
- 23.Lansman RA, Clayton DA. Selective nicking of mammalian mitochondrial DNA in vivo: photosensitization by incorporation of 5-bromodeoxyuridines. J. Mol. Biol. 1975;99:761–776. doi: 10.1016/s0022-2836(75)80183-5. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 25.Kanazawa T, Zappaterra MD, Hasegawa A, Wright AP, Newman-Smith ED, Buttle KF, McDonald K, Mannella CA, van der Bliek AM. The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet. 2008;4:e1000022. doi: 10.1371/journal.pgen.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 27.Sugimoto T, Mori C, Takanami T, Sasagawa Y, Saito R, Ichiishi E, Higashitani A. Caenorhabditis elegans par2.1/mtssb-1 is essential for mitochondrial DNA replication and its defect causes comprehensive transcriptional alterations including a hypoxia response. Exp. Cell Res. 2008;314:103–114. doi: 10.1016/j.yexcr.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 28.Liau WS, Gonzalez-Serricchio AS, Deshommes C, Chin K, LaMunyon CW. A persistent mitochondrial deletion reduces fitness and sperm performance in heteroplasmic populations of C. elegans. BMC Genet. 2007;8:8. doi: 10.1186/1471-2156-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawasaki I, Hanazawa M, Gengyo-Ando K, Mitani S, Maruyama I, Iino Y. ASB-1, a germline-specific isoform of mitochondrial ATP synthase b subunit, is required to maintain the rate of germline development in Caenorhabditis elegans. Mech. Dev. 2007;124:237–251. doi: 10.1016/j.mod.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Artal-Sanz M, Tsang WY, Willems EM, Grivell LA, Lemire BD, van der Spek H, Nijtmans LG. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J. Biol. Chem. 2003;278:32091–32099. doi: 10.1074/jbc.M304877200. [DOI] [PubMed] [Google Scholar]
- 31.Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 32.Tsang WY, Sayles LC, Grad LI, Pilgrim DB, Lemire BD. Mitochondrial respiratory chain deficiency in Caenorhabditis elegans results in developmental arrest and increased life span. J. Biol. Chem. 2001;276:32240–32246. doi: 10.1074/jbc.M103999200. [DOI] [PubMed] [Google Scholar]